1. Introduction
The autopod, the multi-fingered extremity located at the end of a hindlimb or forelimb, is a specific characteristic of tetrapods [
1,
2,
3]. The fossil record suggests that this anatomical structure evolved from sarcopterygian fins by sequential expansion and elaboration of their endochondral bones and concomitant reduction of the apical dermoskeleton [
1,
2,
3]. Regarding the number of digits, both paleontological and developmental data suggest an evolutionary sequence that went from an adactyl state detectable up to the divergence of sarcopterygians [
4], then sidestepping by digit-like radials in stem sarcopterygians [
5], polydactyly in stem tetrapod and culminating in the fixation of pentadactyl [
6,
7,
8]. From a developmental point of view, understanding the molecular mechanisms that enabled the fin-to-limb transition is a puzzling question that has long held the attention of the scientific community and holds the potential of enlightening molecular biologists on how gene regulatory networks and interactions evolved.
The patterning of the skeletal elements, in both fin and limb, largely relies on the action of signaling centers operating from the onset of their outgrowth: the apical ectodermal ridge (AER) and the zone of polarizing activity (ZPA) [
9]. However, there are marked differences in the morphology and development of the most distal part of fins and limbs. In tetrapods, the influence of the AER is maintained throughout limb development, being important for the differentiation of the stylopod, zeugopod and autopod [
10,
11,
12]. In fish, however, this structure is quickly converted into a finfold (FF) during fin development that then gives rise to the apical dermoskeleton [
13]. However, its function seems to be conserved during fish fin development and relates to the coordination of the proximo-distal patterning [
14]. Recent studies performed in zebrafish suggest that the timing of the AER-FF transition may have mediated the differences found between fins and limbs and that the acquisition of a mechanism repressing the formation of the FF may have been crucial in the development of tetrapod limbs [
15].
The patterning of the endoskeleton structure has been proven to rely on the expression of 5’
HoxA and
HoxD genes in both fins and limbs, as demonstrated by different studies performed on a wide variety of model systems [
16,
17,
18,
19,
20]. Indeed, several lines of evidence highlight the role of these transcriptional factors as key intervenients in limb development. For example, loss-of-function mutations in genes located at the 5’ end of the mouse HoxA and HoxD clusters lead to the appearance of limbs with atavistic characters [
8,
18,
20]. The impact that these genes have on the patterning of the limb, proven by loss-of-function assays, led Duboule and colleagues to suggest that modifications on Hox regulation may have been a source of morphological variation during the evolution of tetrapod limbs [
21]. Moreover, they suggest that increased levels of these transcription factors during fin development may have caused extra-proliferation distally, setting the grounds for the formation of additional endoskeleton elements, which occurred concomitantly with the reduction of the apical dermoskeleton. To make the proof of principle of these ideas, Freitas and colleagues overexpressed the zebrafish counterpart of
HoxD13 (
hoxd13a), generating a fin phenotype characterized by additional endochondral tissue and reduction or ablation of the apical finfold [
22]. Thus,
hoxd13a overexpression induces a phenotype that mirrors two of the major morphological alterations marked as crucial in limb evolution by the fossil record: endochondral expansion and dermoskeleton retraction [
1,
22].
Taking into account the results obtained by Zákány
et al. [
8] and Freitas
et al. [
22], it seems that both HoxA and HoxD clusters contributed to the fin-to-limb transition, although at distinct time points and by regulating different cellular events. Zákány and colleagues analyzed the phenotypes of several compound mouse mutants with distal
HoxA and
HoxD loss-of-function alleles and proposed that the
HoxA13 gene was the initial/primordial major contributor for the determination of the autopod identity [
8]. This was probably followed, in the course of evolution, by the recruitment of distal
HoxD genes (
HoxD11,
HoxD12 and
HoxD13), whose expression is associated with finger elongation and fixation of [
8,
23]. Further evidences that supports this sequence of events comes from the observation that
HoxA13 is expressed earlier than
HoxD13 in the distal mesenchyme during limb development [
23], being probably an earlier specifier of the autopod identity. Moreover, the expression of the
Hox13 paralogs in zebrafish is detected in the most distal mesenchyme of the developing fins, closely mirroring the expression patterns described in tetrapods [
17]. This suggests that
HoxA13 was already essential to induce a distal appendage identity even prior to the origin of the autopod in the tetrapod lineage. Taken together, these data suggest that alterations in the expression and function of HoxA13 may have been the primary mechanisms leading to the formation of the autopod in the tetrapod lineage [
24,
25].
