Antagonistic Regulation of ABA Responses by Duplicated Tandemly Repeated DUF538 Protein Genes in Arabidopsis
Abstract
:1. Introduction
2. Results
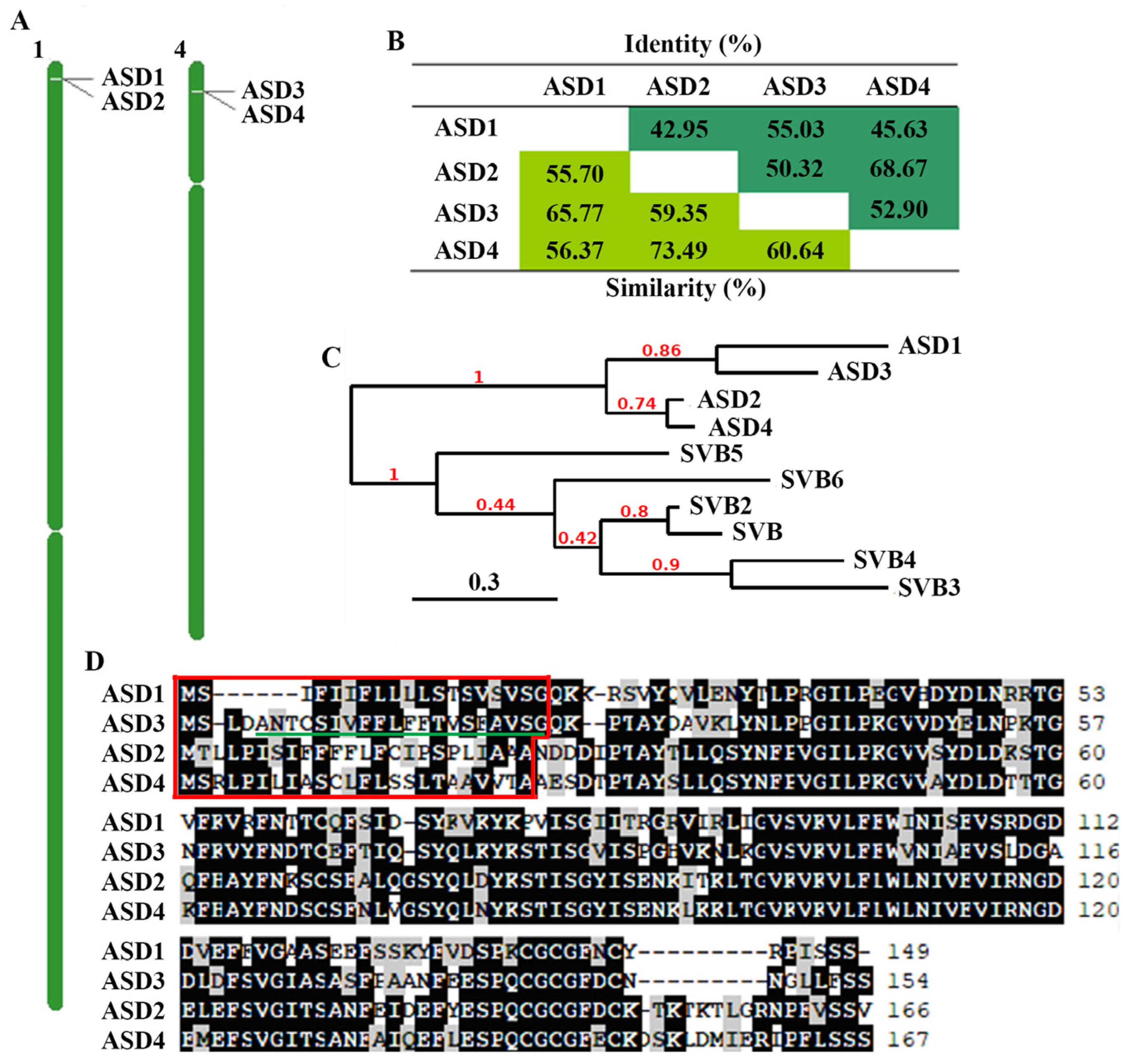
2.1. ASDs in Arabidopsis
2.2. ASDs Are ABA Response Genes
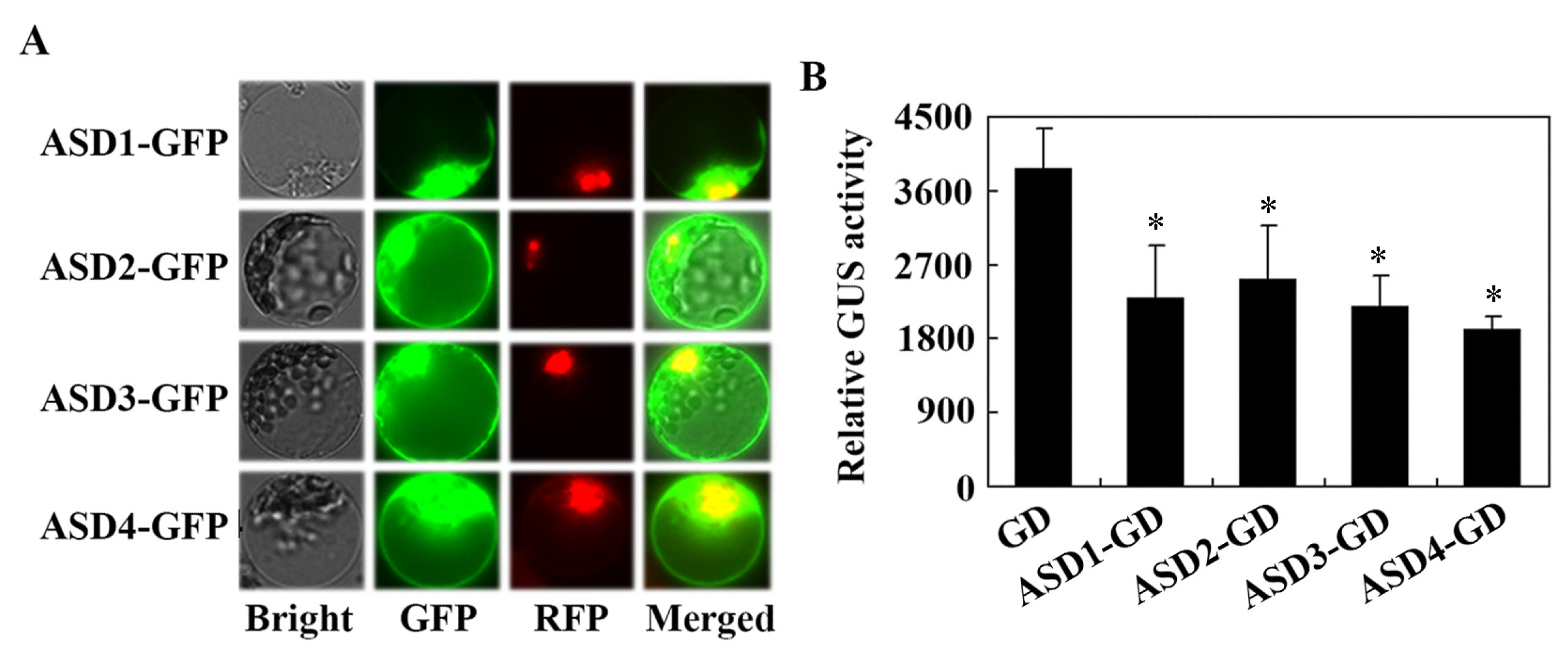
2.3. ASDs Are Associated with the Plasma Membrane but Are Also Located in the Cytosol and Nucleus
2.4. ABA Sensitivity Is Increased in Transgenic Plants Over-Expressing ASD1/ASD3 but Decreased in Transgenic Plants Over-Expressing ASD2/ASD4
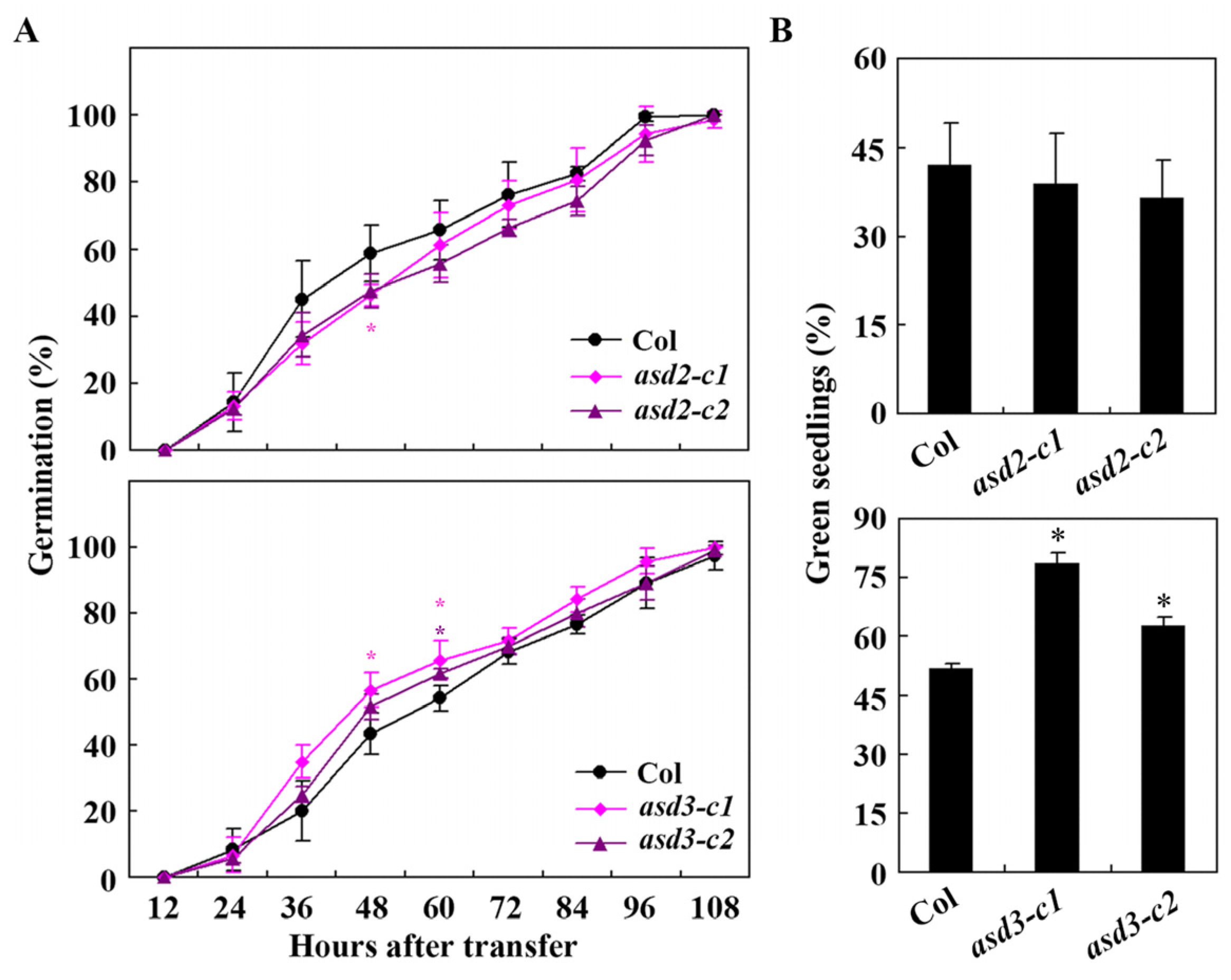
2.5. ABA Sensitivity Is Increased in the asd2 Mutants but Decreased in the asd3 Mutants
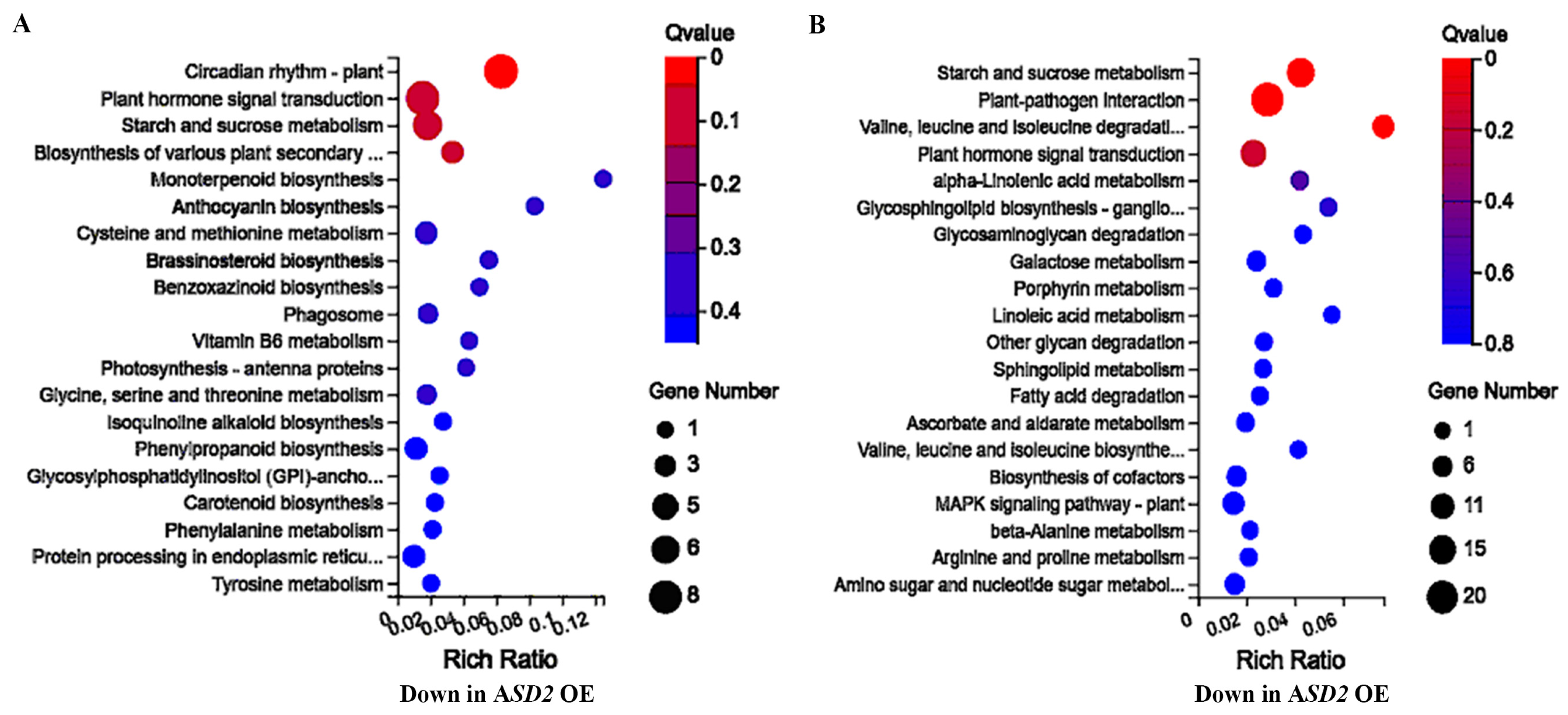
2.6. ASD2 Affected Genes Are Enriched in Different Processes, including Plant Hormone Signaling
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Constructs
4.3. Plant Transformation and Over-Expression in Transgenic Plants and Mutants Selection
4.4. DNA Isolation and PCR
4.5. Plasmid DNA Isolation, Protoplasts Isolation, and Transfection
4.6. RNA Isolation and Quantitative RT-PCR (qRT-PCR)
4.7. Seed Germination and Cotyledon Greening Assays
4.8. Transcriptome Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, G.; Liu, Y.; Ye, N.; Liu, R.; Zhang, J. Involvement of the abscisic acid catabolic gene CYP707A2 in the glucose-induced delay in seed germination and post-germination growth of Arabidopsis. Physiol. Plant. 2011, 143, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Zhu, G.; Liu, Y.; Zhang, A.; Li, Y.; Liu, R.; Shi, L.; Jia, L.; Zhang, J. Ascorbic acid and reactive oxygen species are involved in the inhibition of seed germination by abscisic acid in rice seeds. J. Exp. Bot. 2012, 63, 1809–1822. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Zhu, J.-K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [PubMed]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J.; et al. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signaling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Tian, H.; Chen, S.; Yang, W.; Wang, T.; Zheng, K.; Wang, Y.; Cheng, Y.; Zhang, N.; Liu, S.; Li, D.; et al. A novel family of transcription factors conserved in angiosperms is required for ABA signaling. Plant Cell Environ. 2017, 40, 2958–2971. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, N.; Zhou, G.; Hussain, S.; Ahmed, S.; Tian, H.; Wang, S. Knockout of the entire family of AITR genes in Arabidopsis leads to enhanced drought and salinity tolerance without fitness costs. BMC Plant Biol. 2021, 21, 137. [Google Scholar] [CrossRef]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar] [CrossRef]
- Song, L.; Huang, S.C.; Wise, A.; Castanon, R.; Nery, J.R.; Chen, H.; Watanabe, M.; Thomas, J.; Bar-Joseph, Z.; Ecker, J.R. A transcription factor hierarchy defines an environmental stress response network. Science 2016, 354, 1550. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.I.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.; Rodrigues, A.; Saez, A.; Rubio, S.; Antoni, R.; Dupeux, F.; Park, S.-Y.; Márquez, J.A.; Cutler, S.R.; Rodriguez, P.L. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009, 60, 575–588. [Google Scholar] [CrossRef]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernández, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H.; et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef]
- Antoni, R.; Gonzalez-Guzman, M.; Rodriguez, L.; Peirats-Llobet, M.; Pizzio, G.A.; Fernandez, M.A.; De Winne, N.; De Jaeger, G.; Dietrich, D.; Bennett, M.J.; et al. PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 2013, 161, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.L.; Benning, G.; Grill, E. ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett. 1998, 421, 185–190. [Google Scholar] [CrossRef]
