Phylogenetic Analysis of the PR-4 Gene Family in Euphorbiaceae and Its Expression Profiles in Tung Tree (Vernicia fordii)
Abstract
:1. Introduction
2. Results
2.1. Identification and Characterization Analysis of the PR-4 Family
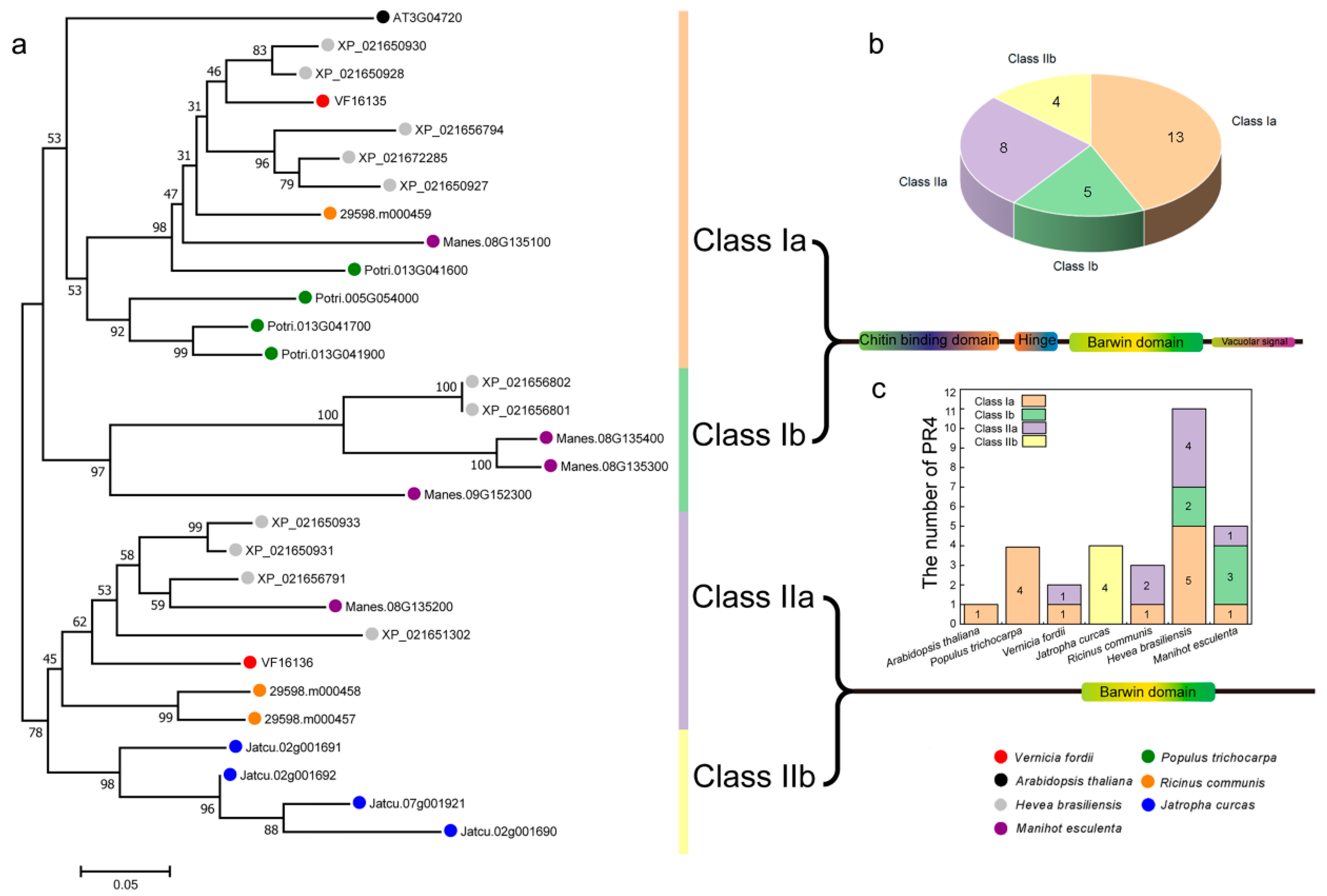
2.2. Evolutionary Relationship Analysis of the PR-4 Family
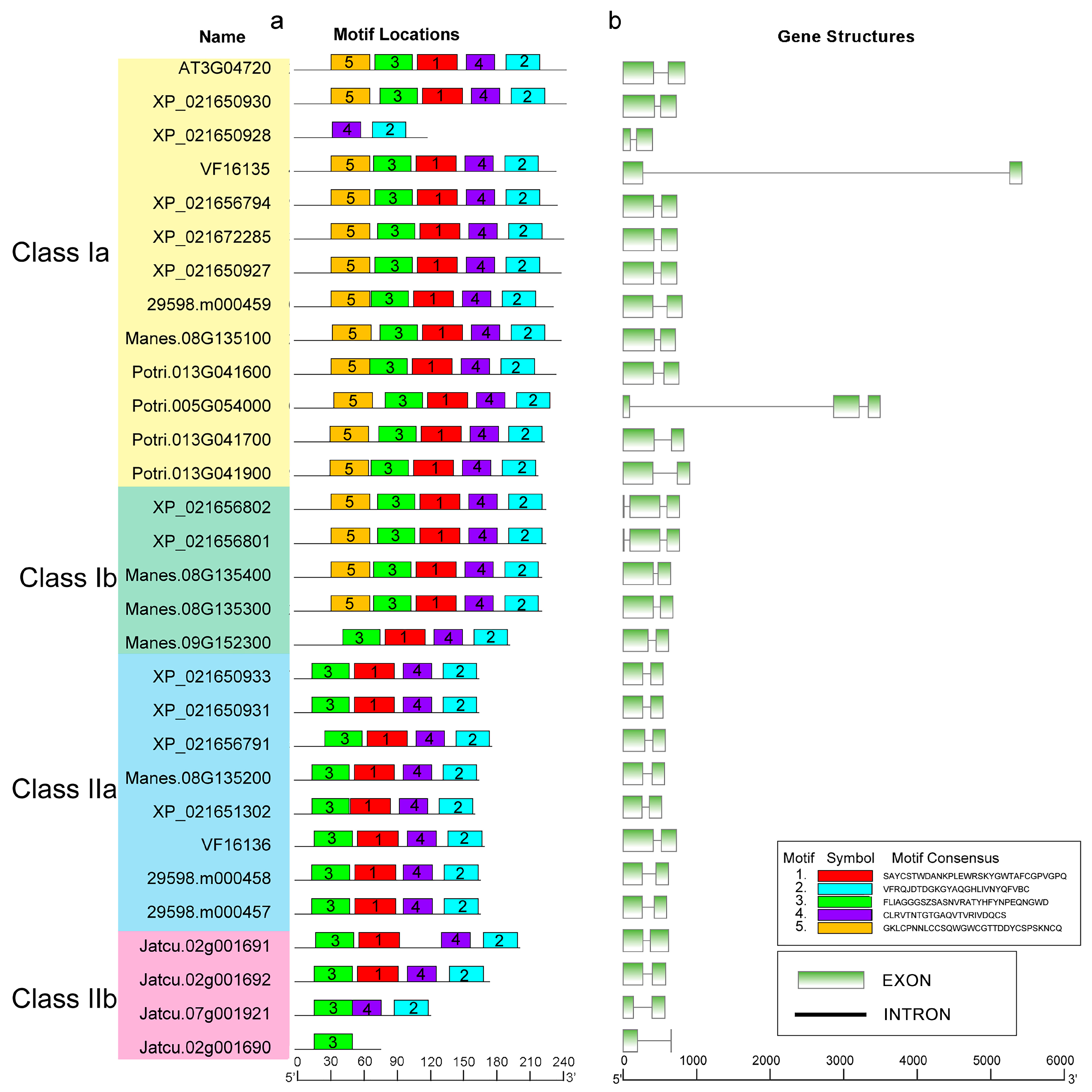
2.3. Conserved Motifs and Gene Structure in the PR-4 Family
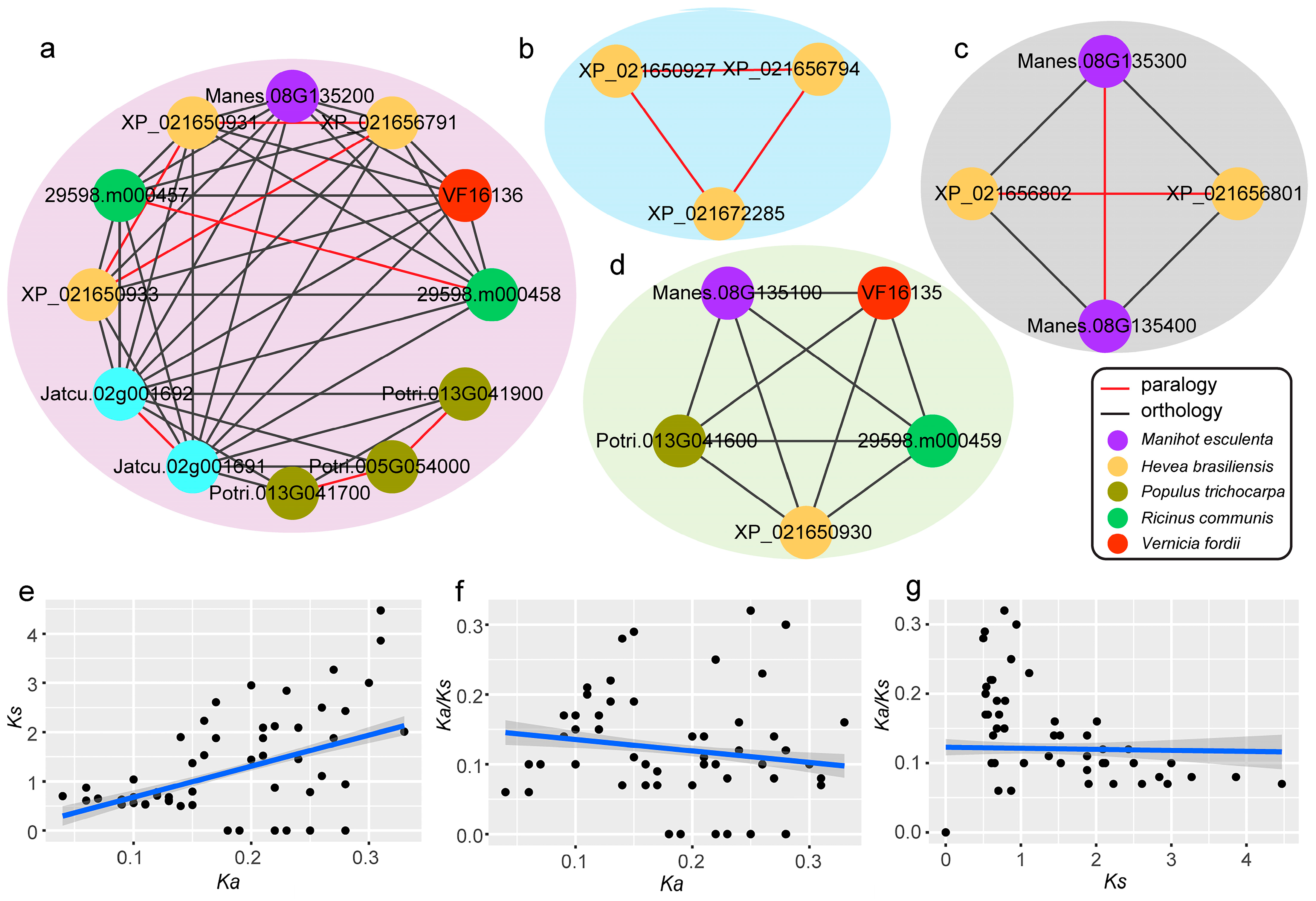
2.4. Orthologous and Paralogous Genes of the PR-4 Gene Family
2.5. Analysis of Cis-Acting Elements in PR-4 Promoters
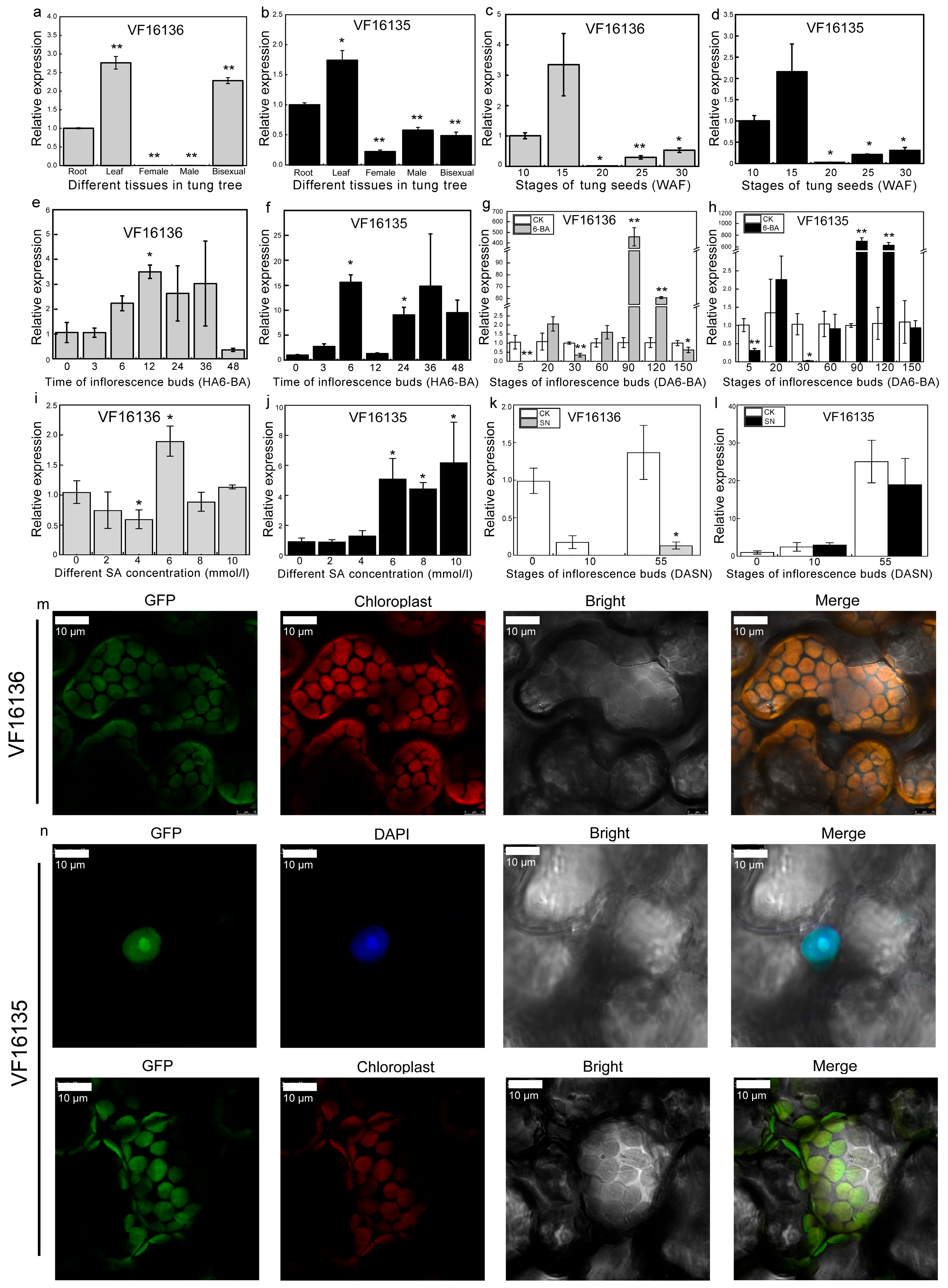
2.6. Expression Profile Analysis of PR-4 Genes in Tung Tree
3. Discussion
3.1. Evolution of PR-4 Gene Family Members in Euphorbiaceae
3.2. Characteristics of PR-4 Gene Family Members in Euphorbiaceae
3.3. VfPR-4 May Participate in the Development of Leaves and Seeds in Tung Tree
3.4. VfPR-4 Proteins Function in Response to 6-BA and SA
4. Materials and Methods
4.1. Plant Materials
4.2. Identification and Sequence Analysis of PR-4 Family
4.3. Phylogenetic Analysis and Sequence Characterization
4.4. Identification of Paralogous and Orthologous Genes
4.5. Analysis of Cis-Acting Elements in PR-4 Genes
4.6. RNA Extraction and Real-Time Quantitative PCR Analysis
4.7. Subcellular Location Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tan, X. Status and suggestion on development of Vernicia fordii. Nonwood For. Res. 2006, 24, 62–64. [Google Scholar]
- Yan, Y.; Youwei, L.; Xiaofeng, T. Discussion on coordinated development of Vernicia fordii industry and future environment-friendly coatings industry in China. Non Wood For. Res. 2018, 36, 188–192. [Google Scholar]
