The Role of Transcription Factors in the Regulation of Plant Shoot Branching
Abstract
:1. Introduction
2. Axillary Meristem Formation
3. Axillary Meristem Outgrowth
4. Application of Branching-Regulating TFs
5. Environmental Pathways Involved in the Control of Shoot Branching
6. Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, C.; Wang, J.; Wenkel, S.; Chandler, J.W.; Werr, W.; Jiao, Y. Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators. Development 2018, 145, dev158352. [Google Scholar] [CrossRef] [Green Version]
- Acker, R.; Weise, S.F.; Swanton, C.J. Influence of interference from a mixed weed species stand on soybean (Glycine max (l.) merr.) growth. Can. J. Plant Sci. 1993, 73, 1293–1304. [Google Scholar] [CrossRef]
- Costes, E.; Lauri, P.E.; Simon, S.; Andrieu, B. Plant architecture, its diversity and manipulation in agronomic conditions, in relation with pest and pathogen attacks. Eur. J. Plant Pathol. 2013, 135, 455–470. [Google Scholar] [CrossRef]
- Wang, Y.; Jiao, Y. Axillary meristem initiation-a way to branch out. Curr. Opin. Plant Biol. 2018, 41, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Greb, T.; Clarenz, O.; Schafer, E.; Muller, D.; Herrero, R.; Schmitz, G.; Theres, K. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003, 17, 1175–1187. [Google Scholar] [CrossRef] [Green Version]
- Long, J.; Barton, M.K. Initiation of axillary and floral meristems in Arabidopsis. Dev. Biol. 2000, 218, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Jiao, Y. Regulation of Axillary Meristem Initiation by Transcription Factors and Plant Hormones. Front. Plant Sci. 2016, 7, 183. [Google Scholar] [CrossRef] [Green Version]
- Rameau, C.; Bertheloot, J.; Leduc, N.; Andrieu, B.; Foucher, F.; Sakr, S. Multiple pathways regulate shoot branching. Front. Plant Sci. 2014, 5, 741. [Google Scholar] [CrossRef] [Green Version]
- Yan, K.; Li, C.C.; Wang, Y.; Wang, X.Q.; Wang, Z.M.; Wei, D.Y.; Tang, Q.L. AGL18-1 delays flowering time through affecting expression of flowering-related genes in Brassica juncea. Plant Biotechnol. 2018, 35, 357–363. [Google Scholar] [CrossRef] [Green Version]
- McSteen, P.; Leyser, O. Shoot branching. Annu. Rev. Plant Biol. 2005, 56, 353–374. [Google Scholar] [CrossRef]
- Groover, A.T.; Mansfield, S.D.; DiFazio, S.P.; Dupper, G.; Fontana, J.R.; Millar, R.; Wang, Y. The Populus homeobox gene ARBORKNOX1 reveals overlapping mechanisms regulating the shoot apical meristem and the vascular cambium. Plant Mol. Biol. 2006, 61, 917–932. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.; Zhang, K.; Li, F.; Huang, Y.; Fan, M.; Huang, T. The AtMYB2 inhibits the formation of axillary meristem in Arabidopsis by repressing RAX1 gene under environmental stresses. Plant Cell Rep. 2020, 39, 1755–1765. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; An, G. Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J. 2012, 69, 445–461. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Zhang, X.; He, J.; Yu, H.; Wang, Y.; Shi, B.; Han, Y.; Wang, G.; Feng, X.; Zhang, C.; et al. An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol. Syst. Biol. 2014, 10, 755. [Google Scholar] [CrossRef]