- Gosti, F.; Beaudoin, N.; Serizet, C.; Webb, A.A.; Vartanian, N.; Giraudat, J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 1999, 11, 1897–1910. [Google Scholar] [CrossRef]
- Fujii, H.; Verslues, P.E.; Zhu, J.-K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar] [CrossRef]
- Guo, J.; Yang, X.; Weston, D.J.; Chen, J.G. Abscisic acid receptors: Past, present and future. J. Integr. Plant Biol. 2011, 53, 469–479. [Google Scholar] [CrossRef]
- Dong, T.; Park, Y.; Hwang, I. Abscisic acid: Biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 2015, 58, 29–48. [Google Scholar]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K.; et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132. [Google Scholar] [CrossRef]
- Takahashi, S.; Yoshikawa, M.; Kamada, A.; Ohtsuki, T.; Uchida, A.; Nakayama, K.; Satoh, H. The photoconvertible water-soluble chlorophyll-binding protein of Chenopodium album is a member of DUF538, a superfamily that distributes in Embryophyta. J. Plant Physiol. 2013, 170, 1549–1552. [Google Scholar] [CrossRef]
- Gholizadeh, A. DUF538 protein superfamily is predicted to be chlorophyll hydrolyzing enzymes in plants. Physiol. Mol. Biol. Plants 2016, 22, 77–85. [Google Scholar] [CrossRef]
- Gholizadeh, A.; Kohnehrouz, B.B. Identification of DUF538 cDNA clone from Celosia cristata expressed sequences of nonstressed and stressed leaves. Russ. J. Plant Physiol. 2010, 57, 247–252. [Google Scholar] [CrossRef]
- Gholizadeh, A.; Kohnehrouz, S.B. DUF538 protein super family is predicted to be the potential homologue of bactericidal/permeability-increasing protein in plant system. Protein J. 2013, 32, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, A. Prediction of tertiary structure homology between bactericidal/permeability increasing protein of innate immune system and hydrolase enzymes. Int. J. Biosci. 2014, 5, 1–6. [Google Scholar]