Taking into account the conserved
hoxa13 expression patterns found distally in developing fish fins, why do they not give rise to autopod-like structures? Information gathered from developmental, phylogenetic and gene function studies suggests that, in the course of evolution, two factors had to meet to allow the formation of an autopod domain, those being the alteration of the proximo-distal patterning leading to the formation of novel distal territories, characterized by a specific transcriptome, that co-occurred with, or was followed by, an increase in the levels of
HoxA13 transcripts [
8,
26].
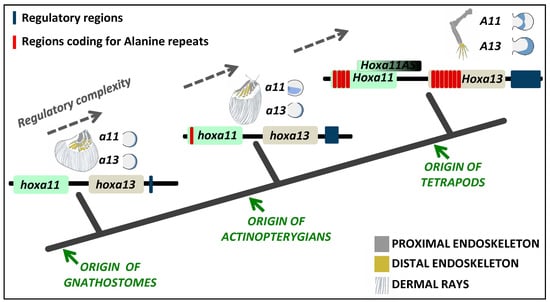
Regarding the proximo-distal patterning, striking differences have been found in the expression of 5’
HoxA genes in fish and tetrapod models that may underlie an increased endoskeletal complexity throughout fin/limb evolution (
Figure 1). This process counts with the combined action of three genes encoding master regulators of transcription:
Meis1, which seems to be essential for stylopod specification;
HoxA11 required for proper zeugopod development; and
HoxA13, which contributes to an autopod identity [
27,
28,
29,
30,
31]. Interestingly, during tetrapod limb development, the initially overlapping expression domains of
HoxA11 and
HoxA13 split up to a point in which
HoxA11 is exclusively expressed in the prospective zeugopod, while
HoxA13 is restricted to the future autopod (
Figure 1) [
16,
32,
33]. The expression patterns of these genes during fish fin development are, however, quite different. In zebrafish, a full separation of the
hoxa11 and
hoxa13 expression domains has never been reported in the developing fins [
17,
21,
34]. In other fish representing distinct phylogenetic positions, such as chondrichthyans (catshark; [
35]) and basal actinopterygians (paddlefish; [
26,
36]), there is only a transient separation of
hoxa11 and
hoxa13 expression domains during early stages of fin development. These data suggest that the decoupling of
HoxA13/HoxA11 expression domains may have occurred in the common ancestor of gnathostomes and was secondarily lost in a particular lineage of actinopterygians, the teleosts.
The origin of a distal domain exclusively expressing
HoxA13 may have influenced the formation of novel endoskeleton structures [
26,
37,
38]. Further support of this hypothesis is provided by regeneration studies in
Xenopus, where non-separation of
HoxA11 and
HoxA13 expression patterns, making distal blastemal cells
HoxA11 and
HoxA13 double positive, appeared to be associated with the inability to regenerate an autopod [
16]. Interestingly,
HoxA11 and
HoxA13 mRNA are co-expressed in the developing limbs of axolotls, but the translated HoxA11 and HoxA13 proteins are distributed in distinct domains consistent with the formation of a zeugopod-autopod border [
39]. Questions that remain to be answered are, for example: How was the separation of HoxA11 and HoxA13 domains achieved, and what were the implications for the evolution of the limb morphology? What were the molecular mechanisms associated with this alterations? Or, how did alterations in HoxA11 and HoxA13 protein levels contribute to the autopod origin?
From a gene regulatory perspective, there are several mechanisms that could have contributed to the establishment of a HoxA13 single positive distal appendage domain that allowed the formation and posterior evolution of the autopod. These may include: (1) alteration in the signaling pathways that act upstream of HoxA13, regulating its expression and/or the expression of HoxA11; (2) remodeling/alteration or elaboration of the cis-regulatory motifs that assume the spatial-temporal control of the expression for these genes; (3) acquisition/loss of elements of the post-transcriptional gene regulatory network; (4) alteration of the protein-protein or protein-DNA interactions through alteration of the coding sequence.