- Chen, J.; Zhang, S.; Zhang, X.; Wang, W.; Deng, M. Current status of Vernicia fordii development at home and abroad, and its development strategy in Jianyang. Subtrop. Agric. Res. 2009, 5, 69–72. [Google Scholar]
- McCann, L.P. Development of the pistillate flower and structure of the fruit of tung (Aleurites fordii). J. Agric. Res. 1942, 65, 361–378. [Google Scholar]
- Li, W.; Liu, M.; Dong, X.; Cao, H.; Wu, Y.; Shang, H.; Huang, H.; Zhang, L. Flower biology and ontogeny of the tung tree (Vernicia fordii Hemsl.). Trees. Struct. Funct. 2020, 34, 1363–1381. [Google Scholar] [CrossRef]
- Mao, Y.; Liu, W.; Chen, X.; Xu, Y.; Lu, W.; Hou, J.; Ni, J.; Wang, Y.; Wu, L. Flower development and sex determination between male and female flowers in Vernicia fordii. Front. Plant Sci. 2017, 8, 1291. [Google Scholar] [CrossRef]
- Liu, M.; Li, W.; Zhao, G.; Fan, X.; Long, H.; Fan, Y.; Shi, M.; Tan, X.; Zhang, L. New insights of salicylic acid into stamen abortion of female flowers in tung tree (Vernicia fordii). Front. Genet. 2019, 10, 316. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Van Strien, E. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol. Mol. Plant Pathol. 1999, 55, 85–97. [Google Scholar] [CrossRef]
- Vlot, A.C.; Dempsey, D.M.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef]
- Liu, J.-J.; Ekramoddoullah, A.K. The family 10 of plant pathogenesis-related proteins: Their structure, regulation, and function in response to biotic and abiotic stresses. Physiol. Mol. Plant Pathol. 2006, 68, 3–13. [Google Scholar] [CrossRef]
- Sels, J.; Mathys, J.; De Coninck, B.M.; Cammue, B.P.; De Bolle, M.F. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Svensson, B.; Svendsen, I.; Hoejrup, P.; Roepstorff, P.; Ludvigsen, S.; Poulsen, F.M. Primary structure of barwin: A barley seed protein closely related to the C-terminal domain of proteins encoded by wound-induced plant genes. Biochemistry 1992, 31, 8767–8770. [Google Scholar] [CrossRef] [PubMed]
