Epigenetic Modifications Related to Potato Skin Russeting
Abstract
:1. Introduction
2. Results
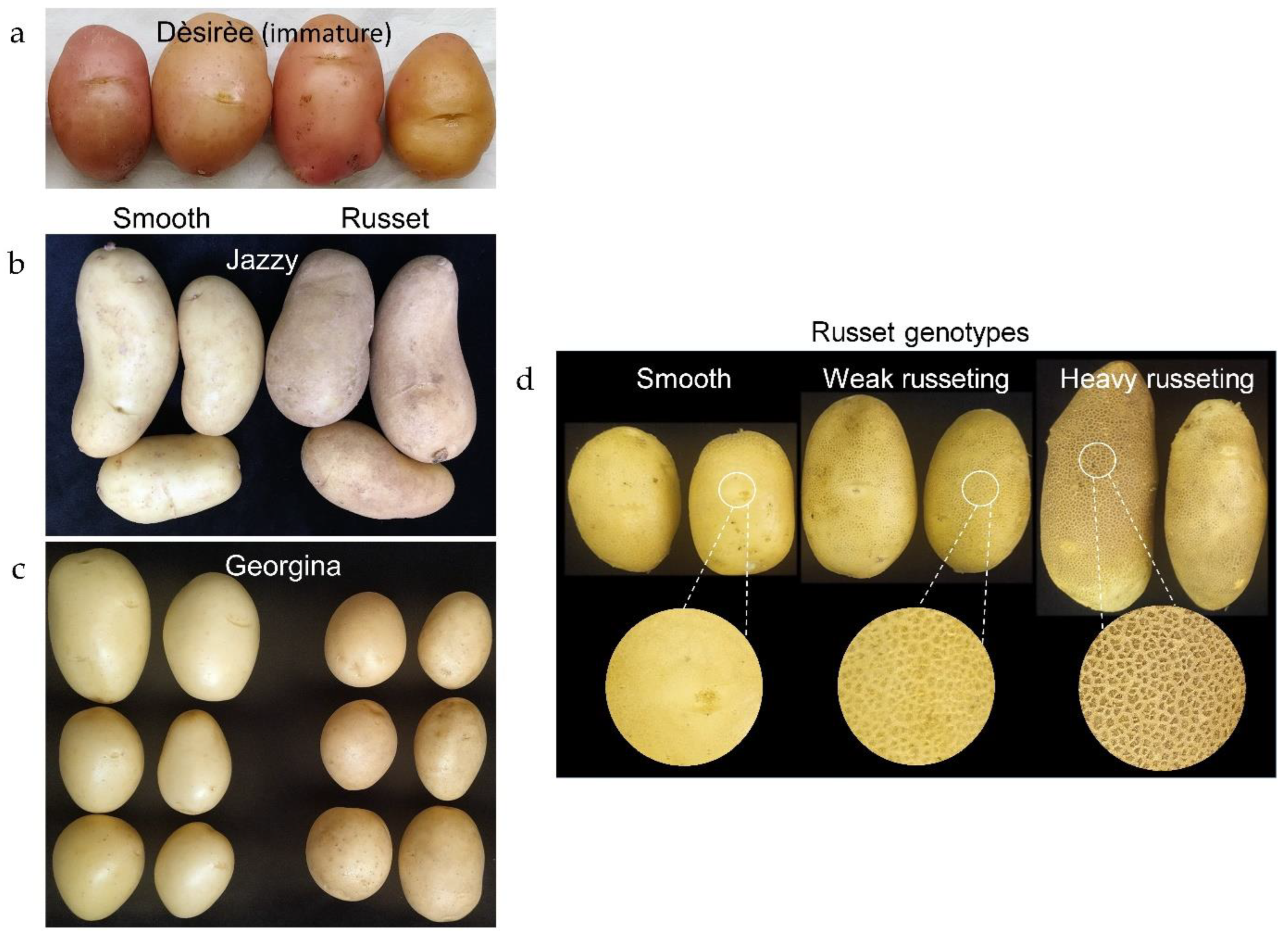
2.1. Description of Russet Skin
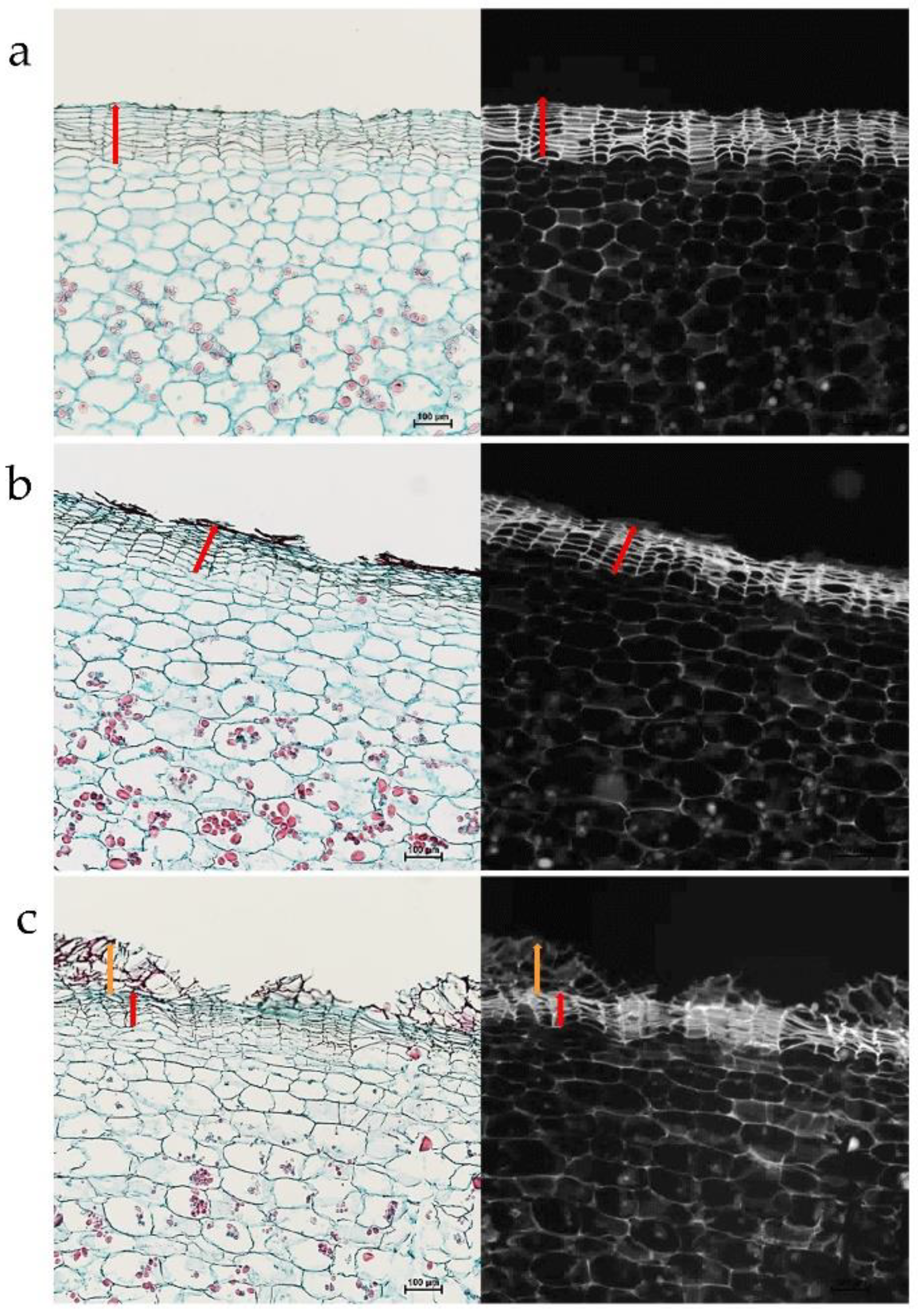
2.2. Anatomical Study of Russeted Skin
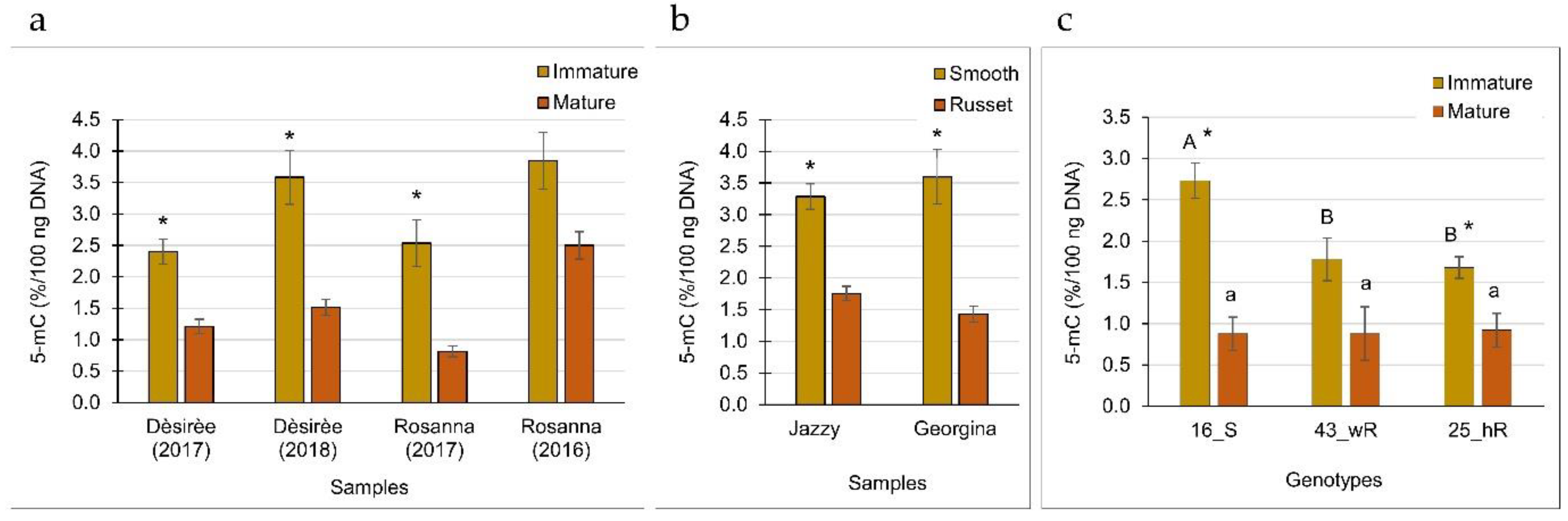
2.3. Global DNA Methylation of Russeted Skin
2.4. Expression of Epigenetic Modifiers in Russeted Skin
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Anatomical Studies
4.3. Total DNA Extraction and Global DNA Methylation (GDM) Assay
4.4. Total RNA Extraction, cDNA Synthesis, and Quantitative Reverse Transcriptase PCR (qRT-PCR)
| Gene Code | Gene | Potato ID | Forward Primers | Reverse Primers |
|---|---|---|---|---|
| DML1 | DEMETER-LIKE DNA GLYCOSYLASE 1 | Soltu.DM.09G004240 | CCACCAAATGTTTCTCGACC | TGTTGTGGCTTCAATACCCT |
| DML3 | DEMETER-LIKE DNA GLYCOSYLASE 3 | Soltu.DM.04G017430 | CTGAGCACACAGTTTTGAGG | ATTGCTCTCAGCCTGTAACC |
| DME | DNA GLYCOSYLASE DEMETER | Soltu.DM.11G005260 | ATTCAAAGGCCTAACCACAG | CTGGCTTTCCTTTTGTCCTA |
| MET1 | DNA (CYTOSINE-5)-METHYLTRANSFERASE 1 | Soltu.DM.11G013230 | TAAGGTGGGGATGTGCTTTC | GTGCATATGCCAAAGGAGGT |
| CMT3 | CHROMOMETHYLASE 3 | Soltu.DM.12G000130 | ACTTGCCTGGAGTTCGTGTT | GTAGTTCCTCAAAACCGTTT |
| DRM2 | DOMAINS REARRANGED | Soltu.DM.02G006560 | GGATCAGGAAAGCTGTGGAG | GGTTCTTTGGAAACCCCAAT |
| NAC | NASCENT POLYPEPTIDE-ASSOCIATED COMPLEX | Soltu.DM.09G002650 | ATATAGAGCTGGTGATGACT | TCCATGATAGCAGAGACTA |
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reeve, R.M.; Hautala, E.; Weaver, M.L. Anatomy and compositional variation within potatoes 1. Developmental histology of the tuber. Am. Potato J. 1969, 46, 361–373. [Google Scholar] [CrossRef]
- Kumar, P.; Ginzberg, I. Potato periderm development and tuber skin quality. Plants 2022, 11, 2099. [Google Scholar] [CrossRef] [PubMed]