By analyzing the reports published to date, there are three mechanisms that may uphold an evolutionary relevance in the context of the fin-to-limb transition, those being the acquisition/expansion of
HoxA11 and
HoxA13 homopolymeric repeats, acquisition of a new long-non-coding RNA with possible
HoxA11 inhibitory function and the origin of novel
cis-regulatory regions acting on
HoxA11 and
HoxA13 [
40,
41,
42,
43]. Here, we will explore the evolutionary/developmental relevance of these three mechanisms, reviewing the current state of the art concerning
HoxA11 and
HoxA13 regulation throughout development in species at different phylogenetic positions and discussing how these developmental regulatory networks may have evolved, being at the core of the morphological diversification found in vertebrate appendages.
2. Polyalanine Repeats and the Fin-to-Limb Transition
Amino acid repeats are common among eukaryote proteins, and their length and frequency cannot be explained solely by random chance, suggesting a biological function and positive selection [
44,
45]. Interestingly, prokaryote proteins are impoverished in amino acid repeats, when compared to their eukaryotic homologues, indicating that evolution of such amino acid repeats happened after establishment of the initial protein structures and possibly concomitantly with the appearance of novel functionalities in eukaryotes [
44]. Indeed, numerous proteins with a variety of different biological functions display amino acid repeats, such as protein kinases, signaling proteins, membrane transporters and transcription factors [
44]. Strikingly, the majority of amino acid repeats detected in eukaryotes are present in proteins involved in the formation of large protein-protein or protein-nucleic acid complexes, such as transcriptional factors, possibly influencing their performance [
44]. Thus, changes in these particular sequences during evolution may have produced a significant impact, leading to the genesis of novel morphological structures, for example.
Interestingly, alterations in the length of amino acid tandem repeats have long been suggested as a major source of phenotypic variation in evolution [
46]. This hypothesis was constructed based on the observation that: (1) amino acid tandem repeats are abundant and frequently conserved in the coding sequence of genes involved in development; (2) repeat length variation, both expansion and contraction, occurs in a locus-specific manner 100,000-times more frequently than point mutations; and (3) transcription, mRNA processing, protein synthesis and morphology are affected by length variation in the amino acid repeats [
46]. There are, indeed, some examples that offer support to this hypothesis. For example, within dog breeds, there is a clear association between the contraction of glutamine repeats in the
Alx-4 gene and the development of bilateral polydactyly that characterizes the Great Pyrenees breed [
46]. Moreover, a correlation was found between the length of the polyglutamine and polyalanine tracts in the
Runx-2 gene and the facial morphology of different dog breeds [
46], showing the potential that amino acid repeat variability may assume in the diversification of phenotypes.
Polyalanine repeats are types of amino acid repeats characterized by tandem repeated alanine residues [
44]. These regions can modulate protein function through various mechanisms, such as enabling or increasing a transcriptional factor capacity to act as a repressor, modifying the interactome of a protein or acting as spacers, merely providing a structural component that enhances the performance of its functional domains [
40,
47,
48,
49]. Interestingly, alterations in the alanine tracts of particular Hox proteins have been proven to impact limb development, to a greater or lesser extent [
50,
51]. For instance, deletion of one of the HoxD13 alanine polymers in mouse caused fusion of the forelimb sesamoid bones [
51,
52]. Moreover, increased length of HOXD13 polyalanine repeats has been associated with several cases of synpolydactylism [
51,
53].
HoxA11 and HoxA13 are among the transcriptional factors with crucial function during limb development, where polyalanine repeats have been identified [
40,
41]. For the human HOXA13, in particular, clear associations were established between the expansion of particular polyalanine tracts (I, III and V) and the development of autopod malformations associated with the human hand-foot-genital syndrome (HFGS) [
54,
55]. Similar phenotypes were identified in mutant mice with an expanded polyalanine tract [
56].