- Ding, L.; Yan, S.; Jiang, L.; Liu, M.; Zhang, J.; Zhao, J.; Zhao, W.; Han, Y.Y.; Wang, Q.; Zhang, X. HANABA TARANU regulates the shoot apical meristem and leaf development in cucumber (Cucumis sativus L.). J. Exp. Bot. 2015, 66, 7075–7087. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.K.; Geisler, M.; Springer, P.S. LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 2009, 136, 2423–2432. [Google Scholar] [CrossRef] [Green Version]
- Ikezaki, M.; Kojima, M.; Sakakibara, H.; Kojima, S.; Ueno, Y.; Machida, C.; Machida, Y. Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J. 2010, 61, 70–82. [Google Scholar] [CrossRef]
- Teichmann, T.; Muhr, M. Shaping plant architecture. Front. Plant Sci. 2015, 6, 233. [Google Scholar] [CrossRef] [Green Version]
- Tsuda, K.; Abraham-Juarez, M.J.; Maeno, A.; Dong, Z.; Aromdee, D.; Meeley, R.; Shiroishi, T.; Nonomura, K.I.; Hake, S. KNOTTED1 Cofactors, BLH12 and BLH14, Regulate Internode Patterning and Vein Anastomosis in Maize. Plant Cell 2017, 29, 1105–1118. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, G.; Tillmann, E.; Carriero, F.; Fiore, C.; Cellini, F.; Theres, K. The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc. Natl. Acad. Sci. USA 2002, 99, 1064–1069. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.; Zhang, J.; Wang, X.; Han, X.; Wei, B.; Wang, J.; Li, B.; Yu, H.; Huang, Q.; Gu, H.; et al. The WRKY Transcription Factor WRKY71/EXB1 Controls Shoot Branching by Transcriptionally Regulating RAX Genes in Arabidopsis. Plant Cell 2015, 27, 3112–3127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, D.; Schmitz, G.; Theres, K. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 2006, 18, 586–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandler, J.W.; Cole, M.; Flier, A.; Grewe, B.; Werr, W. The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 2007, 134, 1653–1662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fambrini, M.; Salvini, M.; Pugliesi, C. Molecular cloning, phylogenetic analysis, and expression patterns of LATERAL SUPPRESSOR-LIKE and REGULATOR OF AXILLARY MERISTEM FORMATION-LIKE genes in sunflower (Helianthus annuus L.). Dev. Genes Evol. 2017, 227, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Keller, T.; Abbott, J.; Moritz, T.; Doerner, P. Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 2006, 18, 598–611. [Google Scholar] [CrossRef] [Green Version]
- Nie, J.; Wen, C.; Xi, L.; Lv, S.; Zhao, Q.; Kou, Y.; Ma, N.; Zhao, L.; Zhou, X. The AP2/ERF transcription factor CmERF053 of chrysanthemum positively regulates shoot branching, lateral root, and drought tolerance. Plant Cell Rep. 2018, 37, 1049–1060. [Google Scholar] [CrossRef]
- Yang, F.; Wang, Q.; Schmitz, G.; Muller, D.; Theres, K. The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J. 2012, 71, 61–70. [Google Scholar] [CrossRef]