- Woolfson, K.N.; Esfandiari, M.; Bernards, M.A. Suberin biosynthesis, assembly, and regulation. Plants 2022, 11, 555. [Google Scholar] [CrossRef]
- Nomberg, G.; Marinov, O.; Arya, G.C.; Manasherova, E.; Cohen, H. The key enzymes in the suberin biosynthetic pathway in plants: An update. Plants 2022, 11, 392. [Google Scholar] [CrossRef] [PubMed]
- De Jong, H. Inheritance of russeting in cultivated diploid potatoes. Potato Res. 1981, 24, 309–313. [Google Scholar] [CrossRef]
- Ginzberg, I.; Barel, G.; Ophir, R.; Tzin, E.; Tanami, Z.; Muddarangappa, T.; De Jong, W.; Fogelman, E. Transcriptomic profiling of heat-stress response in potato periderm. J. Exp. Bot. 2009, 60, 4411–4421. [Google Scholar] [CrossRef]
- Ginzberg, I.; Minz, D.; Faingold, I.; Soriano, S.; Mints, M.; Fogelman, E.; Warshavsky, S.; Zig, U.; Yermiyahu, U. Calcium mitigated potato skin physiological disorder. Am. J. Potato Res. 2012, 89, 351–362. [Google Scholar] [CrossRef]
- Keren-Keiserman, A.; Baghel, R.S.; Fogelman, E.; Faingold, I.; Zig, U.; Yermiyahu, U.; Ginzberg, I. Effects of polyhalite fertilization on skin quality of potato tuber. Front. Plant Sci. 2019, 10, 1379. [Google Scholar] [CrossRef]
- Okazawa, Y.; Iriuda, N. On occurrence of defected potato tubers with rough russeted skin. Jpn. J. Crop Sci. 1980, 49, 58–65. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Timm, H.; Spurr, A.R. Effects of soil temperature on growth and nutrition of potato plants and tuberization, composition, and periderm structure of tubers. Proc. Am. Soc. Hortic. Sci. 1964, 84, 412–423. [Google Scholar]
- Vulavala, V.K.R.; Fogelman, E.; Faigenboim, A.; Shoseyov, O.; Ginzberg, I. The transcriptome of potato tuber phellogen reveals cellular functions of cork cambium and genes involved in periderm formation and maturation. Sci. Rep. 2019, 9, 10216. [Google Scholar] [CrossRef] [PubMed]