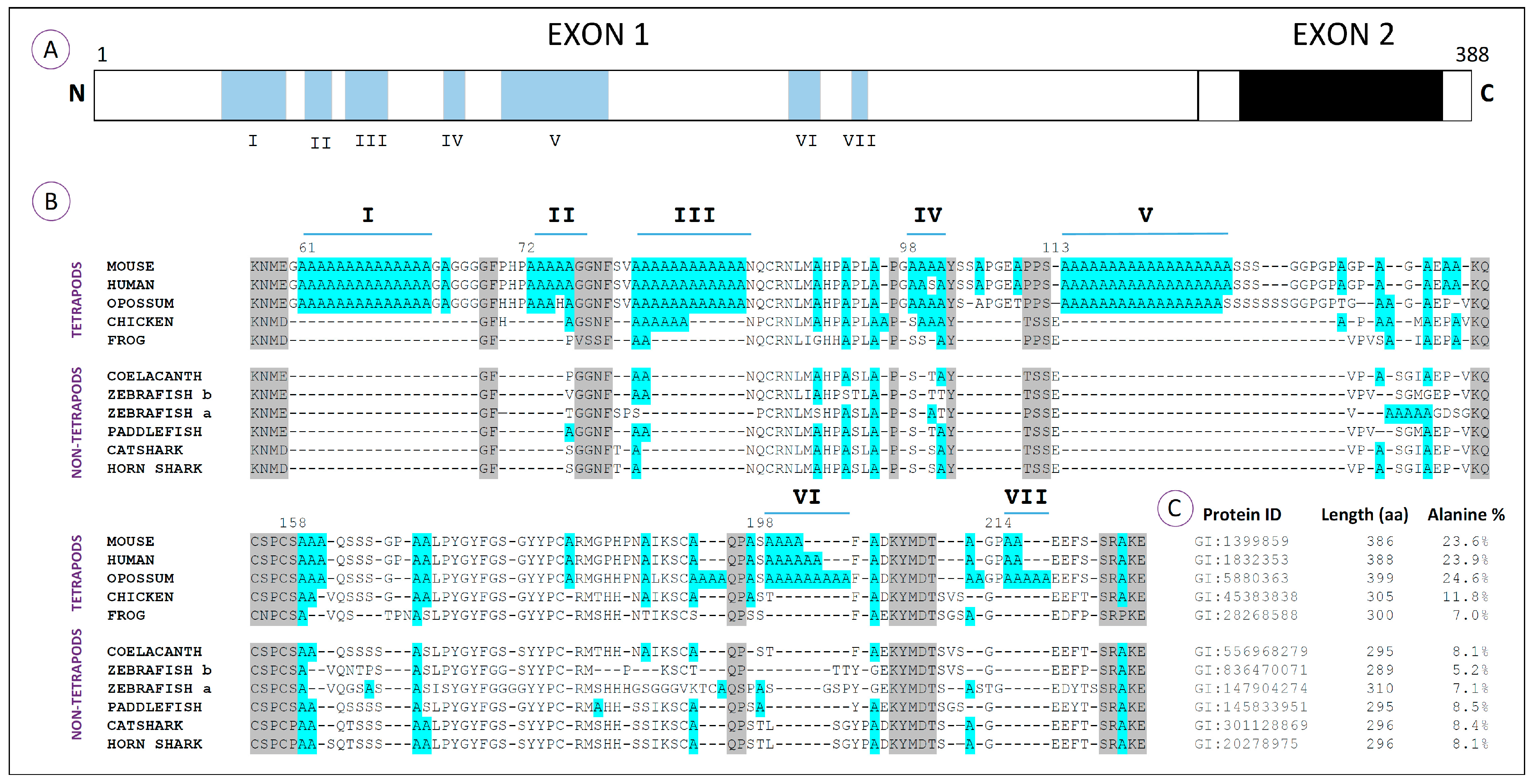
Comparative analyses using HoxA11 amino acid sequences from vertebrates reveal that this protein is composed of three major domains [
41]: Domains I and III correspond to the N- and C-terminal regions respectively; Domain II is encoded by exon 1 and corresponds to a variable and overall hydrophilic region (
Figure 2A). Characteristic reconstructions of Domains I and III suggest that the rates of coding sequence evolution have not changed significantly in tetrapods (frog and chick) relative to lobe-finned fish (coelacanth). However accelerated rates of coding sequence evolution were observed for the mammalian and newt lineages and shown to be a gene-specific phenomenon [
41]. Interestingly, when the hydrophilic region of the HoxA11 proteins is compared, stretches of more than three consecutive alanine residues are identified exclusively in the tetrapod sequences, together with an increase in the length of this region (
Figure 2B). Thus, the molecular evolution of this transcription factor might have involved an increase of the hydrophilic region by the addition of polyalanine tracts. An hypothetical scenario is that the alanine repeats present in the common ancestor of tetrapods may have acted as a “seed” for the expansion of polyalanine stretches that then contributed to an increased plasticity of HoxA11, potentiating novel functionalities in this lineage, ultimately contributing to the developmental events involved in the fin-to-limb transition [
41].
