Genomics for Yield and Yield Components in Durum Wheat
Abstract
:1. Introduction
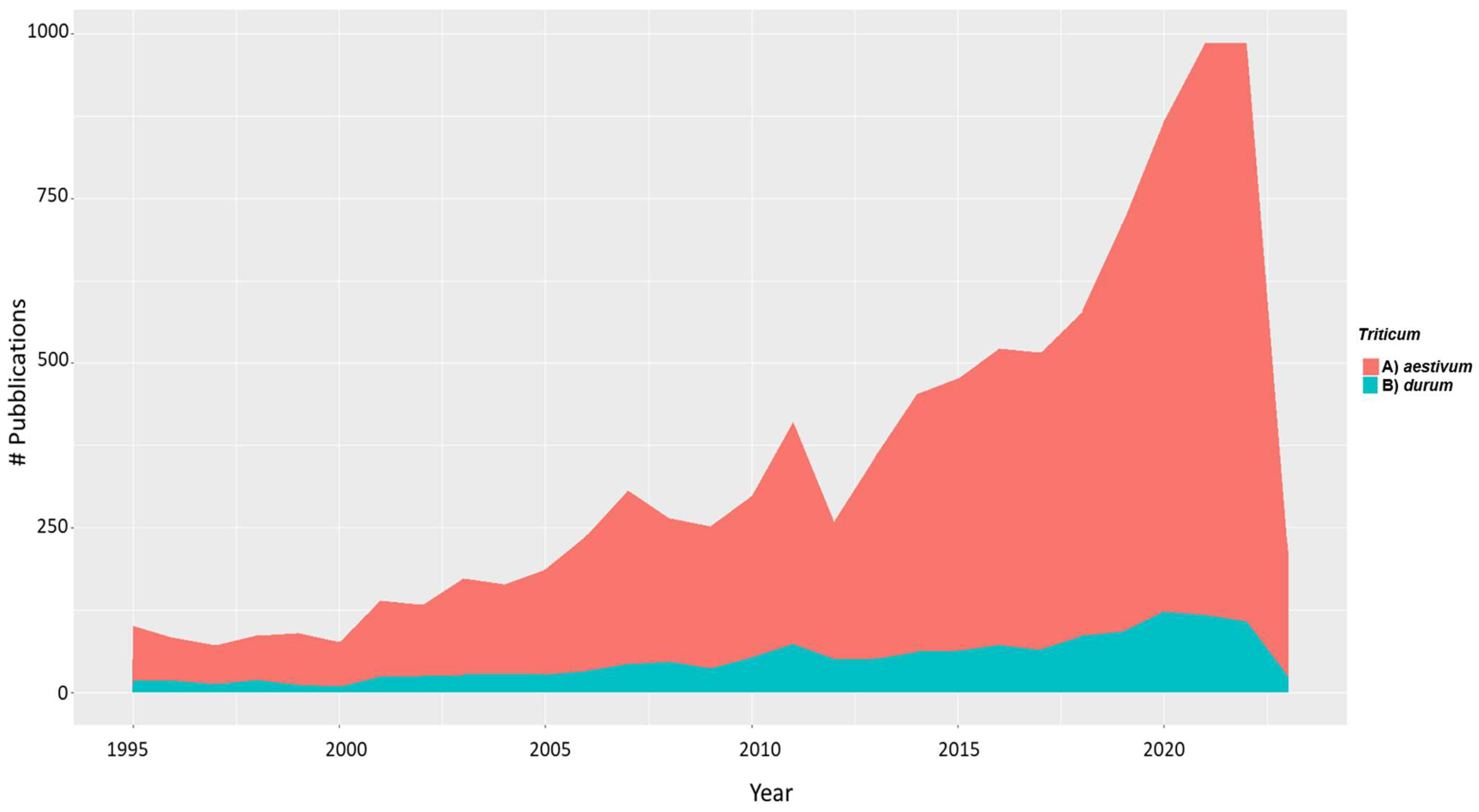
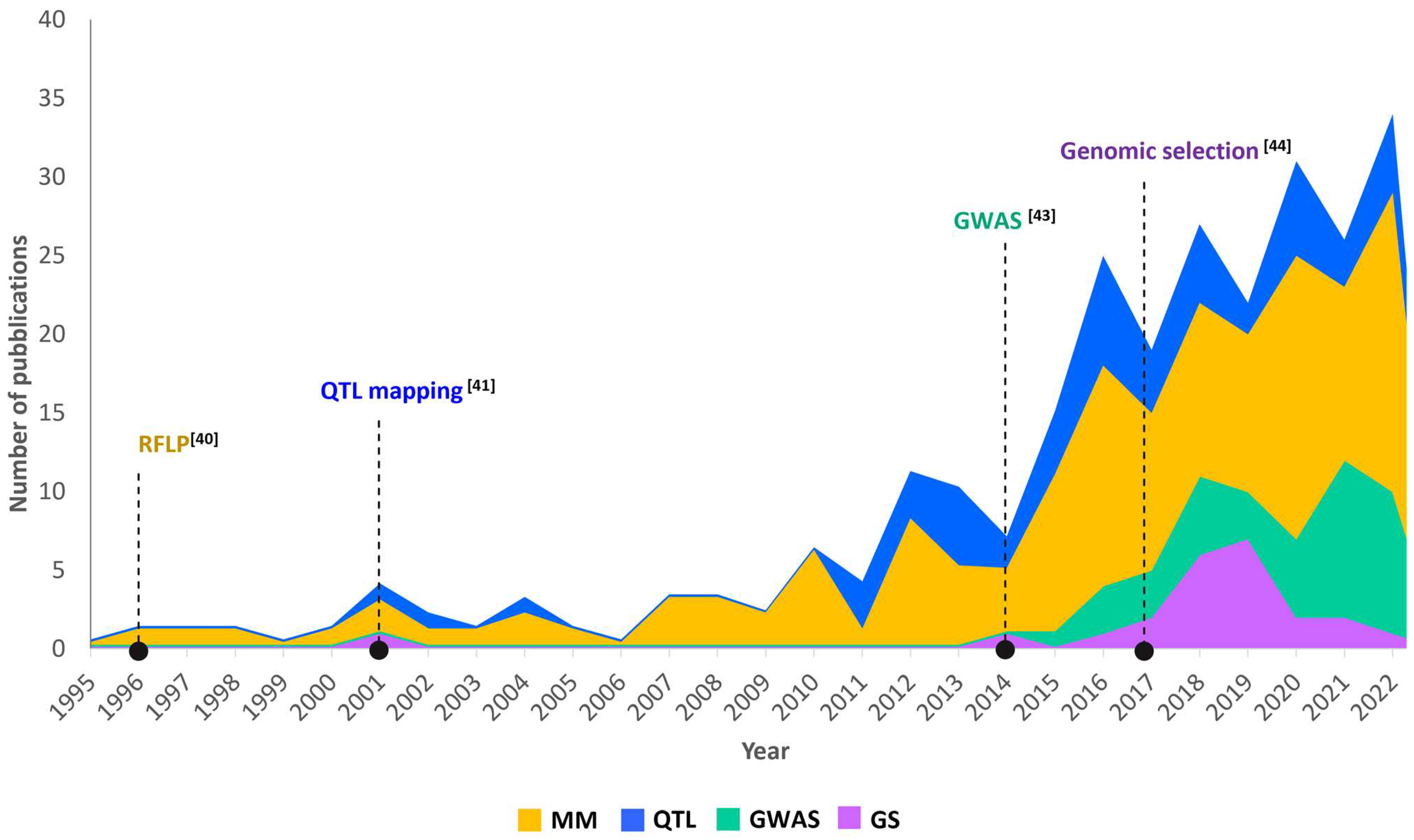
2. Trends of Genomic Technologies to Advance Yield and Yield Components in Durum Wheat
3. QTL Mapping Strategies for Yield and Its Components in Durum Wheat
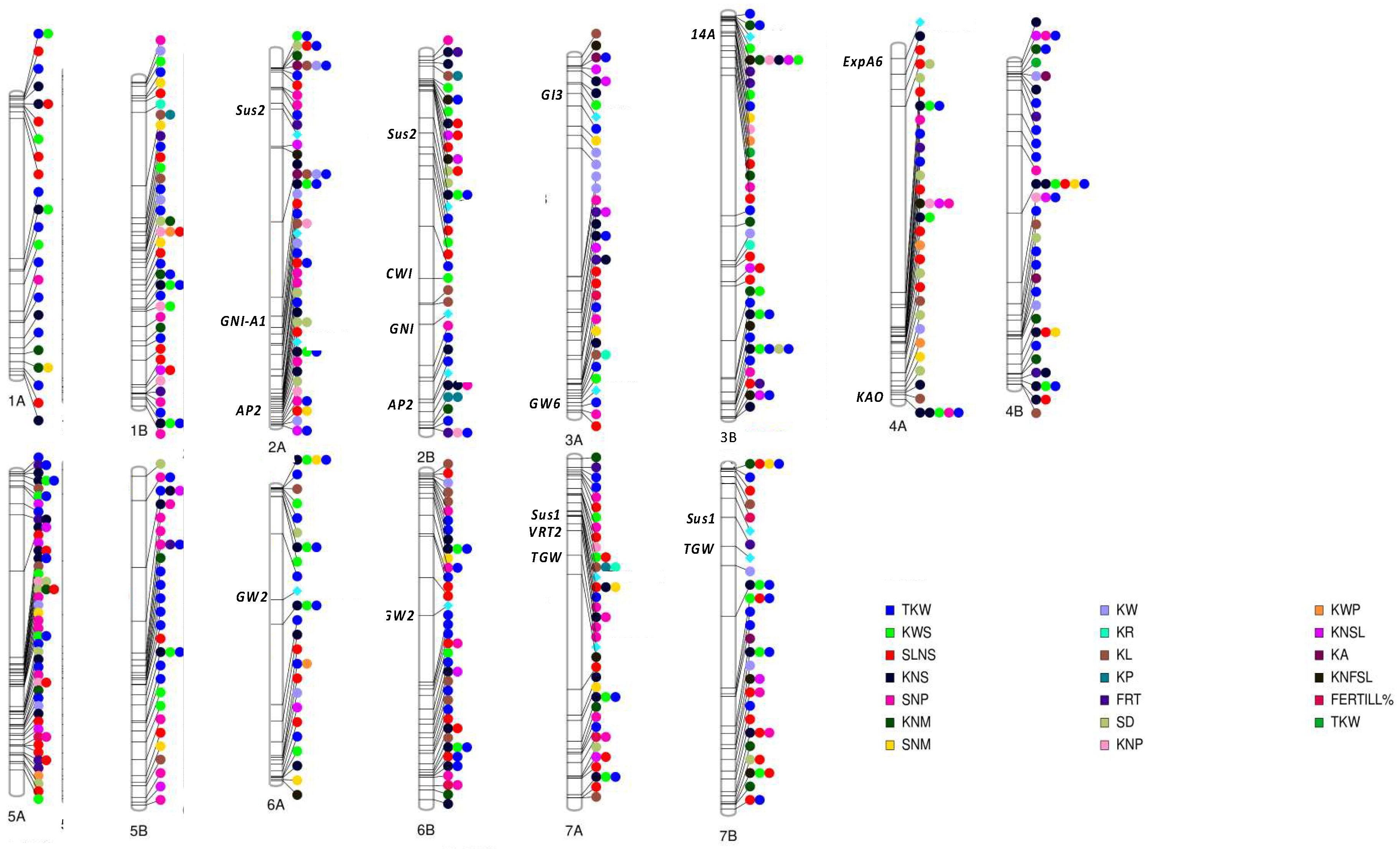
3.1. Genomic Regions Associated with Grain Number (GN)
3.2. Genomic Regions Associated with Grain Weight (GW)
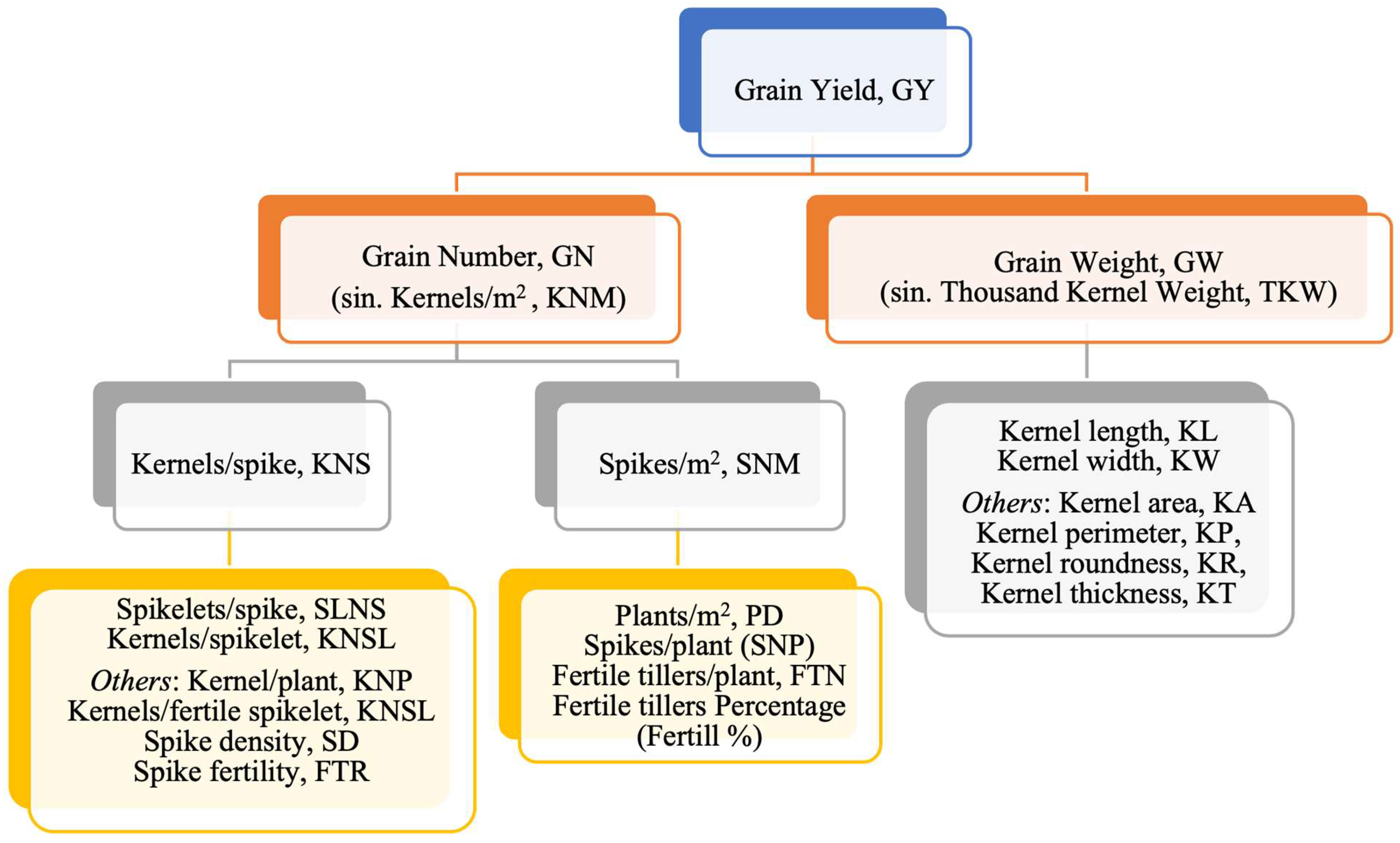
4. Genes Affecting Yield and Its Components in Durum Wheat
5. A Holistic Approach to Studying Yield and Its Components in Post-Genomic Era
6. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beres, B.L.; Rahmani, E.; Clarke, J.M.; Grassini, P.; Pozniak, C.J.; Geddes, C.M.; Porker, K.D.; May, W.E.; Ransom, J.K. A Systematic Review of Durum Wheat: Enhancing Production Systems by Exploring Genotype, Environment, and Management (G × E × M) Synergies. Front. Plant Sci. 2020, 11, 568657. [Google Scholar] [CrossRef]
- Ceglar, A.; Toreti, A.; Zampieri, M.; Royo, C. Global Loss of Suitable Durum Wheat Areas in the Future. Environ. Res. Lett. 2021, 16, 104049. [Google Scholar] [CrossRef]
- De Vita, P.; Taranto, F. Durum Wheat (Triticum turgidum ssp. durum) Breeding to Meet the Challenge of Climate Change. Adv. Plant Breed. Strateg. Cereals 2019, 5, 471–524. [Google Scholar] [CrossRef]
- MacCaferri, M.; Sanguineti, M.C.; Demontis, A.; El-Ahmed, A.; Garcia Del Moral, L.; Maalouf, F.; Nachit, M.; Nserallah, N.; Ouabbou, H.; Rhouma, S.; et al. Association Mapping in Durum Wheat Grown across a Broad Range of Water Regimes. J. Exp. Bot. 2011, 62, 409–438. [Google Scholar] [CrossRef] [Green Version]
- Mengistu, D.K.; Kidane, Y.G.; Catellani, M.; Frascaroli, E.; Fadda, C.; Pè, M.E.; Dell’Acqua, M. High-Density Molecular Characterization and Association Mapping in Ethiopian Durum Wheat Landraces Reveals High Diversity and Potential for Wheat Breeding. Plant Biotechnol. J. 2016, 14, 1800–1812. [Google Scholar] [CrossRef] [Green Version]
- Kidane, Y.G.; Mancini, C.; Mengistu, D.K.; Frascaroli, E.; Fadda, C.; Pè, M.E.; Dell’Acqua, M. Genome Wide Association Study to Identify the Genetic Base of Smallholder Farmer Preferences of Durum Wheat Traits. Front. Plant Sci. 2017, 8, 1230. [Google Scholar] [CrossRef] [Green Version]
- Soriano, J.M.; Malosetti, M.; Roselló, M.; Sorrells, M.E.; Royo, C. Dissecting the Old Mediterranean Durum Wheat Genetic Architecture for Phenology, Biomass and Yield Formation by Association Mapping and QTL Meta-Analysis. PLoS ONE 2017, 12, e0178290. [Google Scholar] [CrossRef] [Green Version]
- Sukumaran, S.; Reynolds, M.P.; Sansaloni, C. Genome-Wide Association Analyses Identify QTL Hotspots for Yield and Component Traits in Durum Wheat Grown under Yield Potential, Drought, and Heat Stress Environments. Front. Plant Sci. 2018, 9, 81. [Google Scholar] [CrossRef] [Green Version]
- Giunta, F.; De Vita, P.; Mastrangelo, A.M.; Sanna, G.; Motzo, R. Environmental and Genetic Variation for Yield-Related Traits of Durum Wheat as Affected by Development. Front. Plant Sci. 2018, 9, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangini, G.; Blanco, A.; Nigro, D.; Signorile, M.A.; Simeone, R. Candidate Genes and Quantitative Trait Loci for Grain Yield and Seed Size in Durum Wheat. Plants 2021, 10, 312. [Google Scholar] [CrossRef] [PubMed]
- Mangini, G.; Gadaleta, A.; Colasuonno, P.; Marcotuli, I.; Signorile, A.M.; Simeone, R.; De Vita, P.; Mastrangelo, A.M.; Laidò, G.; Pecchioni, N.; et al. Genetic Dissection of the Relationships between Grain Yield Components by Genome-Wide Association Mapping in a Collection of Tetraploid Wheats. PLoS ONE 2018, 13, e0190162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colasuonno, P.; Marcotuli, I.; Gadaleta, A.; Soriano, J.M.; Borrelli, M. Plants Review from Genetic Maps to QTL Cloning: An Overview for Durum Wheat. Plants 2021, 10, 315. [Google Scholar] [CrossRef] [PubMed]
- Soriano, J.M.; Colasuonno, P.; Marcotuli, I.; Gadaleta, A. Meta-QTL Analysis and Identification of Candidate Genes for Quality, Abiotic and Biotic Stress in Durum Wheat. Sci. Rep. 2021, 11, 11877. [Google Scholar] [CrossRef] [PubMed]
- Golan, G.; Ayalon, I.; Perry, A.; Zimran, G.; Ade-Ajayi, T.; Mosquna, A.; Distelfeld, A.; Peleg, Z. GNI-A1 Mediates Trade-off between Grain Number and Grain Weight in Tetraploid Wheat. Theor. Appl. Genet. 2019, 132, 2353–2365. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Liu, Y.; Zhang, P.; Chen, T.; Tian, T.; Wang, P.; Che, Z.; Shahinnia, F.; Yang, D. Identification of Quantitative Trait Loci (QTL) and Meta-QTL Analysis for Kernel Size-Related Traits in Wheat (Triticum aestivum L.). BMC Plant Biol. 2022, 22, 607. [Google Scholar] [CrossRef]
- Guo, Z.; Chen, D.; Alqudah, A.M.; Röder, M.S.; Ganal, M.W.; Schnurbusch, T. Genome-wide Association Analyses of 54 Traits Identified Multiple Loci for the Determination of Floret Fertility in Wheat. New Phytol. 2017, 214, 257–270. [Google Scholar] [CrossRef]
- Schierenbeck, M.; Alqudah, A.M.; Lohwasser, U.; Tarawneh, R.A.; Simón, M.R.; Börner, A. Genetic Dissection of Grain Architecture-Related Traits in a Winter Wheat Population. BMC Plant Biol. 2021, 21, 417. [Google Scholar] [CrossRef]
- Ferrante, A.; Savin, R.; Slafer, G.A. Floret Development and Grain Setting Differences between Modern Durum Wheats under Contrasting Nitrogen Availability. J. Exp. Bot. 2013, 64, 169–184. [Google Scholar] [CrossRef]
- Koutroubas, S.D.; Antoniadis, V.; Fotiadis, S.; Damalas, C.A. Growth, Grain Yield and Nitrogen Use Efficiency of Mediterranean Wheat in Soils Amended with Municipal Sewage Sludge. Nutr. Cycl. Agroecosyst. 2014, 100, 227–243. [Google Scholar] [CrossRef]
- Xie, Q.; Mayes, S.; Sparkes, D.L. Carpel Size, Grain Filling, and Morphology Determine Individual Grain Weight in Wheat. J. Exp. Bot. 2015, 66, 6715–6730. [Google Scholar] [CrossRef] [Green Version]