- Murphy, H.; Goven, M. Influence of source of potash on yield, specific gravity, and surface russeting of the Russet Burbank variety in Maine. Am. Potato J. 1965, 42, 192–194. [Google Scholar] [CrossRef]
- Inácio, V.; Barros, P.M.; Costa, A.; Roussado, C.; Gonçalves, E.; Costa, R.; Graça, J.; Oliveira, M.M.; Morais-Cecílio, L. Differential DNA methylation patterns are related to phellogen origin and quality of Quercus suber cork. PLoS ONE 2017, 12, e0169018. [Google Scholar] [CrossRef] [PubMed]
- Inácio, V.; Martins, M.T.; Graça, J.; Morais-Cecílio, L. Cork oak young and traumatic periderms show PCD typical chromatin patterns but different chromatin-modifying genes expression. Front. Plant Sci. 2018, 9, 1194. [Google Scholar] [CrossRef] [PubMed]
- Ramos, M.; Rocheta, M.; Carvalho, L.; Inácio, V.; Graça, J.; Morais-Cecilio, L. Expression of DNA methyltransferases is involved in Quercus suber cork quality. Tree Genet. Genomes 2013, 9, 1481–1492. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.-K. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 2009, 12, 133–139. [Google Scholar] [CrossRef]
- Lieberman-Lazarovich, M.; Kaiserli, E.; Bucher, E.; Mladenov, V. Natural and induced epigenetic variation for crop improvement. Curr. Opin. Plant Biol. 2022, 70, 102297. [Google Scholar] [CrossRef]
- Barel, G.; Ginzberg, I. Potato skin proteome is enriched with plant defense components. J. Exp. Bot. 2008, 59, 3347–3357. [Google Scholar] [CrossRef]
- Kooke, R.; Johannes, F.; Wardenaar, R.; Becker, F.; Etcheverry, M.; Colot, V.; Vreugdenhil, D.; Keurentjes, J.J.B. Epigenetic basis of morphological variation and phenotypic plasticity in Arabidopsis thaliana. Plant Cell 2015, 27, 337–348. [Google Scholar] [CrossRef]