- Parijs, J.; Broekaert, W.F.; Peumans, G.W.J. Hevein: An antifungal protein from rubber-tree (Hevea brasiliensis) latex. Planta 1991, 183, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Guo, Z.; Zhang, Y.; Wang, Y.; Yang, G.; Yang, L.; Wang, R.; Xie, Z. Isolation and characterization of two distinct Class II PR4 genes from the oriental lily hybrid Sorbonne. Russ. J. Plant Physiol. 2017, 64, 707–717. [Google Scholar] [CrossRef]
- Singh, A.; Jain, D.; Tyagi, C.; Singh, S.; Kumar, S.; Singh, I.K. In silico prediction of active site and in vitro DNase and RNase activities of Helicoverpa-inducible pathogenesis related-4 protein from Cicer arietinum. Int. J. Biol. Macromol. Struct. Funct. Interact. 2018, 113, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Caporale, C.; Facchiano, A.; Bertini, L.; Leonardi, L.; Chilosi, G.; Buonocore, V.; Caruso, C. Comparing the modeled structures of PR-4 proteins from wheat. J. Mol. Model. 2003, 9, 9–15. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, D.; Ji, M.; Tian, J.; Ding, H.; Deng, Z. Transcriptome Dynamic Analysis Reveals New Candidate Genes Associated with Resistance to Fusarium Head Blight in Two Chinese Contrasting Wheat Genotypes. Int. J. Mol. Sci. 2023, 24, 4222. [Google Scholar] [CrossRef] [PubMed]
- Fister, A.S.; Mejia, L.C.; Zhang, Y.; Herre, E.A.; Maximova, S.N.; Guiltinan, M.J. Theobroma cacao L. pathogenesis-related gene tandem array members show diverse expression dynamics in response to pathogen colonization. BMC Genom. 2016, 17, 1–16. [Google Scholar]
- Franco, F.P.; Dias, R.O.; Toyama, D.; Henrique-Silva, F.; Moura, D.S.; Silva-Filho, M.C. Structural and functional characterization of PR-4 SUGARWINs from sugarcaneand their role in plant defense. Front. Plant Sci. 2019, 9, 1916. [Google Scholar] [CrossRef]