Moreover, the overall alanine content within the hydrophilic region is higher in tetrapods than in non-tetrapod organisms. This is especially evident in amniotes, animals that develop a paddle-like hand plate prior to digit differentiation. In fact, the amphibian’s hydrophilic region lacks polyalanine tracts that appear to be conserved in other tetrapod lineages, and coincidently, the formation of their autopods is not preceded by the development of a hand plate [
59,
60]. Thus, the addition of alanine residues in the hydrophilic region may have led to novel functionalities of the HoxA11 protein that may have contributed to the origin of the hand plate. Alternatively, the lower alanine content found in the hydrophilic region of amphibians, when compared to other tetrapod linages, may reflect their own evolutionary history that seems to have led to a highly derived morphology [
61].
Interestingly, in non-tetrapod organisms, the higher percentage of alanine residues in the hydrophilic region is found in the paddlefish, which in fact presents a subtle separation of
HoxA11 and
HoxA13 expression domains at early stages of fin development (
Figure 1A; [
36]). Thus, hypothetically, higher alanine content in the HoxA11 hydrophilic regions may influence the expression pattern of this protein. Evolutionarily, this could have been the process by which
HoxA11 expression patterns evolved, probably contributing to the fin-to-limb transition.
Given that the repressor domains of transcription factors are commonly small, unstructured, hydrophilic regions with high alanine content [
62], these observations suggest that expanded alanine stretches might be enhancers of repressor domains that were instrumental in setting tetrapod-specific features, such as the autopods, as proposed by Chiu and colleagues [
41]. However, the search of such a repressor function found no evidence in mouse [
63]. Nevertheless, there is still grounds for Chiu and colleagues’ hypotheses, given that the studies performed in mouse just revealed results for a partial deletion of the alanine residues outside the homeodomain, and there is no indication of their location. Thus, just a complete deletion of the alanine residues within the hydrophilic region would be informative. Moreover, the alanine stretches have other predictable roles, such as increasing the plasticity of HoxA11 function, modifying its interactome and/or acting as spacers, providing a structural component crucial to the performance of its functional domains, as suggested by several authors with respect to the function of polyalanine tracts [
40,
47,
48,
49].
Taking together these analyses, we hypothesize that a sequential increase of the alanine content within the HoxA11 hydrophilic regions may have contributed to: (1) the evolution of HoxA11 expression patterns during fin development; (2) the establishment of zeugopod/autopod borders; and (3) the formation of the hand plate in the common ancestor of amniotes.
Regarding the HoxA13 proteins, a total of seven polyalanine tracts (I–VII), characterized by a minimum of four clustered alanine residues, has been identified in a variety of vertebrate species at distinct phylogenetic positions (
Figure 3A,B; [
40,
47]). Comparative amino acid analyses reveal that these sequences are characteristic of amniotes, being particularly conserved in mammals (
Figure 3B). In fact, Tract III seems to be the only one conserved between mammalian and avian genomes, which suggests that most polyalanine tracts from the HoxA13 proteins arose after the divergence of birds from the lineage that would culminate in the appearance of mammals [
40,
47]. The incorporation of polyalanine tracts together with flanking segments rich in proline, serine and glycine seems to have increased the N-terminus of mammals approximately 35%, when compared to the fish counterparts [
40]. Indeed, while tetrapod HoxA13 proteins have more than 20% alanine content, the zebrafish orthologs have less than 10% (
Figure 3C). Within other organisms representing groups that derived prior to the origin of amniotes (frog, coelacanth, zebrafish, paddlefish and sharks), there are alanine residues that align with the polyalanine tracts of amniotes and may represent the primordial elements that gave rise to them. In agreement with this idea, Mortlock and colleagues suggested that the origin of large alanine repeats in mammals might be explained by a dynamic process of recurring replication slippage and point mutation within alanine repeat codons [
40].