- Secco, D.; Whelan, J.; Rouached, H.; Lister, R. Nutrient stress-induced chromatin changes in plants. Curr. Opin. Plant Biol. 2017, 39, 1–7. [Google Scholar] [CrossRef]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Goll, M.G.; Bestor, T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005, 74, 481–514. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhong, X.; Bernatavichute, Y.V.; Stroud, H.; Feng, S.; Caro, E.; Vashisht, A.A.; Terragni, J.; Chin, H.G.; Tu, A. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 2012, 151, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Stroud, H.; Do, T.; Du, J.; Zhong, X.; Feng, S.; Johnson, L.; Patel, D.J.; Jacobsen, S.E. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 2014, 21, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; Kim, M.Y.; Hsieh, P.-H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef]
- Yaari, R.; Katz, A.; Domb, K.; Harris, K.D.; Zemach, A.; Ohad, N. RdDM-independent de novo and heterochromatin DNA methylation by plant CMT and DNMT3 orthologs. Nat. Commun. 2019, 10, 1613. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef]
- Zhu, J.-K. Active DNA Demethylation Mediated by DNA Glycosylases. Annu. Rev. Genet. 2009, 43, 143–166. [Google Scholar] [CrossRef]
- Ginzberg, I.; Gerchikov, N.; Ziv, E.; Fogelman, E.; Tanami, Z.; Warschavsky, S. Potato tuber skin development: The effect of hot climate and plant desiccation. Acta Hortic. 2005, 684, 93–98. [Google Scholar] [CrossRef]
- Vulavala, V.K.; Elbaum, R.; Yermiyahu, U.; Fogelman, E.; Kumar, A.; Ginzberg, I. Silicon fertilization of potato: Expression of putative transporters and tuber skin quality. Planta 2016, 243, 217–229. [Google Scholar] [CrossRef]
- Fogelman, E.; Tanami, S.; Ginzberg, I. Anthocyanin synthesis in native and wound periderms of potato. Physiol. Plant. 2015, 153, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Vulavala, V.K.R.; Fogelman, E.; Rozental, L.; Faigenboim, A.; Tanami, Z.; Shoseyov, O.; Ginzberg, I. Identification of genes related to skin development in potato. Plant Mol. Biol. 2017, 94, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Sun, L.; Huang, R.