- Li, Y.; Xia, T.; Gao, F.; Li, Y. Control of Plant Branching by the CUC2/CUC3-DA1-UBP15 Regulatory Module. Plant Cell 2020, 32, 1919–1932. [Google Scholar] [CrossRef]
- Hibara, K.; Karim, M.R.; Takada, S.; Taoka, K.; Furutani, M.; Aida, M.; Tasaka, M. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 2006, 18, 2946–2957. [Google Scholar] [CrossRef] [Green Version]
- Raman, S.; Greb, T.; Peaucelle, A.; Blein, T.; Laufs, P.; Theres, K. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 2008, 55, 65–76. [Google Scholar] [CrossRef]
- Wai, A.H.; An, G. Axillary meristem initiation and bud growth in rice. J. Plant Biol. 2017, 60, 440–451. [Google Scholar] [CrossRef]
- Katayama, N.; Tanaka, R.; Fujinami, R.; Imaichi, R. Expression pattern of CUC3 ortholog in Zeylanidium tailichenoides (Podostemaceae) infers organization of a unique distichous shoot in Podostemoideae. J. Plant Res. 2019, 132, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, G.M.; Ramakrishna, K.; Chongloi, G.L.; Vijayraghavan, U. Functions for rice RFL in vegetative axillary meristem specification and outgrowth. J. Exp. Bot. 2015, 66, 2773–2784. [Google Scholar] [CrossRef] [Green Version]
- Rao, N.N.; Prasad, K.; Kumar, P.R.; Vijayraghavan, U. Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc. Natl. Acad. Sci. USA 2008, 105, 3646–3651. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.; Qin, G. EXB1/WRKY71 transcription factor regulates both shoot branching and responses to abiotic stresses. Plant Signal Behav. 2016, 11, e1150404. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Jiang, Y.; Liang, G.; Yang, S.; Yu, D. Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 2014, 26, 230–245. [Google Scholar] [CrossRef] [Green Version]
- Muller, D.; Leyser, O. Auxin, cytokinin and the control of shoot branching. Ann. Bot. 2011, 107, 1203–1212. [Google Scholar] [CrossRef] [Green Version]
- Domagalska, M.A.; Leyser, O. Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 2011, 12, 211–221. [Google Scholar] [CrossRef]
- Yu, S.; Ligang, C.; Liping, Z.; Diqiu, Y. Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. J. Biosci. 2010, 35, 459–471. [Google Scholar] [PubMed]
- Huang, X.; Effgen, S.; Meyer, R.C.; Theres, K.; Koornneef, M. Epistatic natural allelic variation reveals a function of AGAMOUS-LIKE6 in axillary bud formation in Arabidopsis. Plant Cell 2012, 24, 2364–2379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nardmann, J.; Werr, W. The shoot stem cell niche in angiosperms: Expression patterns of WUS orthologues in rice and maize imply major modifications in the course of mono- and dicot evolution. Mol. Biol. Evol. 2006, 23, 2492–2504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, W.; Ohmori, Y.; Ushijima, T.; Matsusaka, H.; Matsushita, T.; Kumamaru, T.; Kawano, S.; Hirano, H.Y. Axillary Meristem Formation in Rice Requires the WUSCHEL Ortholog TILLERS ABSENT1. Plant Cell 2015, 27, 1173–1184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohmori, Y.; Tanaka, W.; Kojima, M.; Sakakibara, H.; Hirano, H.Y. WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in rice. Plant Cell 2013, 25, 229–241. [Google Scholar] [CrossRef] [Green Version]