Taking into consideration the potential impact of these polyalanine tracts in the functional plasticity of the proteins, it is possible that their incorporation within the HoxA13 transcription factor may have enabled novel developmental roles contributing to the evolution of the limb structure. However, the absence of such sequences in the HoxA13 of amphibians is intriguing in this scenario. As for HoxA11, we hypothesize that the substantial difference found in the alanine content of the amphibian HoxA13 proteins may explain the lack of a hand plate prior to digit differentiation, which typically characterize the development of amphibian limbs [
59]. In order to test this idea, it would be ideal to gain insight into the functional role of Region III, which is conserved in the remaining tetrapod lineages. Several experimental approaches might be relevant to test this hypothesis, including harboring an equivalent region into zebrafish HoxA13, evaluating its capacity to generate a hand plate-like phenotype. Deletions of this particular region in mouse may also be informative to link its functional role to the formation of the hand plate structure.
As for HoxA11, a possible outcome of the modifications in the alanine content of HoxA13 proteins may be the enhancement of their role as repressors. An example of such a type of repressive function is well described for HoxA7, in which N-terminal polyalanine repeats are fundamental to enhance its repressive function [
65]. This might have enabled this protein to act as a repressor of the
HoxA11 gene during limb development, leading to the establishment of an ancestral zeugopod/autopod boundary and the development of a hand plate. Moreover, dissociation of
HoxA11 and
HoxA13 expression domains may have potentiated the expansion of the skeletogenic process that culminated in the formation of additional endoskeleton elements, such as digits (
Figure 4).
Thus, there is in fact grounds to speculate that the incorporation of polyalanine repeats in HoxA11 and HoxA13 proteins may have been an important step for the evolution of limbs. Although a remark has to be made regarding the alanine content of HoxA11 and HoxA13 proteins between fish, amphibians and birds as the comparative alanine content does not support a clear correlation between higher alanine percentage and the presence of an autopod (
Figure 2 and
Figure 3). Two hypotheses may explain this observation. The first one is that the relatively similar alanine content of amphibians and birds in comparison with fish, but lower in comparison with mammals, results from the different evolutionary paths that those groups experienced [
68]. Indeed, some reptiles and amphibians have a remarkable evolutionary history with reference to the evolution of autopod, with squamates having multiple autopod losses, estimated to have occurred at least 25 times independently [
69]. Thus, for amphibians, reptiles and birds, a different molecular signature may be correlated with the evolution of the autopod [
68]. The second hypothesis is that the contribution of alanines for autopod evolution is not merely quantitative, but also qualitative. However, functional assays are mandatory to make the proof of principle of these hypotheses, as the importance of alanine repeats for limb evolution remains largely unexplored. Taking advantage of the genome editing capacity of the CRISPR-Cas9 technology [
70] and the lack of polyalanine repeats in zebrafish [
40,
41], it would be possible to introduce polyalanine repeats within the zebrafish HoxA11 or HoxA13 and assess how such sequences impact fin development and/or alter the way these transcription factors interact with their downstream targets. The realization of such experiments holds the potential of exploring how homopolymeric amino acid repeats may have impacted the fin-to-limb transition at quantitative and qualitative levels and, on a broader scale, provides evidence of how such repeat expansions may have contributed to the morphological plasticity in vertebrates.
3. Non-Coding RNAs and the Fin-to-Limb Transition
In the past decade, the scientific community has discovered that RNA’s biological function is far more complex than the initially proposed role as simple mediators in the DNA-to-protein information flow [
71]. Recently, several classes of non-coding RNAs (ncRNAs) with proven biological function have been discovered, such as: long-non-coding RNAs (lncRNAs), microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs), small interfering RNAs (siRNAs), enhancer RNAs (eRNAs) and promoter-associated RNAs (PARs) [
72]. Those ncRNAs have various functions, such as suppression of transposon activity and post-transcriptional gene silencing. They may also act as gene transcriptional activators or repressors and be involved in gene epigenetic control [
72,
73,
74].