-Z.; Liang, W.; Liu, X.; He, H.; Fukuda, H.; He, X.-Q.; Qian, W. Active DNA demethylation regulates tracheary element differentiation in Arabidopsis. Sci. Adv. 2020, 6, eaaz2963. [Google Scholar] [CrossRef] [PubMed]
- Ai, Y.; Jing, S.; Cheng, Z.; Song, B.; Xie, C.; Liu, J.; Zhou, J. DNA methylation affects photoperiodic tuberization in potato (Solanum tuberosum L.) by mediating the expression of genes related to the photoperiod and GA pathways. Hortic. Res. 2021, 8, 181. [Google Scholar] [CrossRef]
- Dutta, M.; Raturi, V.; Gahlaut, V.; Kumar, A.; Sharma, P.; Verma, V.; Gupta, V.K.; Sood, S.; Zinta, G. The interplay of DNA methyltransferases and demethylases with tuberization genes in potato (Solanum tuberosum L.) genotypes under high temperature. Front. Plant Sci. 2022, 13, 933740. [Google Scholar] [CrossRef]
- Sabbah, S.; Raise, M.; Tal, M. Methylation of DNA in NaCl-adapted cells of potato. Plant Cell Rep. 1995, 14, 467–470. [Google Scholar] [CrossRef]
- Ruzin, S.E. Plant Microtechnique and Microscopy; Oxford University Press: New York, NY, USA, 1999. [Google Scholar]
- Johansen, D.A. Plant Microtechniques; McGraw-Hill Book Company, Inc.: New York, NY, USA, 1940. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, P.; Kaplan, Y.; Endelman, J.B.; Ginzberg, I. Epigenetic Modifications Related to Potato Skin Russeting. Plants 2023, 12, 2057. https://doi.org/10.3390/plants12102057
Kumar P, Kaplan Y, Endelman JB, Ginzberg I. Epigenetic Modifications Related to Potato Skin Russeting. Plants. 2023; 12(10):2057. https://doi.org/10.3390/plants12102057
Chicago/Turabian StyleKumar, Pawan, Yulia Kaplan, Jeffrey B. Endelman, and Idit Ginzberg. 2023. "Epigenetic Modifications Related to Potato Skin Russeting" Plants 12, no. 10: 2057. https://doi.org/10.3390/plants12102057