- Tabuchi, H.; Zhang, Y.; Hattori, S.; Omae, M.; Shimizu-Sato, S.; Oikawa, T.; Qian, Q.; Nishimura, M.; Kitano, H.; Xie, H.; et al. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 2011, 23, 3276–3287. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.; Shao, G.; Xiong, J.; Jiao, Y.; Wang, J.; Liu, G.; Meng, X.; Liang, Y.; Xiong, G.; Wang, Y.; et al. MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J. Genet. Genom. 2015, 42, 71–78. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S. AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol. 2011, 156, 1612–1619. [Google Scholar] [CrossRef] [Green Version]
- Bai, X.; Huang, Y.; Mao, D.; Wen, M.; Zhang, L.; Xing, Y. Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice. Sci. Rep. 2016, 6, 19022. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, K.; Maekawa, M.; Ujiie, S.; Satake, Y.; Furutani, I.; Okamoto, H.; Shimamoto, K.; Kyozuka, J. LAX and SPA: Major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA 2003, 100, 11765–11770. [Google Scholar] [CrossRef] [Green Version]
- Gallavotti, A.; Zhao, Q.; Kyozuka, J.; Meeley, R.; Ritter, M.; Doebley, J.; Pe, M.; Schmidt, R. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature 2004, 432, 630–635. [Google Scholar] [CrossRef]
- Meng, Q.; Li, X.; Zhu, W.; Yang, L.; Liang, W.; Dreni, L.; Zhang, D. Regulatory network and genetic interactions established by OsMADS34 in rice inflorescence and spikelet morphogenesis. J. Integr. Plant Biol. 2017, 59, 693–707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C. Toward understanding the stem-cell origin and molecular regulation of rice tillering. J. Genet. Genom. 2015, 42, 47–48. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, Y.; Yu, Y.; Duan, J.; Liao, Z.; Xiong, G.; Meng, X.; Liu, G.; Qian, Q.; Li, J. Degradation of MONOCULM 1 by APC/C(TAD1) regulates rice tillering. Nat. Commun. 2012, 3, 750. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Basile, A.; Fambrini, M.; Tani, C.; Shukla, V.; Licausi, F.; Pugliesi, C. The Ha-ROXL gene is required for initiation of axillary and floral meristems in sunflower. Genesis 2019, 57, e23307. [Google Scholar] [CrossRef] [PubMed]
- Lewis, M.W.; Bolduc, N.; Hake, K.; Htike, Y.; Hay, A.; Candela, H.; Hake, S. Gene regulatory interactions at lateral organ boundaries in maize. Development 2014, 141, 4590–4597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizzotti, C.; Galliani, B.M.; Dreni, L.; Sommer, H.; Bombarely, A.; Masiero, S. ERAMOSA controls lateral branching in snapdragon. Sci. Rep. 2017, 7, 41319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, Z.; Chen, L.; Zhang, Y.; Li, Z.Y.; Liu, M.; Li, W.P.; Ju, Y.L.; Fang, Y.L. VvBRC inhibits shoot branching in grapevine. Sci. Hortic. 2021, 289, 110370. [Google Scholar] [CrossRef]