- Irigoyen, M.L.; Garceau, D.C.; Bohorquez-Chaux, A.; Lopez-Lavalle, L.A.B.; Perez-Fons, L.; Fraser, P.D.; Walling, L.L. Genome-wide analyses of cassava Pathogenesis-related (PR) gene families reveal core transcriptome responses to whitefly infestation, salicylic acid and jasmonic acid. BMC Genom. 2020, 21, 1–18. [Google Scholar] [CrossRef]
- Bai, S.; Dong, C.; Li, B.; Dai, H. A PR-4 gene identified from Malus domestica is involved in the defense responses against Botryosphaeria dothidea. Plant Physiol. Biochem. 2013, 62, 23–32. [Google Scholar] [CrossRef]
- Bravo, J.M.; Campo, S.; Murillo, I.; Coca, M.; Segundo, B.S. Fungus- and wound-induced accumulation of mRNA containing a class II chitinase of the pathogenesis-related protein 4 (PR-4) family of maize. Plant Mol. Biol. 2003, 52, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Bertini, L.; Leonardi, L.; Caporale, C.; Tucci, M.; Cascone, N.; Di Berardino, I.; Buonocore, V.; Caruso, C. Pathogen-responsive wheat PR4 genes are induced by activators of systemic acquired resistance and wounding. Plant Sci. 2003, 164, 1067–1078. [Google Scholar] [CrossRef]
- Sultana, N.; Islam, S.; Juhasz, A.; Yang, R.; Ma, W. Transcriptomic Study for Identification of Major Nitrogen Stress Responsive Genes in Australian Bread Wheat Cultivars. Front. Genet. 2020, 11, 583785. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.-C.; Lin, J.-H.; Chua, A.C.; Chung, T.-Y.; Tsai, I.-C.; Tzen, J.T.; Chou, W.-M. Cloning and expression of pathogenesis-related protein 4 from jelly fig (Ficus awkeotsang Makino) achenes associated with ribonuclease, chitinase and anti-fungal activities. Plant Physiol. Biochem. 