- Huang, N.; Dang, H.; Mu, W.; Ma, J.; Ma, Y.; Wang, L.; Shi, M.; Tian, H.; Liu, J.; Chen, Y.; et al. High Yield with Efficient Nutrient Use: Opportunities and Challenges for Wheat. iScience 2023, 26, 106135. [Google Scholar] [CrossRef]
- Slafer, G.A.; Elia, M.; Savin, R.; García, G.A.; Terrile, I.I.; Ferrante, A.; Miralles, D.J.; González, F.G. Fruiting Efficiency: An Alternative Trait to Further Rise Wheat Yield. Food Energy Secur. 2015, 4, 92–109. [Google Scholar] [CrossRef]
- Brinton, J.; Uauy, C. A Reductionist Approach to Dissecting Grain Weight and Yield in Wheat OO. Plant Biol. 2018, 61, 337–358. [Google Scholar] [CrossRef] [Green Version]
- Peltonen-Sainio, P.; Kangas, A.; Salo, Y.; Jauhiainen, L. Grain Number Dominates Grain Weight in Temperate Cereal Yield Determination: Evidence Based on 30 Years of Multi-Location Trials. Field Crops Res. 2007, 100, 179–188. [Google Scholar] [CrossRef]
- Abeledo, L.G.; Savin, R.; Slafer, G.A. Wheat Productivity in the Mediterranean Ebro Valley: Analyzing the Gap between Attainable and Potential Yield with a Simulation Model. Eur. J. Agron. 2008, 28, 541–550. [Google Scholar] [CrossRef]
- Lizana, X.C.; Calderini, D.F. Yield and Grain Quality of Wheat in Response to Increased Temperatures at Key Periods for Grain Number and Grain Weight Determination: Considerations for the Climatic Change Scenarios of Chile. J. Agric. Sci. 2013, 151, 209–221. [Google Scholar] [CrossRef]
- Zhou, M.; Butterbach-Bahl, K. Assessment of Nitrate Leaching Loss on a Yield-Scaled Basis from Maize and Wheat Cropping Systems. Plant Soil 2014, 374, 977–991. [Google Scholar] [CrossRef]
- De Vita, P.; Di Paolo, E.; Fecondo, G.; Di Fonzo, N.; Pisante, M. No-Tillage and Conventional Tillage Effects on Durum Wheat Yield, Grain Quality and Soil Moisture Content in Southern Italy. Soil Tillage Res. 2007, 92, 69–78. [Google Scholar] [CrossRef]
- De Vita, P.; Mastrangelo, A.M.; Matteu, L.; Mazzucotelli, E.; Virzì, N.; Palumbo, M.; Storto, M.L.; Rizza, F.; Cattivelli, L. Genetic Improvement Effects on Yield Stability in Durum Wheat Genotypes Grown in Italy. Field Crops Res. 2010, 119, 68–77. [Google Scholar] [CrossRef]
- Reynolds, M.P.; Slafer, G.A.; Foulkes, J.M.; Griffiths, S.; Murchie, E.H.; Carmo-Silva, E.; Asseng, S.; Chapman, S.C.; Sawkins, M.; Gwyn, J.; et al. A Wiring Diagram to Integrate Physiological Traits of Wheat Yield Potential. Nat. Food 2022, 3, 318–324. [Google Scholar] [CrossRef]
- Slafer, G.A.; John Foulkes, M.; Reynolds, M.P.; Murchie, E.H.; Carmo-Silva, E.; Flavell, R.; Gwyn, J.; Sawkins, M.; Griffiths, S.; Mexico, C.; et al. A ‘Wiring Diagram’ for Sink Strength Traits Impacting Wheat Yield Potential. J. Exp. Bot. 2023, 74, 40–71. [Google Scholar] [CrossRef] [PubMed]
- Bustos, D.V.; Hasan, A.K.; Reynolds, M.P.; Calderini, D.F. Combining High Grain Number and Weight through a DH-Population to Improve Grain Yield Potential of Wheat in High-Yielding Environments. Field Crops Res. 2013, 145, 106–115. [Google Scholar] [CrossRef]
- Quintero, A.; Molero, G.; Reynolds, M.P.; Calderini, D.F. Trade-off between Grain Weight and Grain Number in Wheat Depends on GxE Interaction: A Case Study of an Elite CIMMYT Panel (CIMCOG). Eur. J. Agron. 2018, 92, 17–29. [Google Scholar] [CrossRef]
- Molero, G.; Joynson, R.; Pinera-Chavez, F.J.; Gardiner, L.J.; Rivera-Amado, C.; Hall, A.; Reynolds, M.P. Elucidating the Genetic Basis of Biomass Accumulation and Radiation Use Efficiency in Spring Wheat and Its Role in Yield Potential. Plant Biotechnol. J. 2019, 17, 1276–1288. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Yang, Y.; Lin, X.; Xiao, J. Deciphering Spike Architecture Formation towards Yield Improvement in Wheat. J. Genet. Genom. 2023; in press. [Google Scholar] [CrossRef]
- Alam, I.; Batool, K.; Huang, Y.; Liu, J.; Ge, L. Developing Genetic Engineering Techniques for Control of Seed Size and Yield. Int. J. Mol. Sci. 2022, 23, 13256. [Google Scholar] [CrossRef]
- Rigatti, A.; de Pelegrin, A.J.; Meier, C.; Lunkes, A.; Klein, L.A.; da Silva, A.F.; Bellé, E.P.; Silva, A.D.B.; Marchioro, V.S.; de Souza, V.Q. Combination Capacity and Association Among Traits of Grain Yield in Wheat (Triticum aestivum L.): A Review. J. Agric. Sci. 2018, 10, 179. [Google Scholar] [CrossRef] [Green Version]
- Tshikunde, N.M.; Mashilo, J.; Shimelis, H.; Odindo, A. Agronomic and Physiological Traits, and Associated Quantitative Trait Loci (QTL) Affecting Yield Response in Wheat (Triticum aestivum L.): A Review. Front. Plant Sci. 2019, 10, 1428. [Google Scholar] [CrossRef] [Green Version]
- Hadley, W. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Blanco, A.; De Giovanni, C.; Laddomada, B.; Sciancalepore, A.; Simeone, R.; Devos, K.M.; Gale, M.D. Quantitative Trait Loci Influencing Grain Protein Content in Tetraploid Wheats. Plant Breed. 1996, 115, 310–316. [Google Scholar] [CrossRef]
- Blanco, A.; Lotti, C.; Simeone, R.; Signorile, A.; De Santis, V.; Pasqualone, A.; Troccoli, A.; Di Fonzo, N. Detection of Quantitative Trait Loci for Grain Yield and Yield Components across Environments in Durum Wheat. Cereal Res. Commun. 2001, 29, 237–244. [Google Scholar] [CrossRef]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of Polyploid Wheat Genomic Diversity Using a High-Density 90,000 Single Nucleotide Polymorphism Array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canè, M.A.; Maccaferri, M.; Nazemi, G.; Salvi, S.; Francia, R.; Colalongo, C.; Tuberosa, R.; Nazemi, G. Association Mapping for Root Architectural Traits in Durum Wheat Seedlings as Related to Agronomic Performance Mean of Low-Yielding Environments. Mol. Breed. 2014, 34, 1629–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiedler, J.D.; Salsman, E.; Liu, Y.; Michalak de Jiménez, M.; Hegstad, J.B.; Chen, B.; Manthey, F.A.; Chao, S.; Xu, S.; Elias, E.M.; et al. Genome-Wide Association and Prediction of Grain and Semolina Quality Traits in Durum Wheat Breeding Populations. Plant Genome 2017, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Taranto, F.; Nicolia, A.; Pavan, S.; De Vita, P.; D’Agostino, N. Biotechnological and Digital Revolution for Climate-Smart Plant Breeding. Agronomy 2018, 8, 277. [Google Scholar] [CrossRef] [Green Version]