- Yang, Y.; Nicolas, M.; Zhang, J.; Yu, H.; Guo, D.; Yuan, R.; Zhang, T.; Yang, J.; Cubas, P.; Qin, G. The TIE1 transcriptional repressor controls shoot branching by directly repressing BRANCHED1 in Arabidopsis. PLoS Genet. 2018, 14, e1007296. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Smith, S.M.; Li, J. Genetic Regulation of Shoot Architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468. [Google Scholar] [CrossRef]
- Tsuji, H.; Tachibana, C.; Tamaki, S.; Taoka, K.; Kyozuka, J.; Shimamoto, K. Hd3a promotes lateral branching in rice. Plant J. 2015, 82, 256–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashiguchi, K.; Sasaki, E.; Shimada, Y.; Nagae, M.; Ueno, K.; Nakano, T.; Yoneyama, K.; Suzuki, Y.; Asami, T. Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Biosci. Biotechnol. Biochem. 2009, 73, 2460–2465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, R.; Gruber, M.Y.; Amyot, L.; Hannoufa, A. SPL13 regulates shoot branching and flowering time in Medicago sativa. Plant Mol. Biol. 2018, 96, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Hu, X.; Yang, Y.; He, C.; Hu, J.; Wang, X. Strigolactone signaling regulates cambial activity through repression of WOX4 by transcription factor BES1. Plant Physiol. 2022, 188, 255–267. [Google Scholar] [CrossRef]
- Bemer, M.; van Mourik, H.; Muino, J.M.; Ferrandiz, C.; Kaufmann, K.; Angenent, G.C. FRUITFULL controls SAUR10 expression and regulates Arabidopsis growth and architecture. J. Exp. Bot. 2017, 68, 3391–3403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Ljung, K.; Breton, G.; Schmitz, R.J.; Pruneda-Paz, J.; Cowing-Zitron, C.; Cole, B.J.; Ivans, L.J.; Pedmale, U.V.; Jung, H.S.; et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012, 26, 785–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holalu, S.V.; Reddy, S.K.; Blackman, B.K.; Finlayson, S.A. Phytochrome interacting factors 4 and 5 regulate axillary branching via bud abscisic acid and stem auxin signalling. Plant Cell Environ. 2020, 43, 2224–2238. [Google Scholar] [CrossRef]
- Mehrnia, M.; Balazadeh, S.; Zanor, M.I.; Mueller-Roeber, B. EBE, an AP2/ERF transcription factor highly expressed in proliferating cells, affects shoot architecture in Arabidopsis. Plant Physiol. 2013, 162, 842–857. [Google Scholar] [CrossRef] [Green Version]
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003, 33, 513–520. [Google Scholar] [CrossRef]
- Guo, S.; Xu, Y.; Liu, H.; Mao, Z.; Zhang, C.; Ma, Y.; Zhang, Q.; Meng, Z.; Chong, K. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat. Commun. 2013, 4, 1566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minakuchi, K.; Kameoka, H.; Yasuno, N.; Umehara, M.; Luo, L.; Kobayashi, K.; Hanada, A.; Ueno, K.; Asami, T.; Yamaguchi, S.; et al. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant Cell Physiol. 2010, 51, 1127–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, L.; Li, W.; Miura, K.; Ashikari, M.; Kyozuka, J. Control of tiller growth of rice by OsSPL14 and Strigolactones, which work in two independent pathways. Plant Cell Physiol. 2012, 53, 1793–1801. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Liu, K.; Song, W.; Zhang, J.; Yao, Y.; Xin, M.; Hu, Z.; Peng, H.; Ni, Z.; Sun, Q.; et al. Pleiotropic function of the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE gene TaSPL14 in wheat plant architecture. Planta 2021, 253, 44. [Google Scholar] [CrossRef] [PubMed]