2012, 56, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.P.; Crabtree, J.; Zhao, Q.; Lorenzi, H.; Orvis, J.; Puiu, D.; Melake-Berhan, A.; Jones, K.M.; Redman, J.; Chen, G. Draft genome sequence of the oilseed species Ricinus communis. Nature Biotechnol. 2010, 28, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Feng, B.; Xiao, J.; Xia, Z.; Zhou, X.; Li, P.; Zhang, W.; Wang, Y.; Møller, B.L.; Zhang, P. Cassava genome from a wild ancestor to cultivated varieties. Nat. Commun. 2014, 5, 5110. [Google Scholar] [CrossRef]
- Ha, J.; Shim, S.; Lee, T.; Kang, Y.J.; Hwang, W.J.; Jeong, H.; Laosatit, K.; Lee, J.; Kim, S.K.; Satyawan, D. Genome sequence of Jatropha curcas L., a non-edible biodiesel plant, provides a resource to improve seed-related traits. Plant Biotechnol. J. 2019, 17, 517–530. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, M.; Long, H.; Dong, W.; Tan, X. Tung Tree (Vernicia fordii) Genome Provides A Resource for Understanding Genome Evolution and Improved Oil Production. Genom Proteom. Bioinform. 2020, 17, 558–575. [Google Scholar] [CrossRef]
- Liu, J. The Chromosome-Based Rubber Tree Genome Provides New Insights into Spurge Genome Evolution and Rubber Biosynthesis. Mol. Plant 2020, 13, 15. [Google Scholar] [CrossRef]
- Wang, J.B.; Li, W.Y.; Ge, C.; Zhang, L.; Liu, M. Effects of exogenous hormones on the fiowering and fruit characteristics ofmale tung trees of ‘putaotong’ (Vernicia fordii Hemsl.). J. Central South Univ. For. Technol. 2023, 20–32+110. [Google Scholar]
- Ellegren, H. Comparative genomics and the study of evolution by natural selection. Mol. Ecol. 2008, 17, 4586–4596. [Google Scholar] [CrossRef]
- Liu, Y.; Cui, J.; Zhou, X.; Luan, Y.; Luan, F. Genome-wide identification, characterization and expression analysis of the TLP gene family in melon (Cucumis melo L.). Genomics 2020, 112, 2499–2509. [Google Scholar] [CrossRef] [PubMed]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef]