- Esposito, S.; Taranto, F.; Vitale, P.; Ficco, D.B.M.; Colecchia, S.A.; Stevanato, P.; De Vita, P. Unlocking the Molecular Basis of Wheat Straw Composition and Morphological Traits through Multi-Locus GWAS. BMC Plant Biol. 2022, 22, 519. [Google Scholar] [CrossRef]
- Maccaferri, M.; Harris, N.S.; Twardziok, S.O.; Pasam, R.K.; Gundlach, H.; Spannagl, M.; Ormanbekova, D.; Lux, T.; Prade, V.M.; Milner, S.G.; et al. Durum Wheat Genome Highlights Past Domestication Signatures and Future Improvement Targets. Nat. Genet. 2019, 51, 885–895. [Google Scholar] [CrossRef] [Green Version]
- Foulkes, M.J.; Slafer, G.A.; Davies, W.J.; Berry, P.M.; Sylvester-Bradley, R.; Martre, P.; Calderini, D.F.; Griffiths, S.; Reynolds, M.P. Raising Yield Potential of Wheat. III. Optimizing Partitioning to Grain While Maintaining Lodging Resistance. J. Exp. Bot. 2011, 62, 469–486. [Google Scholar] [CrossRef] [Green Version]
- Naruoka, Y.; Talbert, L.E.; Lanning, S.P.; Blake, N.K.; Martin, J.M.; Sherman, J.D. Identification of Quantitative Trait Loci for Productive Tiller Number and Its Relationship to Agronomic Traits in Spring Wheat. Theor. Appl. Genet. 2011, 123, 1043–1053. [Google Scholar] [CrossRef]
- Vitale, P.; Fania, F.; Esposito, S.; Pecorella, I.; Pecchioni, N.; Palombieri, S.; Sestili, F.; Lafiandra, D.; Taranto, F.; De Vita, P. QTL Analysis of Five Morpho-Physiological Traits in Bread Wheat Using Two Mapping Populations Derived from Common Parents. Genes 2021, 12, 604. [Google Scholar] [CrossRef]
- Kato, K.; Miura, H.; Sawada, S. Mapping QTLs Controlling Grain Yield and Its Components on Chromosome 5A of Wheat. Theor. Appl. Genet. 2000, 101, 1114–1121. [Google Scholar] [CrossRef]
- Li, H.; Jiang, D.; Wollenweber, B.; Dai, T.; Cao, W. Effects of Shading on Morphology, Physiology and Grain Yield of Winter Wheat. Eur. J. Agron. 2010, 33, 267–275. [Google Scholar] [CrossRef]
- Huang, X.Q.; Kempf, H.; Canal, M.W.; Röder, M.S. Advanced Backcross QTL Analysis in Progenies Derived from a Cross between a German Elite Winter Wheat Variety and a Synthetic Wheat (Triticum aestivum L.). Theor. Appl. Genet. 2004, 109, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Kulwal, P.L.; Balyan, H.S.; Gupta, P.K. QTL Mapping for Yield and Yield Contributing Traits in Two Mapping Populations of Bread Wheat. Mol. Breed. 2007, 19, 163–177. [Google Scholar] [CrossRef]
- Jia, H.; Wan, H.; Yang, S.; Zhang, Z.; Kong, Z.; Xue, S.; Zhang, L.; Ma, Z. Genetic Dissection of Yield-Related Traits in a Recombinant Inbred Line Population Created Using a Key Breeding Parent in China’s Wheat Breeding. Theor. Appl. Genet. 2013, 126, 2123–2139. [Google Scholar] [CrossRef]
- Yang, X.; Asseng, S.; Wong, M.T.F.; Yu, Q.; Li, J.; Liu, E. Quantifying the Interactive Impacts of Global Dimming and Warming on Wheat Yield and Water Use in China. Agric. For. Meteorol. 2013, 182–183, 342–351. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, J.; Zhang, L.; Rong, C.; Zhao, F.; Peng, T.; Li, H.; Cheng, D.; Liu, X.; Qin, H.; et al. Association Analysis of Genomic Loci Important for Grain Weight Control in Elite Common Wheat Varieties Cultivated with Variable Water and Fertiliser Supply. PLoS ONE 2013, 8, e57853. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wang, S.; Sun, H.; Chen, S.; Shao, L.; Liu, X. Contribution of Cultivar, Fertilizer and Weather to Yield Variation of Winter Wheat over Three Decades: A Case Study in the North China Plain. Eur. J. Agron. 2013, 50, 52–59. [Google Scholar] [CrossRef]
- Graziani, M.; Maccaferri, M.; Royo, C.; Salvatorelli, F.; Tuberosa, R. QTL Dissection of Yield Components and Morpho-Physiological Traits in a Durum Wheat Elite Population Tested in Contrasting Thermo-Pluviometric Conditions. Crop Pasture Sci. 2014, 65, 80–95. [Google Scholar] [CrossRef]
- Reynolds, M.P.; Pask, A.J.D.; Hoppitt, W.J.E.; Sonder, K.; Sukumaran, S.; Molero, G.; Pierre, C.S.; Payne, T.; Singh, R.P.; Braun, H.J.; et al. Strategic Crossing of Biomass and Harvest Index—Source and Sink—Achieves Genetic Gains in Wheat. Euphytica 2017, 213, 257. [Google Scholar] [CrossRef] [Green Version]
- Anuarbek, S.; Id, S.A.; Pecchioni, N.; Laidò, G.; Maccaferri, M.; Tuberosa, R.; Turuspekovid, Y. Quantitative Trait Loci for Agronomic Traits in Tetraploid Wheat for Enhancing Grain Yield in Kazakhstan Environments. PLoS ONE 2020, 15, e0234863. [Google Scholar] [CrossRef]
- Roncallo, P.F.; Akkiraju, P.C.; Cervigni, G.L.; Echenique, V.C. QTL Mapping and Analysis of Epistatic Interactions for Grain Yield and Yield-Related Traits in Triticum turgidum L. var. Durum. Euphytica 2017, 213, 277. [Google Scholar] [CrossRef]
- Thi Thanh, P.; Ioan Vladutu, C.; Kianian, S.F.; Thien Thanh, P.; Ishii, T.; Nitta, M.; Nasuda, S.; Mori, N. Molecular Genetic Analysis of Domestication Traits in Emmer Wheat. I: Map Construction and QTL Analysis Using an F 2 Pupulation. Biotechnol. Biotechnol. Equip. 2013, 27, 3627–3637. [Google Scholar] [CrossRef] [Green Version]
- Würschum, T.; Leiser, W.L.; Langer, S.M.; Tucker, M.R.; Longin, C.F.H. Phenotypic and Genetic Analysis of Spike and Kernel Characteristics in Wheat Reveals Long-Term Genetic Trends of Grain Yield Components. Theor. Appl. Genet. 2018, 131, 2071–2084. [Google Scholar] [CrossRef]
- Iannucci, A.; Marone, D.; Russo, M.A.; De Vita, P.; Miullo, V.; Ferragonio, P.; Blanco, A.; Gadaleta, A.; Mastrangelo, A.M. Mapping QTL for Root and Shoot Morphological Traits in a Durum Wheat × T. dicoccum Segregating Population at Seedling Stage. Int. J. Genom. 2017, 2017, 6876393. [Google Scholar] [CrossRef] [Green Version]