- Gholizadeh, A. Pectin methylesterase activity of plant DUF538 protein superfamily. Physiol. Mol. Biol. Plants 2020, 26, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, A. Heterologous expression of stress-responsive DUF538 domain containing protein and its morpho-biochemical consequences. Protein J. 2011, 30, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Du, Y.; He, C.; Dietrich, C.R.; Li, J.; Ma, X.; Wang, R.; Liu, Q.; Liu, S.; Wang, G.; et al. Maize glossy6 is involved in cuticular wax deposition and drought tolerance. J. Exp. Bot. 2019, 70, 3089–3099. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jin, Z.; Huang, R.; Zhou, J.; Song, F.; Yao, L.; Li, P.; Lu, W.; Xiao, L.; Quan, M.; et al. Leaf physiology variations are modulated by natural variations that underlie stomatal morphology in Populus. Plant Cell Environ. 2023, 46, 150–170. [Google Scholar] [CrossRef] [PubMed]
- Marks, M.D.; Wenger, J.P.; Gilding, E.; Jilk, R.; Dixon, R.A. Transcriptome analysis of Arabidopsis wild-type and gl3-sst sim trichomes identifies four additional genes required for trichome development. Mol. Plant 2009, 2, 803–822. [Google Scholar] [CrossRef]
- Yu, C.Y.; Kanehara, K. The unfolded protein response modulates a phosphoinositide-binding protein through the IRE1-bZIP60 pathway. Plant Physiol. 2020, 183, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Oxley, D.; Ktistakis, N.; Farmaki, T. Differential isolation and identification of PI(3)P and PI(3,5)P2 binding proteins from Arabidopsis thaliana using an agarose-phosphatidylinositol-phosphate affinity chromatography. J. Proteom. 2013, 91, 580–594. [Google Scholar] [CrossRef]
- Hussain, S.; Zhang, N.; Wang, W.; Ahmed, S.; Cheng, Y.; Chen, S.; Wang, X.; Wang, Y.; Hu, X.; Wang, T.; et al. Involvement of ABA Responsive SVB Genes in the Regulation of Trichome Formation in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 6790. [Google Scholar] [CrossRef]