- Chao, M.; Yin, Z.; Hao, D.; Zhang, J.; Song, H.; Ning, A.; Xu, X.; Yu, D. Variation in Rubisco activase (RCAβ) gene promoters and expression in soybean [Glycine max (L.) Merr.]. J. Exp. Bot. 2014, 65, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Olive, M.R.; Walker, J.C.; Singh, K.; Dennis, E.S.; Peacock, W.J. Functional properties of the anaerobic responsive element of the maize Adh1 gene. Plant Mol. Biol. 1990, 15, 593–604. [Google Scholar] [CrossRef]
- Caporale, C.; Di Berardino, I.; Leonardi, L.; Bertini, L.; Cascone, A.; Buonocore, V.; Caruso, C. Wheat pathogenesis-related proteins of class 4 have ribonuclease activity. Febs Lett. 2004, 575, 71–76. [Google Scholar] [CrossRef]
- Feng, Y.; Ren, Y.; Zhang, H.; Heng, Y.; Wang, Z.; Wang, Y. Halostachys caspica pathogenesis-related protein 10 acts as a cytokinin reservoir to regulate plant growth and development. Front. Plant Sci. 2023, 14, 1116985. [Google Scholar] [CrossRef]
- Kragh, K.M.; Hendriks, T.; de Jong, A.J.; Lo Schiavo, F.; Bucherna, N.; Hojrup, P.; Mikkelsen, J.D.; de Vries, S.C. Characterization of chitinases able to rescue somatic embryos of the temperature-sensitive carrot variant ts 11. Plant Mol. Biol. 1996, 31, 631–645. [Google Scholar] [CrossRef]
- Altenbach, S.B.; Kothari, K.M.; Tanaka, C.K.; Hurkman, W.J. Genes encoding the PR-4 protein wheatwin are developmentally regulated in wheat grains and respond to high temperatures during grainfill. Plant Sci. 2007, 173, 135–143. [Google Scholar] [CrossRef]