- Deng, N.; Ling, X.; Sun, Y.; Zhang, C.; Fahad, S.; Peng, S.; Cui, K.; Nie, L.; Huang, J. Influence of Temperature and Solar Radiation on Grain Yield and Quality in Irrigated Rice System. Eur. J. Agron. 2015, 64, 37–46. [Google Scholar] [CrossRef]
- Zaïm, M.; Kabbaj, H.; Kehel, Z.; Gorjanc, G.; Filali-Maltouf, A.; Belkadi, B.; Nachit, M.M.; Bassi, F.M. Combining QTL Analysis and Genomic Predictions for Four Durum Wheat Populations Under Drought Conditions. Front. Genet. 2020, 11, 316. [Google Scholar] [CrossRef]
- Wang, Z.; Dhakal, S.; Cerit, M.; Wang, S.; Rauf, Y.; Yu, S.; Maulana, F.; Huang, W.; Anderson, J.D.; Ma, X.F.; et al. QTL mapping of yield components and kernel traits in wheat cultivars TAM 112 and Duster. Front Plant Sci. 2022, 13, 1057701. [Google Scholar] [CrossRef]
- Ramya, P.; Chaubal, A.; Kulkarni, K.; Gupta, L.; Kadoo, N.; Dhaliwal, H.S.; Chhuneja, P.; Lagu, M.; Gupta, V. QTL Mapping of 1000-Kernel Weight, Kernel Length, and Kernel Width in Bread Wheat (Triticum aestivum L.). J. Appl. Genet. 2010, 51, 421–429. [Google Scholar] [CrossRef]
- Cui, F.; Ding, A.; Zhao, C.; Feng, D.; Wang, X.; Wang, L.; Gao, J.; Wang, H. Wheat Kernel Dimensions: How Do They Contribute to Kernel Weight at an Individual QTL Level? J. Genet. 2011, 90, 409–425. [Google Scholar] [CrossRef]
- Peng, J.; Ronin, Y.; Fahima, T.; Röder, M.S.; Li, Y.; Nevo, E.; Korol, A. Domestication Quantitative Trait Loci in Triticum Dicoccoides, the Progenitor of Wheat. Proc. Natl. Acad. Sci. USA 2003, 100, 2489–2494. [Google Scholar] [CrossRef]
- Elouafi, I.; Nachit, M.M. A Genetic Linkage Map of the Durum × Triticum Dicoccoides Backcross Population Based on SSRs and AFLP Markers, and QTL Analysis for Milling Traits. Theor. Appl. Genet. 2004, 108, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, M.; Sanguineti, M.C.; Corneti, S.; Ortega, J.L.A.; Salem, M.B.; Bort, J.; DeAmbrogio, E.; Del Moral, L.F.G.; Demontis, A.; El-Ahmed, A.; et al. Quantitative Trait Loci for Grain Yield and Adaptation of Durum Wheat (Triticum durum Desf.) across a Wide Range of Water Availability. Genetics 2008, 178, 489–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanco, A.; Mangini, G.; Giancaspro, A.; Giove, S.; Colasuonno, P.; Simeone, R.; Signorile, A.; De Vita, P.; Mastrangelo, A.M.; Cattivelli, L.; et al. Relationships between Grain Protein Content and Grain Yield Components through Quantitative Trait Locus Analyses in a Recombinant Inbred Line Population Derived from Two Elite Durum Wheat Cultivars. Mol. Breed. 2012, 30, 79–92. [Google Scholar] [CrossRef]
- Patil, R.M.; Tamhankar, S.A.; Oak, M.D.; Raut, A.L.; Honrao, B.K.; Rao, V.S.; Misra, S.C. Mapping of QTL for Agronomic Traits and Kernel Characters in Durum Wheat (Triticum durum Desf.). Euphytica 2013, 190, 117–129. [Google Scholar] [CrossRef]
- Russo, M.A.; Ficco, D.B.M.; Laidò, G.; Marone, D.; Papa, R.; Blanco, A.; Gadaleta, A.; De Vita, P.; Mastrangelo, A.M. A Dense Durum Wheat × T. dicoccum Linkage Map Based on SNP Markers for the Study of Seed Morphology. Mol. Breed. 2014, 34, 1579–1597. [Google Scholar] [CrossRef]
- Fatiukha, A.; Filler, N.; Lupo, I.; Lidzbarsky, G.; Klymiuk, V.; Korol, A.B.; Pozniak, C.; Fahima, T.; Krugman, T. Grain Protein Content and Thousand Kernel Weight QTLs Identified in a Durum × Wild Emmer Wheat Mapping Population Tested in Five Environments. Theor. Appl. Genet. 2020, 133, 119–131. [Google Scholar] [CrossRef]
- Huang, F.; Li, X.; Du, X.; Li, S.; Li, N.; Lv, Y.; Zou, S.; Zhang, Q.; Wang, L.; Ni, Z.; et al. SNP-Based Identification of QTLs for Thousand-Grain Weight and Related Traits in Wheat 8762/Keyi 5214 DH Lines. J. Integr. Agric. 2023; in press. [Google Scholar] [CrossRef]
- Abbo, S.; Pinhasi van-Oss, R.; Gopher, A.; Saranga, Y.; Ofner, I.; Peleg, Z. Plant Domestication versus Crop Evolution: A Conceptual Framework for Cereals and Grain Legumes. Trends Plant Sci. 2014, 19, 351–360. [Google Scholar] [CrossRef]
- Lin, Y.; Zhou, S.; Liang, X.; Han, B.; Yang, J.; Guo, B.; Zhang, J.; Han, H.; Liu, W.; Yang, X.; et al. Introgression of Chromosome 6PL Terminal Segment from Agropyron Cristatum to Increase Both Grain Number and Grain Weight in Wheat. Crop J. 2023, 11, 878–886. [Google Scholar] [CrossRef]
- Börner, A.; Schumann, E.; Fürste, A.; Cöster, H.; Leithold, B.; Röder, M.S.; Weber, W.E. Mapping of Quantitative Trait Loci Determining Agronomic Important Characters in Hexaploid Wheat (Triticum aestivum L.). Theor. Appl. Genet. 2002, 105, 921–936. [Google Scholar] [CrossRef]
- Sun, L.; Huang, S.; Sun, G.; Zhang, Y.; Hu, X.; Nevo, E.; Peng, J.; Sun, D. SNP-Based Association Study of Kernel Architecture in a Worldwide Collection of Durum Wheat Germplasm. PLoS ONE 2020, 15, e0229159. [Google Scholar] [CrossRef]
- Ji, G.; Xu, Z.; Fan, X.; Zhou, Q.; Chen, L.; Yu, Q.; Liao, S.; Jiang, C.; Feng, B.; Wang, T. Identification and Validation of Major QTL for Grain Size and Weight in Bread Wheat (Triticum aestivum L.). Crop J. 2023, 11, 564–572. [Google Scholar] [CrossRef]
- Liao, S.; Xu, Z.; Fan, X.; Zhou, Q.; Liu, X.; Jiang, C.; Chen, L.; Lin, D.; Feng, B.; Wang, T. Genetic Dissection and Validation of a Major QTL for Grain Weight on Chromosome 3B in Bread Wheat (Triticum aestivum L.). J. Integr. Agric. 2023; in press. [Google Scholar] [CrossRef]
- Peleg, Z.; Fahima, T.; Korol, A.B.; Abbo, S.; Saranga, Y. Genetic Analysis of Wheat Domestication and Evolution under Domestication. J. Exp. Bot. 2011, 62, 5051–5061. [Google Scholar] [CrossRef] [Green Version]
- Golabadi, M.; Arzani, A.; Maibody, S.A.M.M.; Sayed Tabatabaei, B.E.; Mohammadi, S.A.; Arzani, A.; Sayed Tabatabaei, B.E. Identification of Microsatellite Markers Linked with Yield Components under Drought Stress at Terminal Growth Stages in Durum Wheat. Euphytica 2011, 177, 207–221. [Google Scholar] [CrossRef]