- Yu, C.Y.; Sharma, O.; Nguyen, P.H.T.; Hartono, C.D.; Kanehara, K. A pair of DUF538 domain-containing proteins modulates plant growth and trichome development through the transcriptional regulation of GLABRA1 in Arabidopsis thaliana. Plant J. 2021, 108, 992–1004. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, D.; Kim, Y.; Hwang, I. Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 2001, 13, 2175–2190. [Google Scholar] [CrossRef]
- Tiwari, S.; Hagen, G.; Guilfoyle, T. Aux/IAA protein contain a potent transcriptional repression domain. Plant Cell 2004, 16, 533–543. [Google Scholar] [CrossRef]
- Wang, S.; Chang, Y.; Guo, J.; Chen, J.G. Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 2007, 50, 858–872. [Google Scholar] [CrossRef]
- Tiwari, S.; Hagen, G.; Guilfoyle, T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef]
- Wang, S.; Tiwari, S.; Hagen, G.; Guilfoyle, T. AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 2005, 17, 1979–1993. [Google Scholar] [CrossRef]
- Hajdukiewicz, P.; Svab, Z.; Maliga, P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 1994, 25, 989–994. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhang, N.; Hussain, S.; Ahmed, S.; Yang, W.; Wang, S. Integration of a FT expression cassette into CRISPR/Cas9 construct enables fast generation and easy identification of transgene-free mutants in Arabidopsis. PLoS ONE 2019, 14, e0218583. [Google Scholar] [CrossRef]
- Wang, Z.; Xing, H.; Dong, L.; Zhang, H.; Han, C.; Wang, X.; Chen, Q. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015, 16, 144. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.; Bent, A. Floral Dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, N.; Zhang, Q.; Zhou, G.; Tian, H.; Hussain, S.; Ahmed, S.; Wang, T.; Wang, S. Genome editing to integrate seed size and abiotic stress tolerance traits in Arabidopsis reveals a role for DPA4 and SOD7 in the regulation of inflorescence architecture. Int. J. Mol. Sci. 2019, 20, 2695. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhou, L.; Zhang, W.; Cai, L.; Guo, H.; Tian, H.; Schiefelbein, J.; Wang, S. A single amino acid substitution in the R3 domain of GLABRA1 leads to inhibition of trichome formation in Arabidopsis without affecting its interaction with GLABRA3. Plant Cell Environ. 2016, 39, 897–907. [Google Scholar] [CrossRef]
- Liu, S.; Hu, Q.; Luo, S.; Li, Q.; Yang, X.; Wang, X.; Wang, S. Expression of wild-type PtrIAA14.1, a poplar Aux/IAA gene causes morphological changes in Arabidopsis. Front. Plant Sci. 2015, 6, 388–399. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Abdi, H. The Bonferonni and Šidák corrections for multiple comparisons. In Encyclopedia of Measurement and Statistics; Salkind, N.J., Ed.; Sage: Thousand Oaks, CA, USA, 2007; pp. 103–107. [Google Scholar]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Wang, W.; Zhang, N.; Cheng, Y.; Hussain, S.; Wang, Y.; Tian, H.; Hussain, H.; Lin, R.; Yuan, Y.; et al. Antagonistic Regulation of ABA Responses by Duplicated Tandemly Repeated DUF538 Protein Genes in Arabidopsis. Plants 2023, 12, 2989. https://doi.org/10.3390/plants12162989
Li Y, Wang W, Zhang N, Cheng Y, Hussain S, Wang Y, Tian H, Hussain H, Lin R, Yuan Y, et al. Antagonistic Regulation of ABA Responses by Duplicated Tandemly Repeated DUF538 Protein Genes in Arabidopsis. Plants. 2023; 12(16):2989. https://doi.org/10.3390/plants12162989
Chicago/Turabian StyleLi, Yingying, Wei Wang, Na Zhang, Yuxin Cheng, Saddam Hussain, Yating Wang, Hainan Tian, Hadia Hussain, Rao Lin, Yuan Yuan, and et al. 2023. "Antagonistic Regulation of ABA Responses by Duplicated Tandemly Repeated DUF538 Protein Genes in Arabidopsis" Plants 12, no. 16: 2989. https://doi.org/10.3390/plants12162989