- Kerr, S.C.; Beveridge, C.A. IPA1: A direct target of SL signaling. Cell Res. 2017, 27, 1191–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, X.; Lu, Z.; Yu, H.; Shao, G.; Xiong, J.; Meng, X.; Jing, Y.; Liu, G.; Xiong, G.; Duan, J.; et al. IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 2017, 27, 1128–1141. [Google Scholar] [CrossRef]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Li, G.; Tan, M.; Ma, J.; Cheng, F.; Li, K.; Liu, X.; Zhao, C.; Zhang, D.; Xing, L.; Ren, X.; et al. Molecular mechanism of MdWUS2-MdTCP12 interaction in mediating cytokinin signaling to control axillary bud outgrowth. J. Exp. Bot. 2021, 72, 4822–4838. [Google Scholar] [CrossRef]
- Moreno-Cortes, A.; Hernandez-Verdeja, T.; Sanchez-Jimenez, P.; Gonzalez-Melendi, P.; Aragoncillo, C.; Allona, I. CsRAV1 induces sylleptic branching in hybrid poplar. New Phytol. 2012, 194, 83–90. [Google Scholar] [CrossRef]
- Lewis, J.M.; Mackintosh, C.A.; Shin, S.; Gilding, E.; Kravchenko, S.; Baldridge, G.; Zeyen, R.; Muehlbauer, G.J. Overexpression of the maize Teosinte Branched1 gene in wheat suppresses tiller development. Plant Cell Rep. 2008, 27, 1217–1225. [Google Scholar] [CrossRef]
- Leduc, N.; Roman, H.; Barbier, F.; Péron, T.; Huché-Thélier, L.; Lothier, J.; Demotes-Mainard, S.; Sakr, S. Light Signaling in Bud Outgrowth and Branching in Plants. Plants 2014, 23, 223–250. [Google Scholar] [CrossRef] [Green Version]
- Evers, J.B.; Vos, J.; Andrieu, B.; Struik, P.C. Cessation of tillering in spring wheat in relation to light interception and red: Far-red ratio. Ann. Bot. 2006, 97, 649–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girault, T.; Bergougnoux, V.; Combes, D.; Viemont, J.-D.; Leduc, N. Light controls shoot meristem organogenic activity and leaf primordia growth during bud burst in Rosa sp. Plant Cell Environ. 2008, 31, 1534–1544. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A. Shade avoidance. New Phytol. 2008, 179, 930–944. [Google Scholar] [CrossRef] [PubMed]
- Ballaré, C.L.; Casal, J.J. Light signals perceived by crop and weed plants. Field Crops Res. 2000, 67, 149–160.83. [Google Scholar] [CrossRef]
- Abidi, F.; Girault, T.; Douillet, O.; Guillemain, G.; Sintes, G.; Laffaire, M.; Ahmed, H.B.; Smiti, S.; Huché-Thélier, L.; Leduc, N. Blue light effects on rose photosynthesis and photomorphogenesis. Plant Biol. (Stuttg.) 2013, 15, 67–74. [Google Scholar] [CrossRef]
- Kuhlmann, F.; Muller, C. MüllerDevelopment-dependent effects of UV radiation exposure on broccoli plants and interactions with herbivorous insects Environ. Exp. Bot. 2009, 66, 61–68. [Google Scholar] [CrossRef]