- Faris, J.D.; Zhang, Q.; Chao, S.; Zhang, Z.; Xu, S.S. Analysis of Agronomic and Domestication Traits in a Durum × Cultivated Emmer Wheat Population Using a High-Density Single Nucleotide Polymorphism-Based Linkage Map. Theor. Appl. Genet. 2014, 127, 2333–2348. [Google Scholar] [CrossRef] [PubMed]
- Soriano, J.M.; Royo, C. Dissecting the Genetic Architecture of Leaf Rust Resistance in Wheat by QTL Meta-Analysis. Phytopathology 2015, 105, 1585–1593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avni, R.; Oren, L.; Shabtay, G.; Assili, S.; Pozniak, C.; Hale, I.; Ben-David, R.; Peleg, Z.; Distelfeld, A. Genome Based Meta-QTL Analysis of Grain Weight in Tetraploid Wheat Identifies Rare Alleles of GRF4 Associated with Larger Grains. Genes 2018, 9, 636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, N.; Rana, M.; Kumar, B.; Chand, S.; Shiv, A.; Wani, S.H.; Kumar, S. Genomic Selection for Wheat Improvement. In Physiological, Molecular, and Genetic Perspectives of Wheat Improvement; Springer: Cham, Switzerland, 2020; pp. 175–207. [Google Scholar] [CrossRef]
- Liu, Y.; Salsman, E.; Wang, R.; Galagedara, N.; Zhang, Q.; Fiedler, J.D.; Liu, Z.; Xu, S.; Faris, J.D.; Li, X. Meta-QTL Analysis of Tan Spot Resistance in Wheat. Theor. Appl. Genet. 2020, 133, 2363–2375. [Google Scholar] [CrossRef]
- Bin Safdar, L.; Andleeb, T.; Latif, S.; Umer, M.J.; Tang, M.; Li, X.; Liu, S.; Quraishi, U.M. Genome-Wide Association Study and QTL Meta-Analysis Identified Novel Genomic Loci Controlling Potassium Use Efficiency and Agronomic Traits in Bread Wheat. Front. Plant Sci. 2020, 11, 70. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Amo, A.; Wei, D.; Chai, Y.; Zheng, J.; Qiao, P.; Cui, C.; Lu, S.; Chen, L.; Hu, Y.G. Large-Scale Integration of Meta-QTL and Genome-Wide Association Study Discovers the Genomic Regions and Candidate Genes for Yield and Yield-Related Traits in Bread Wheat. Theor. Appl. Genet. 2021, 134, 3083–3109. [Google Scholar] [CrossRef]
- Tyagi, S.; Mir, R.R.; Balyan, H.S.; Gupta, P.K. Interval Mapping and Meta-QTL Analysis of Grain Traits in Common Wheat (Triticum aestivum L.). Euphytica 2015, 201, 367–380. [Google Scholar] [CrossRef]
- Maccaferri, M.; Ricci, A.; Salvi, S.; Milner, S.G.; Noli, E.; Martelli, P.L.; Casadio, R.; Akhunov, E.; Scalabrin, S.; Vendramin, V.; et al. A High-Density, SNP-Based Consensus Map of Tetraploid Wheat as a Bridge to Integrate Durum and Bread Wheat Genomics and Breeding. Plant Biotechnol. J. 2015, 13, 648–663. [Google Scholar] [CrossRef] [PubMed]
- Chardon, F.; Virlon, B.; Moreau, L.; Falque, M.; Joets, J.; Decousset, L.; Murigneux, A.; Charcosset, A. Genetic Architecture of Flowering Time in Maize As Inferred From Quantitative Trait Loci Meta-Analysis and Synteny Conservation with the Rice Genome. Genetics 2004, 168, 2169–2185. [Google Scholar] [CrossRef] [Green Version]
- Saini, D.K.; Srivastava, P.; Pal, N.; Gupta, P.K. Meta-QTLs, Ortho-Meta-QTLs and Candidate Genes for Grain Yield and Associated Traits in Wheat (Triticum aestivum L.). Theor. Appl. Genet. 2022, 135, 1049–1081. [Google Scholar] [CrossRef] [PubMed]
- Miralles, D.J.; Richards, R.A.; Slafer, G.A. Duration of the Stem Elongation Period Influences the Number of Fertile Florets in Wheat and Barley. Funct. Plant Biol. 2000, 27, 931–940. [Google Scholar] [CrossRef]
- González, F.G.; Slafer, G.A.; Miralles, D.J. Pre-Anthesis Development and Number of Fertile Florets in Wheat as Affected by Photoperiod Sensitivity Genes Ppd-D1 and Ppd-B1. Euphytica 2005, 146, 253–269. [Google Scholar] [CrossRef]
- Sanna, G.; Giunta, F.; Motzo, R.; Mastrangelo, A.M.; De Vita, P. Genetic Variation for the Duration of Pre-Anthesis Development in Durum Wheat and Its Interaction with Vernalization Treatment and Photoperiod. J. Exp. Bot. 2014, 65, 3177–3188. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Liu, D.; Li, Q.; Mao, X.; Li, A.; Wang, J.; Chang, X.; Jing, R. Overexpression of Wheat Gene TaMOR Improves Root System Architecture and Grain Yield in Oryza Sativa. J. Exp. Bot. 2016, 67, 4155–4167. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Li, M.; Ashraf, U.; Liu, S.; Zhang, J. Exploring the Relationships between Yield and Yield-Related Traits for Rice Varieties Released in China from 1978 to 2017. Front. Plant Sci. 2019, 10, 543. [Google Scholar] [CrossRef] [Green Version]
- Nadolska-Orczyk, A.; Rajchel, I.K.; Orczyk, W.; Gasparis, S. Major Genes Determining Yield-Related Traits in Wheat and Barley. Theor. Appl. Genet. 2017, 130, 1081–1098. [Google Scholar] [CrossRef] [Green Version]
- Xie, G.; Li, Z.; Ran, Q.; Wang, H.; Zhang, J. Over-Expression of Mutated ZmDA1 or ZmDAR1 Gene Improves Maize Kernel Yield by Enhancing Starch Synthesis. Plant Biotechnol. J. 2018, 16, 234–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Lawit, S.J.; Weers, B.; Sun, J.; Mongar, N.; van Hemert, J.; Melo, R.; Meng, X.; Rupe, M.; Clapp, J.; et al. Overexpression of Zmm28 Increases Maize Grain Yield in the Field. Proc. Natl. Acad. Sci. USA 2019, 116, 23850–23858. [Google Scholar] [CrossRef] [Green Version]
- Su, Z.; Hao, C.; Wang, L.; Dong, Y.; Zhang, X. Identification and Development of a Functional Marker of TaGW2 Associated with Grain Weight in Bread Wheat (Triticum aestivum L.). Theor. Appl. Genet. 2011, 122, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhao, Y.L.; Gao, L.F.; Zhao, G.Y.; Zhou, R.H.; Zhang, B.S.; Jia, J.Z. TaCKX6-D1, the Ortholog of Rice OsCKX2, Is Associated with Grain Weight in Hexaploid Wheat. New Phytol. 2012, 195, 574–584. [Google Scholar] [CrossRef]
- Dong, C.; Fu, Y.; Liu, G.; Liu, H. Growth, Photosynthetic Characteristics, Antioxidant Capacity and Biomass Yield and Quality of Wheat (Triticum aestivum L.) Exposed to LED Light Sources with Different Spectra Combinations. J. Agron. Crop Sci. 2014, 200, 219–230. [Google Scholar] [CrossRef]
- Jiang, Y.; Jiang, Q.; Hao, C.; Hou, J.; Wang, L.; Zhang, H.; Zhang, S.; Chen, X.; Zhang, X. A Yield-Associated Gene TaCWI, in Wheat: Its Function, Selection and Evolution in Global Breeding Revealed by Haplotype Analysis. Theor. Appl. Genet. 2015, 128, 131–143. [Google Scholar] [CrossRef]