- Van Damme, E.J.; Charels, D.; Menu-Bouaouiche, L.; Proost, P.; Barre, A.; Rougé, P.; Peumans, W.J. Biochemical, molecular and structural analysis of multiple thaumatin-like proteins from the elderberry tree (Sambucus nigra L.). Planta 2002, 214, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Dastmalchi, M. Elusive partners: A review of the auxiliary proteins guiding metabolic flux in flavonoid biosynthesis. Plant J. 2021, 108, 314–329. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, S.; Piçarra-Pereira, M.A.; Loureiro, V.B.; Teixeira, A.R.; Ferreira, R.B. The diversity of pathogenesis-related proteins decreases during grape maturation. Phytochemistry 2007, 68, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Fraser, R. Evidence for the occurrence of the “pathogenesis-related” proteins in leaves of healthy tobacco plants during flowering. Physiol. Plant Pathol. 1981, 19, 69–76. [Google Scholar] [CrossRef]
- Ma, Z.; Liu, J.-J.; Zamany, A. Identification and functional characterization of an effector secreted by Cronartium ribicola. Phytopathology 2019, 109, 942–951. [Google Scholar] [CrossRef] [PubMed]
- Van Kan, J.A.; Cozijnsen, T.; Danhash, N.; De Wit, P.J. Induction of tomato stress protein mRNAs by ethephon, 2,6-dichloroisonicotinic acid and salicylate. Plant Mol. Biol. 1995, 27, 1205–1213. [Google Scholar] [CrossRef]
- Ward, E.R.; Uknes, S.J.; Williams, S.C.; Dincher, S.S.; Wiederhold, D.L.; Alexander, D.C.; Ahl-Goy, P.; Metraux, J.P.; Ryals, J.A. Coordinate Gene Activity in Response to Agents That Induce Systemic Acquired Resistance. The Plant Cell 1991, 3, 1085–1094. [Google Scholar] [CrossRef]
- Lingmin, D.; Dan, W.; Xiaoqing, X.; Chaohong, Z.; Xiping, W.; Yan, X.; Yuejin, W.; Jianxia, Z. The Novel Gene VpPR4-1 from Vitis pseudoreticulata Increases Powdery Mildew Resistance in Transgenic Vitis vinifera L. Front. Plant Sci. 2016, 7, 695. [Google Scholar]