- Fustec, J.; Beaujard, F. Effect of photoperiod and nitrogen supply on basal shoots development in Rhododendron Catawbiense. Biol. Plant 2000, 43, 511–515. [Google Scholar] [CrossRef]
- Martin, C.A.; Stutz, J.C.; Kimball, B.A.; Idso, S.B.; Akey, D.H. Growth and topological changes of citrus limon (l.) burm. f. ‘eureka’ in response to high temperatures and elevated atmospheric carbon dioxide. Am. Soc. Hortic. Sci. 1995, 120, 1025–1031. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.Y.; Zhao, N.; Tang, H.M.; Gong, B.; Shi, Q.H. Shoot branching regulation and signaling. Plant Growth Regul. 2020, 92, 131–140. [Google Scholar] [CrossRef]

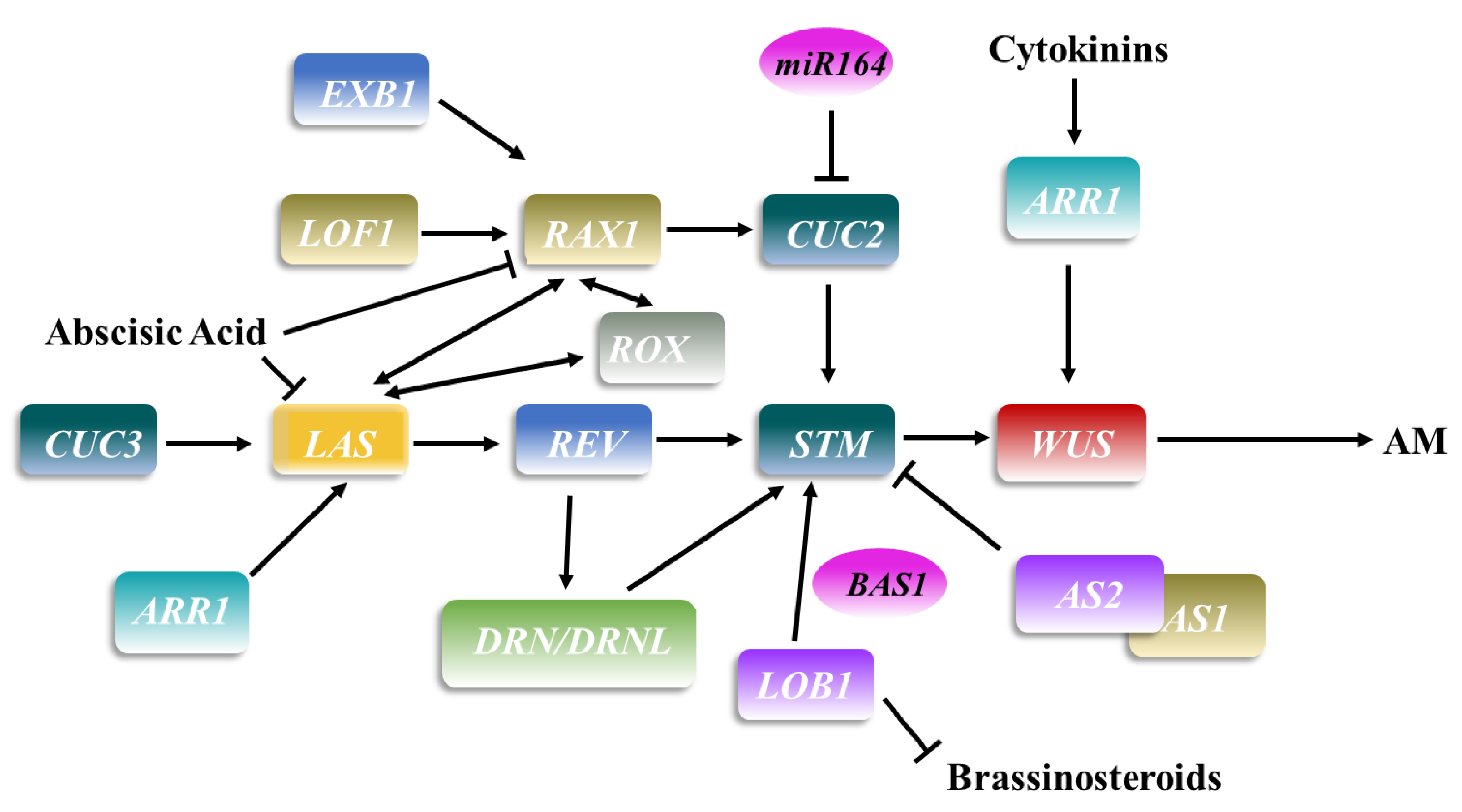
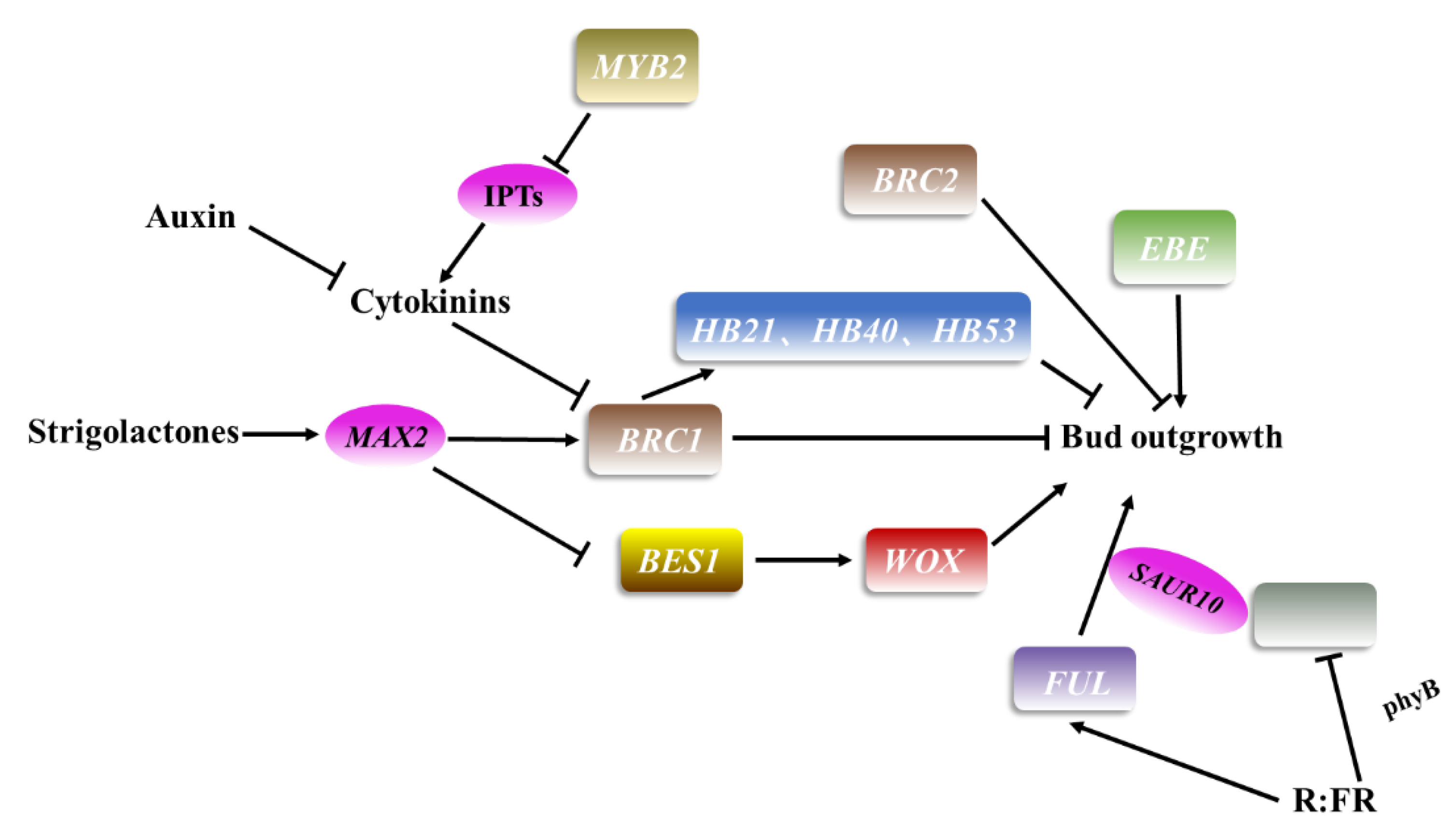
| Name | Homologs in Other Species | Family | Function |
|---|---|---|---|
| AtAGL6 (AGAMOUS-LIKE6) | MADS | Facilitates the formation of axillary meristems | |
| OsMADS34 | Coordinates with LAX1 to regulate the number of primary branches | ||
| OsMADS57 | Is expressed predominantly in the SAM and axillary buds and is involved in SL signaling to enhance axillary bud growth and subsequent tillering | ||
| AtFUL (FRUITFULL) | Is involved in development of the axillary meristem, the expression of which is controlled by auxin | ||
| AtCUC1-3 (CUP SHAPED COTYLEDON1-3) | NAC | Is negatively regulated by BRs and involved in AM initiation | |
| AtSTM (SHOOT MERISTEMLESS) | OsOSH1 (O. sativa homeobox1) | HB-KNOX | Is involved in initiation or maintenance of undifferentiated cell fate in very early stages of AM formation |
| AtLOF1 (LATERAL ORGAN FUSION1) | MYB | Is involved in lateral organ separation and axillary meristem formation | |
| AtLOF2 (LATERAL ORGAN FUSION2) | |||
| AtAS1 (ASYMMETRIC LEAVES1) | Inhibits branching and downregulates STM when cells start to differentiate | ||
| AtRAX1 (REGULATOR OF AXILLARY MERISTEMS 1) | Bl (Blind), S. lycopersicum | Is involved in the early steps of AM initiation and development | |
| AtRAX2-3 | |||
| MsMYB112 | Inhibits collateral growth | ||
| AtMYB2 | Inhibits branching and reduces cytokinin concentrations by inhibiting expression of IPTs in Arabidopsis | ||
| AtWUS (WUSCHEL) | OsTAB1 (TILLERS ABSENT1); OsMOC3 (MONOCULM 3) | WOX | Promotes branching and is involved in maintenance of meristematic stem cell function and regulation of cell division |
| OsWOX4 | Is involved in AM initiation | ||
| AtWOX4 | Regulates Arabidopsis secondary growth by SL signaling | ||
| MdWUS2 (WUSCHEL 2) | Regulates branching by inhibiting the activity of MdTCP12 (BRC2 homolog) | ||
| AtLAS (LATERAL SUPPRESSOR) | Ls, S. lycopersicum; ERA (ERAMOSA, A. majus); OsMOC1; HaLSL (LATERAL SUPPRESSOR LIKE) | GRAS | Is necessary for maintenance of the meristematic potential of the cells in the axils of leaf primordia |
| HaROXL (REGULATOR OF AXILLARY MERISTEM FORMATION LIKE) | ZmBA1 (BARREN STALK1); OsLAX1 (Lax Panicle 1); AtROX (REGULATOR OF AXILLARY MERISTEM FORMATION) | bHLH | Is involved in development of the SAM and lateral young leaf primordia |