- Feng, Y.; Zhai, R.R.; Lin, Z.C.; Cao, L.Y.; Wei, X.H.; Cheng, S.H. Quantitative Trait Locus Analysis for Rice Yield Traits under Two Nitrogen Levels. Rice Sci. 2015, 22, 108–115. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Li, T.; Hao, C.; Wang, Y.; Chen, X.; Zhang, X. TaGS5-3A, a Grain Size Gene Selected during Wheat Improvement for Larger Kernel and Yield. Plant Biotechnol. J. 2016, 14, 1269–1280. [Google Scholar] [CrossRef]
- Wang, S.; Yan, X.; Wang, Y.; Liu, H.; Cui, D.; Chen, F. Haplotypes of the TaGS5-A1 Gene Are Associated with Thousand-Kernel Weight in Chinese Bread Wheat. Front. Plant Sci. 2016, 7, 783. [Google Scholar] [CrossRef] [Green Version]
- Simmonds, J.; Scott, P.; Brinton, J.; Mestre, T.C.; Bush, M.; del Blanco, A.; Dubcovsky, J.; Uauy, C. A Splice Acceptor Site Mutation in TaGW2-A1 Increases Thousand Grain Weight in Tetraploid and Hexaploid Wheat through Wider and Longer Grains. Theor. Appl. Genet. 2016, 129, 1099–1112. [Google Scholar] [CrossRef] [Green Version]
- Sajjad, M.; Ma, X.; Habibullah Khan, S.; Shoaib, M.; Song, Y.; Yang, W.; Zhang, A.; Liu, D. TaFlo2-A1, an Ortholog of Rice Flo2, Is Associated with Thousand Grain Weight in Bread Wheat (Triticum aestivum L.). BMC Plant Biol. 2017, 17, 164. [Google Scholar] [CrossRef] [Green Version]
- Cao, P.; Liang, X.; Zhao, H.; Feng, B.; Xu, E.; Wang, L.; Hu, Y. Identification of the Quantitative Trait Loci Controlling Spike-Related Traits in Hexaploid Wheat (Triticum aestivum L.). Planta 2019, 250, 1967–1981. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhao, D.; Meng, Z.; Xu, K.; Yan, J.; Xia, X.; Cao, S.; Tian, Y.; He, Z.; Zhang, Y. QTL Mapping for Grain Yield-Related Traits in Bread Wheat via SNP-Based Selective Genotyping. Theor. Appl. Genet. 2020, 133, 857–872. [Google Scholar] [CrossRef] [PubMed]
- Marcotuli, I.; Gadaleta, A.; Mangini, G.; Signorile, A.M.; Zacheo, S.A.; Blanco, A.; Simeone, R.; Colasuonno, P. Development of a High-Density SNP-Based Linkage Map and Detection of QTL for β-Glucans, Protein Content, Grain Yield per Spike and Heading Time in Durum Wheat. Int. J. Mol. Sci. 2017, 18, 1329. [Google Scholar] [CrossRef] [Green Version]
- Murai, K.; Miyamae, M.; Kato, H.; Takumi, S.; Ogihara, Y. WAP1, a Wheat APETALA1 Homolog, Plays a Central Role in the Phase Transition from Vegetative to Reproductive Growth. Plant Cell Physiol. 2003, 44, 1255–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, Y.Z.; Mao, S.L.; Jia, R.L.; Chun, M.G.; Xian, S.Z. The Wheat TaGI1, Involved in Photoperiodic Flowering, Encodes an Arabidopsis GI Ortholog. Plant Mol. Biol. 2005, 58, 53–64. [Google Scholar] [CrossRef]
- Guo, J.; Dai, S.; Li, H.; Liu, A.; Liu, C.; Cheng, D.; Cao, X.; Chu, X.; Zhai, S.; Liu, J.; et al. Identification and Expression Analysis of Wheat TaGF14 Genes. Front. Genet. 2018, 9, 12. [Google Scholar] [CrossRef]
- Sakuma, S.; Schnurbusch, T. Of Floral Fortune: Tinkering with the Grain Yield Potential of Cereal Crops. New Phytol. 2020, 225, 1873–1882. [Google Scholar] [CrossRef] [Green Version]
- Adamski, N.M.; Simmonds, J.; Brinton, J.F.; Backhaus, A.E.; Chen, Y.; Smedley, M.; Hayta, S.; Florio, T.; Crane, P.; Scott, P.; et al. Ectopic Expression of Triticum Polonicum VRT-A2 Underlies Elongated Glumes and Grains in Hexaploid Wheat in a Dosage-Dependent Manner. Plant Cell 2021, 33, 2296–2319. [Google Scholar] [CrossRef]
- Li, J.; Zhang, S.; Zhang, R.; Gao, J.; Qi, Y.; Song, G.; Li, W.; Li, Y.; Li, G. Efficient Multiplex Genome Editing by CRISPR/Cas9 in Common Wheat. Plant Biotechnol. J. 2021, 19, 427–429. [Google Scholar] [CrossRef]
- Dong, C.; Zhang, L.; Zhang, Q.; Yang, Y.; Li, D.; Xie, Z.; Cui, G.; Chen, Y.; Wu, L.; Li, Z.; et al. Tiller Number1 Encodes an Ankyrin Repeat Protein That Controls Tillering in Bread Wheat. Nat. Commun. 2023, 14, 836. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Hao, C.; Hou, J.; Wang, Y.; Li, T.; Wang, L.; Ma, Z.; Zhang, X. Homologous Haplotypes, Expression, Genetic Effects and Geographic Distribution of the Wheat Yield Gene TaGW2. BMC Plant Biol. 2014, 14, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sestili, F.; Pagliarello, R.; Zega, A.; Saletti, R.; Pucci, A.; Botticella, E.; Masci, S.; Tundo, S.; Moscetti, I.; Foti, S.; et al. Enhancing Grain Size in Durum Wheat Using RNAi to Knockdown GW2 Genes. Theor. Appl. Genet. 2019, 132, 419–429. [Google Scholar] [CrossRef]
- Hanif, M.; Gao, F.; Liu, J.; Wen, W.; Zhang, Y.; Rasheed, A.; Xia, X.; He, Z.; Cao, S. TaTGW6-A1, an Ortholog of Rice TGW6, Is Associated with Grain Weight and Yield in Bread Wheat. Mol. Breed. 2016, 36, 1. [Google Scholar] [CrossRef]
- Hu, M.J.; Zhang, H.P.; Cao, J.J.; Zhu, X.F.; Wang, S.X.; Jiang, H.; Wu, Z.Y.; Lu, J.; Chang, C.; Sun, G.L.; et al. Characterization of an IAA-Glucose Hydrolase Gene TaTGW6 Associated with Grain Weight in Common Wheat (Triticum aestivum L.). Mol. Breed. 2016, 36, 25. [Google Scholar] [CrossRef]
- Khan, N.; Zhang, Y.; Wang, J.; Li, Y.; Chen, X.; Yang, L.; Zhang, J.; Li, C.; Li, L.; Ur Rehman, S.; et al. TaGSNE, a WRKY Transcription Factor, Overcomes the Trade-off between Grain Size and Grain Number in Common Wheat and Is Associated with Root Development. J. Exp. Bot. 2022, 73, 6678–6696. [Google Scholar] [CrossRef]
- Tillett, B.J.; Hale, C.O.; Martin, J.M.; Giroux, M.J. Genes Impacting Grain Weight and Number in Wheat (Triticum aestivum L. ssp. aestivum). Plants 2022, 11, 1772. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Kong, Z.; Xie, Q.; Jia, H.; Huang, W.; Zhang, L.; Cheng, R.; Yang, Z.; Qi, X.; Lv, G.; et al. Fine Mapping of KLW1 That Conditions Kernel Weight Mainly through Regulating Kernel Length in Wheat (Triticum aestivum L.). Theor. Appl. Genet. 2023, 136, 110. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Jiang, Q.; Hao, C.; Wang, Y.; Zhang, H.; Zhang, X. Global Selection on Sucrose Synthase Haplotypes during a Century of Wheat Breeding 1. Plant Physiol. 2014, 164, 1918–1929. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Q.; Hou, J.; Hao, C.; Wang, L.; Ge, H.; Dong, Y.; Zhang, X. The Wheat (T. aestivum) Sucrose Synthase 2 Gene (TaSus2) Active in Endosperm Development Is Associated with Yield Traits. Funct. Integr. Genom. 2011, 11, 49–61. [Google Scholar] [CrossRef]