- Liu, X.; Huang, B.; Lin, J.; Fei, J.; Chen, Z.; Pang, Y.; Sun, X.; Tang, K. A novel pathogenesis-related protein (SsPR10) from Solanum surattense with ribonucleolytic and antimicrobial activity is stress-and pathogen-inducible. J. Plant Physiol. 2006, 163, 546–556. [Google Scholar] [CrossRef]
- Morris, J.S.; Caldo, K.M.P.; Liang, S.; Facchini, P.J. PR10/Bet v1-like Proteins as Novel Contributors to Plant Biochemical Diversity. ChemBioChem 2021, 22, 264–287. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Zdobnov, E.M.; Rolf, A. InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Ling, X.; Zhaobin, D.; Lu, F.; Yongjiang, L.; Zhaoyuan, W.; Hailong, G.; Guoqing, Z.; Gu, Y.Q.; Devin, C.D.; Qingyou, X. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nuclc Acids Res. 2019, 47, W52–W58. [Google Scholar]
- Lescot; M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Han, X.; Lu, M.; Chen, Y.; Zhan, Z.; Cui, Q.; Wang, Y. Selection of reliable reference genes for gene expression studies using real-time PCR in tung tree during seed development. PLoS ONE 2012. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Yi, Y.; Wang, J.; Ge, L.; Zhang, L.; Liu, M. Phylogenetic Analysis of the PR-4 Gene Family in Euphorbiaceae and Its Expression Profiles in Tung Tree (Vernicia fordii). Plants 2023, 12, 3154. https://doi.org/10.3390/plants12173154
Yang C, Yi Y, Wang J, Ge L, Zhang L, Liu M. Phylogenetic Analysis of the PR-4 Gene Family in Euphorbiaceae and Its Expression Profiles in Tung Tree (Vernicia fordii). Plants. 2023; 12(17):3154. https://doi.org/10.3390/plants12173154
Chicago/Turabian StyleYang, Chengbo, Yaqi Yi, Jiabei Wang, Liu Ge, Lin Zhang, and Meilan Liu. 2023. "Phylogenetic Analysis of the PR-4 Gene Family in Euphorbiaceae and Its Expression Profiles in Tung Tree (Vernicia fordii)" Plants 12, no. 17: 3154. https://doi.org/10.3390/plants12173154