| OsLAX2 (LAX PANICLE2) | Is involved in development of the SAM and lateral young leaf primordia | ||
| AtPIF4/5 (PHYTOCHROME INTERACTING FACTORs 4/5) | Inhibits the branching caused by phyB dysfunction and low R:FR | ||
| OsFZP (FRIZZLE PANICLE) | ZmBD1; COM2, H. vulgare | AP2/ERF-ERF | Represses axillary meristem formation |
| AtEBE (ERF BUD ENHANCER) | Is involved in cell proliferation and axillary bud growth | ||
| AtDRN (DORNRÖSCHEN) | Regulates STM expression and AM initiation | ||
| AtDRNL (DORNRÖSCHEN-LIKE) | |||
| AtERF053 | Is involved in cytokinin control of stem branching | ||
| OsRFL (RICE FLORICULA/LEAFY) | OsAPO2 (PANICLE ORGANIZATION 2) | Promotes AM specificity through its action on LAX1 and CUC genes | |
| ZmBAD1 (BRANCH ANGLE DEFECTIVE1) | TCP | Promotes the formation of lateral meristems (e.g., branches) and axillary organs (e.g., leaf pillows) in wild-type maize | |
| AtBRC1 (BRANCHED1) | OsTB1 (TEOSINTE BRANCHED 1); OsFC1 (FINECULM1); VvBRC | Negatively regulates axillary bud growth | |
| AtBRC2 (BRANCHED2) | MdTCP12 | Has a redundant role with BRC1 in regulation of axillary bud growth | |
| AtAS2 (ASYMMETRIC LEAVES1) | LOB | Inhibits branching and downregulates STM when cells start to differentiate | |
| AtLOB1 (LATERAL ORGAN BOUNDARIES 1) | Is negatively regulated by BRs to reduce cell division and expansion in the border zone | ||
| AtWRKY71/EXB1 | WRKY | Is expressed in tissues surrounding the AM start site | |
| WRKY72 | Positively regulates bud branching | ||
| AtREV (REVOLUTA) | HD-ZIP | Upregulates STM expression and promotes AM initiation | |
| HB21 (Homeobox21) | Inhibits branching, directly downstream of BRC1 | ||
| HB40 (Homeobox40) | |||
| HB53 (Homeobox53) | |||
| AtSPL13 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 13) | SBP | Inhibits the growth of axillary buds | |
| AtIPA1 (IDEAL PLANT ARCHITECTUREL1) | OsSPL14 | Acts with D53 to mediate SL-regulated tiller development in rice | |
| AtARR1 (ARABIDOPSIS RESPONSE REGULATOR 1) | GARP-ARR-B | Promotes branching, acts Downstream of cytokinins and promotes LAS expression by binding to their promoters | |
| AtBES1 (BRI1-EMS-SUPPRESSOR1) | BES1 | Inhibits branching and negatively regulates cambium activity in the SL signaling pathway in Arabidopsis |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Fang, W.; Chen, F.; Song, A. The Role of Transcription Factors in the Regulation of Plant Shoot Branching. Plants 2022, 11, 1997. https://doi.org/10.3390/plants11151997
Zhang L, Fang W, Chen F, Song A. The Role of Transcription Factors in the Regulation of Plant Shoot Branching. Plants. 2022; 11(15):1997. https://doi.org/10.3390/plants11151997
Chicago/Turabian StyleZhang, Lingling, Weimin Fang, Fadi Chen, and Aiping Song. 2022. "The Role of Transcription Factors in the Regulation of Plant Shoot Branching" Plants 11, no. 15: 1997. https://doi.org/10.3390/plants11151997