- Ma, D.; Yan, J.; He, Z.; Wu, L.; Xia, X. Characterization of a Cell Wall Invertase Gene TaCwi-A1 on Common Wheat Chromosome 2A and Development of Functional Markers. Mol. Breed. 2012, 29, 43–52. [Google Scholar] [CrossRef]
- Rustgi, S.; Shafqat, M.N.; Kumar, N.; Baenziger, P.S.; Ali, M.L.; Dweikat, I.; Campbell, B.T.; Gill, K.S. Genetic Dissection of Yield and Its Component Traits Using High-Density Composite Map of Wheat Chromosome 3A: Bridging Gaps between QTLs and Underlying Genes. PLoS ONE 2013, 8, e0070526. [Google Scholar] [CrossRef] [Green Version]
- Pien, S.; Wyrzykowska, J.; McQueen-Mason, S.; Smart, C.; Fleming, A. Local Expression of Expansin Induces the Entire Process of Leaf Development and Modifies Leaf Shape. Proc. Natl. Acad. Sci. USA 2001, 98, 11812–11817. [Google Scholar] [CrossRef]
- Rochange, S.F.; Wenzel, C.L.; Mcqueen-Mason, S.J. Impaired Growth in Transgenic Plants Over-Expressing an Expansin Isoform. Plant Mol. Biol. 2001, 46, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.; Lee, Y.; Cho, H.-T.; Kende, H. Regulation of Expansin Gene Expression Affects Growth and Development in Transgenic Rice Plants. Plant Cell 2003, 15, 1386–1398. [Google Scholar] [CrossRef] [Green Version]
- Calderini, D.F.; Castillo, F.M.; Arenas-M, A.; Molero, G.; Reynolds, M.P.; Craze, M.; Bowden, S.; Milner, M.J.; Wallington, E.J.; Dowle, A.; et al. Overcoming the Trade-off between Grain Weight and Number in Wheat by the Ectopic Expression of Expansin in Developing Seeds Leads to Increased Yield Potential. New Phytol. 2021, 230, 629–640. [Google Scholar] [CrossRef]
- Milner, M.J.; Bowden, S.; Craze, M.; Wallington, E.J. Ectopic Expression of TaBG1 Increases Seed Size and Alters Nutritional Characteristics of the Grain in Wheat but Does Not Lead to Increased Yields. BMC Plant Biol. 2021, 21, 524. [Google Scholar] [CrossRef]
- Guo, L.; Ma, M.; Wu, L.; Zhou, M.; Li, M.; Wu, B.; Li, L.; Liu, X.; Jing, R.; Chen, W.; et al. Modified Expression of TaCYP78A5 Enhances Grain Weight with Yield Potential by Accumulating Auxin in Wheat (Triticum aestivum L.). Plant Biotechnol. J. 2022, 20, 168–182. [Google Scholar] [CrossRef]
- van Esse, G.W. The Quest for Optimal Plant Architecture. Science 2022, 376, 133–134. [Google Scholar] [CrossRef] [PubMed]
- Beral, A.; Girousse, C.; Le Gouis, J.; Allard, V.; Slafer, G.A. Physiological Bases of Cultivar Differences in Average Grain Weight in Wheat: Scaling down from Plot to Individual Grain in Elite Material. Field Crops Res. 2022, 289, 108713. [Google Scholar] [CrossRef]
- Rodríguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 2017, 171, 470–480.e8. [Google Scholar] [CrossRef] [Green Version]
- Esposito, S.; D’Agostino, N.; Taranto, F.; Sonnante, G.; Sestili, F.; Lafiandra, D.; De Vita, P. Whole-Exome Sequencing of Selected Bread Wheat Recombinant Inbred Lines as a Useful Resource for Allele Mining and Bulked Segregant Analysis. Front. Genet. 2022, 13, 1058471. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, J.; Li, Y.; Ma, L. Current Strategies and Advances in Wheat Biology. Crop J. 2020, 8, 879–891. [Google Scholar] [CrossRef]
- N’Diaye, A.; Haile, J.K.; Nilsen, K.T.; Walkowiak, S.; Ruan, Y.; Singh, A.K.; Clarke, F.R.; Clarke, J.M.; Pozniak, C.J. Haplotype Loci under Selection in Canadian Durum Wheat Germplasm over 60 Years of Breeding: Association with Grain Yield, Quality Traits, Protein Loss, and Plant Height. Front. Plant Sci. 2018, 9, 1589. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.W.; Kaplan, N.L. On the Advantage of Haplotype Analysis in the Presence of Multiple Disease Susceptibility Alleles. Genet. Epidemiol. 2002, 23, 221–233. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Aranzana, M.J.; Kim, S.; Lister, C.; Shindo, C.; Tang, C.; Toomajian, C.; Zheng, H.; Dean, C.; Marjoram, P.; et al. An Arabidopsis Example of Association Mapping in Structured Samples. PLoS Genet. 2007, 3, e4. [Google Scholar] [CrossRef] [Green Version]
- Hamblin, M.T.; Jannink, J.-L. Factors Affecting the Power of Haplotype Markers in Association Studies; Factors Affecting the Power of Haplotype Markers in Association Studies. Plant Genome 2011, 4, 145–153. [Google Scholar] [CrossRef] [Green Version]
- Schoen, A.; Yadav, I.; Wu, S.; Poland, J.; Rawat, N.; Tiwari, V. Identification and High-Resolution Mapping of a Novel Tiller Number Gene (Tin6) by Combining Forward Genetics Screen and MutMap Approach in Bread Wheat. Funct. Integr. Genom. 2023, 23, 157. [Google Scholar] [CrossRef]
- Sánchez-Martín, J.; Steuernagel, B.; Ghosh, S.; Herren, G.; Hurni, S.; Adamski, N.; Vrána, J.; Kubaláková, M.; Krattinger, S.G.; Wicker, T.; et al. Rapid Gene Isolation in Barley and Wheat by Mutant Chromosome Sequencing. Genome Biol. 2016, 17, 221. [Google Scholar] [CrossRef] [Green Version]
- Thind, A.K.; Wicker, T.; Šimková, H.; Fossati, D.; Moullet, O.; Brabant, C.; Vrána, J.; Doležel, J.; Krattinger, S.G. Rapid Cloning of Genes in Hexaploid Wheat Using Cultivar-Specific Long-Range Chromosome Assembly. Nat. Biotechnol. 2017, 35, 793–796. [Google Scholar] [CrossRef] [Green Version]
- Bentley, A.R.; Scutari, M.; Gosman, N.; Faure, S.; Bedford, F.; Howell, P.; Cockram, J.; Rose, G.A.; Barber, T.; Irigoyen, J.; et al. Applying Association Mapping and Genomic Selection to the Dissection of Key Traits in Elite European Wheat. Theor. Appl. Genet. 2014, 127, 2619–2633. [Google Scholar] [CrossRef] [PubMed]
- Bassi, F.M.; Bentley, A.R.; Charmet, G.; Ortiz, R.; Crossa, J. Breeding Schemes for the Implementation of Genomic Selection in Wheat (Triticum spp.). Plant Sci. 2015, 242, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.K.; Balyan, H.S.; Sharma, S.; Kumar, R. Genetics of Yield, Abiotic Stress Tolerance and Biofortification in Wheat (Triticum aestivum L.). Theor. Appl. Genet. 2020, 133, 1569–1602. [Google Scholar] [CrossRef] [PubMed]
- Cappetta, E.; Andolfo, G.; Guadagno, A.; Di Matteo, A.; Barone, A.; Frusciante, L.; Ercolano, M.R. Tomato Genomic Prediction for Good Performance under High-Temperature and Identification of Loci Involved in Thermotolerance Response. Hortic. Res. 2021, 8, 212. [Google Scholar] [CrossRef] [PubMed]
- Cappetta, E.; Andolfo, G.; Di Matteo, A.; Barone, A.; Frusciante, L.; Ercolano, M.R. Accelerating Tomato Breeding by Exploiting Genomic Selection Approaches. Plants 2020, 9, 1236. [Google Scholar] [CrossRef] [PubMed]
- Massman, J.M.; Jung, H.J.G.; Bernardo, R. Genomewide Selection versus Marker-Assisted Recurrent Selection to Improve Grain Yield and Stover-Quality Traits for Cellulosic Ethanol in Maize. Crop Sci. 2013, 53, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Beyene, Y.; Semagn, K.; Mugo, S.; Tarekegne, A.; Babu, R.; Meisel, B.; Sehabiague, P.; Makumbi, D.; Magorokosho, C.; Oikeh, S.; et al. Genetic Gains in Grain Yield Through Genomic Selection in Eight Bi-Parental Maize Populations under Drought Stress. Crop Sci. 2015, 55, 154–163. [Google Scholar] [CrossRef] [Green Version]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Contaldi, F.; Cappetta, E.; Esposito, S. Practical Workflow from High-Throughput Genotyping to Genomic Estimated Breeding Values (GEBVs). Methods Mol. Biol. 2021, 2264, 119–135. [Google Scholar] [CrossRef]
- Battenfield, S.D.; Guzmán, C.; Gaynor, R.C.; Singh, R.P.; Peña, R.J.; Dreisigacker, S.; Fritz, A.K.; Poland, J.A. Genomic Selection for Processing and End-Use Quality Traits in the CIMMYT Spring Bread Wheat Breeding Program. Plant Genome 2016, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Crossa, J.; De Los Campos, G.; Pérez, P.; Gianola, D.; Burgueño, J.; Araus, J.L.; Makumbi, D.; Singh, R.P.; Dreisigacker, S.; Yan, J.; et al. Prediction of Genetic Values of Quantitative Traits in Plant Breeding Using Pedigree and Molecular Markers. Genetics 2010, 186, 713–724. [Google Scholar] [CrossRef] [Green Version]
- González-Camacho, J.M.; De Los Campos, G.; Pérez, P.; Gianola, D.; Cairns, J.E.; Mahuku, G.; Babu, R.; Crossa, J. Genome-Enabled Prediction of Genetic Values Using Radial Basis Function Neural Networks. Theor. Appl. Genet. 2012, 125, 759–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, S.; Schulthess, A.W.; Mirdita, V.; Zhao, Y.; Korzun, V.; Bothe, R.; Ebmeyer, E.; Reif, J.C.; Jiang, Y. Genomic Selection in a Commercial Winter Wheat Population. Theor. Appl. Genet. 2016, 129, 641–651. [Google Scholar] [CrossRef]
- Belamkar, V.; Guttieri, M.J.; Hussain, W.; Jarquín, D.; El-basyoni, I.; Poland, J.; Lorenz, A.J.; Baenziger, P.S. Genomic Selection in Preliminary Yield Trials in a Winter Wheat Breeding Program. G3: Genes Genomes Genet. 2018, 8, 2735–2747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juliana, P.; Singh, R.P.; Braun, H.J.; Huerta-Espino, J.; Crespo-Herrera, L.; Govindan, V.; Mondal, S.; Poland, J.; Shrestha, S. Genomic Selection for Grain Yield in the CIMMYT Wheat Breeding Program—Status and Perspectives. Front. Plant Sci. 2020, 11, 1418. [Google Scholar] [CrossRef]
- Crossa, J.; Jarquín, D.; Franco, J.; Pérez-Rodríguez, P.; Burgueño, J.; Saint-Pierre, C.; Vikram, P.; Sansaloni, C.; Petroli, C.; Akdemir, D.; et al. Genomic Prediction of Gene Bank Wheat Landraces. G3 2016, 6, 1819–1834. [Google Scholar] [CrossRef] [Green Version]
- Haile, J.K.; N’diaye, A.; Clarke, F.; Clarke, J.; Knox, R.; Rutkoski, J.; Bassi, F.M.; Pozniak, C.J.; Haile, J.K.; N’diaye, A.; et al. Genomic Selection for Grain Yield and Quality Traits in Durum Wheat. Mol. Breed. 2018, 38, 75. [Google Scholar] [CrossRef]
- Rapp, M.; Lein, V.; Lacoudre, F.; Lafferty, J.; Müller, E.; Vida, G.; Bozhanova, V.; Ibraliu, A.; Thorwarth, P.; Piepho, H.P.; et al. Simultaneous Improvement of Grain Yield and Protein Content in Durum Wheat by Different Phenotypic Indices and Genomic Selection. Theor. Appl. Genet. 2018, 131, 1315–1329. [Google Scholar] [CrossRef] [PubMed]
- Mérida-García, R.; Liu, G.; He, S.; Gonzalez-Dugo, V.; Dorado, G.; Gálvez, S.; Solís, I.; Zarco-Tejada, P.J.; Reif, J.C.; Hernandez, P. Genetic Dissection of Agronomic and Quality Traits Based on Association Mapping and Genomic Selection Approaches in Durum Wheat Grown in Southern Spain. PLoS ONE 2019, 14, e0211718. [Google Scholar] [CrossRef] [Green Version]
- Sarup, P.; Jensen, J.; Ostersen, T.; Henryon, M.; Sørensen, P. Increased Prediction Accuracy Using a Genomic Feature Model Including Prior Information on Quantitative Trait Locus Regions in Purebred Danish Duroc Pigs. BMC Genet. 2016, 17, 11. [Google Scholar] [CrossRef] [Green Version]
- Michel, S.; Löschenberger, F.; Ametz, C.; Pachler, B.; Sparry, E.; Bürstmayr, H. Simultaneous Selection for Grain Yield and Protein Content in Genomics-Assisted Wheat Breeding. Theor. Appl. Genet. 2019, 132, 1745–1760. [Google Scholar] [CrossRef] [PubMed]
- Jannink, J.-L.; Lorenz, A.J.; Iwata, H. Genomic Selection in Plant Breeding: From Theory to Practice. Brief. Funct. Genom. 2010, 9, 166–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taranto, F.; Esposito, S.; De Vita, P. Genomics for Yield and Yield Components in Durum Wheat. Plants 2023, 12, 2571. https://doi.org/10.3390/plants12132571
Taranto F, Esposito S, De Vita P. Genomics for Yield and Yield Components in Durum Wheat. Plants. 2023; 12(13):2571. https://doi.org/10.3390/plants12132571
Chicago/Turabian StyleTaranto, Francesca, Salvatore Esposito, and Pasquale De Vita. 2023. "Genomics for Yield and Yield Components in Durum Wheat" Plants 12, no. 13: 2571. https://doi.org/10.3390/plants12132571