Although exceptions are known, overall, those RNAs seem to have two characteristics in common: a low transcriptional level and a high evolutionary divergence, with their numbers increasing in the genomes along with the species’ evolutionary history [
72,
75]. For example, a recent study demonstrates that the majority of long-non-coding RNAs in each species are lineage specific [
75]. This finding, along with the discovery that ncRNAs are important for proper gene regulation during development in some species [
74,
76], designates the ncRNAs as possible drivers of species morphological innovation [
74].
Interestingly, the
HOX loci produce a considerable repertoire of ncRNAs belonging to two major classes: miRNAs (e.g.,
mir-10,
mir-196,
mir-99,
mir-126) and lncRNAs (e.g.,
HOTAIR,
HOTTIP,
Mistral,
HOTATRM1) [
77]. Those ncRNAs have been implicated in the regulation of
HOX gene expression at the transcriptional, post-transcriptional and translational level [
77], and a particularly interesting example of the contribution of such RNA species in the modulation of morphological design comes from mir-196, a well-conserved miRNA in vertebrates [
76,
78]. McGlinn and colleagues reported that knocking down
mir-196 expression in chicken embryos caused a morphologic alteration of the last cervical vertebrae into a thoracic-like identity [
76]. The phenotypic alteration seems to result partially from an expansion of the
Hoxb8 expression domain, caused by the reduction of
mir-196 transcripts [
76]. This example shows how relevant miRNAs, and on a broader scale the ncRNAs, are for the regulation of important patterning genes during development [
76], with possible evolutionary implications in the establishment of specific morphological traits.
In addition, these ncRNAs may also participate in the epigenetic control of transcription. The lncRNAs HOTAIR was the first one to be discovered to play that role, alerting the importance of RNAs as controllers of the transcriptional activity that may have been highly influential in the determination of the cell fate and, ultimately, in the evolution of animal morphology [
79,
80]. However, the versatility of functions that these lncRNAs perform during development and disease remains largely unknown. Distinct sources of information (such as conservation, coding potential patterns and anatomical properties) are currently being used to identify the main lncRNA gene families [
80]. Regarding the anatomical properties, lncRNAs have been grouped, for example, as: (1) antisense lncRNAs that overlap protein-coding genes; (2) intronic lncRNAs that are encoded within introns of protein-coding genes; (3) divergent lncRNAs encoded in the opposite direction of a protein coding gene, but sharing its promoter; (4) intergenic lncRNAs that are encoded within the intergenic space between protein-coding loci (lincRNAs).
Among the Hox genes found to be involved in limb development, the
HoxA11 locus stands out as capable of generating several antisense lncRNAs (
HoxA11AS) in a variety of tetrapods [
42,
57,
74,
81] that were found to be polyadenylated and alternately processed [
42,
57]. Moreover, comparison of these antisense
HoxA11 transcripts revealed tissue-specific differences, as well as aspects of RNA processing conserved during evolution, which strongly suggest that the antisense RNAs from the
Hoxa 11 locus are functionally significant [
42].
Two of these
HoxA11AS have overlapping complementary sequences with the first exon of HoxA11 (ENST00000520360.4 and ENST00000522863.1;
Figure 4). In contrast, others seem to have sequence complementary with the enhancer/promoter region of
HoxA11 and appear to lack sequence conservation between fish and mammals (e.g., ENST00000522674.1). In order to evaluate the antisense transcript expression of
HoxA11AS, Hsieh-Li and colleagues performed
in situ hybridizations using sense and antisense probes designed for the first exon of
HoxA11 [
57]. Surprisingly, they found complementary expression patterns of these probes in mouse embryonic limbs. At E9.5, the
HoxA11 gene is transcribed throughout the distal limb bud region and then gets progressively proximally restricted, up to the point where the
HoxA11 gene is only expressed in the prospective zeugopod region [
42,
57]. This progressive distal repression of the
HoxA11 gene coincides with the appearance of the representatives of
HoxA11AS transcripts that begin to be expressed in the most distal limb bud region at E10.5. This antisense expression domain then expands proximally, occupying the prospective autopod region by E11.5 [
42,
57]. The complementary expression patterns found between the sense and antisense probes designed against the
HoxA11 first exon suggest that production of
HoxA11AS transcripts during limb development may conduct expression inhibition of
HoxA11 [
42,
57].
Cross species alignments using the “Comparative Genomic” tool from Ensemble (release 80, May 2015, [
82]) suggest that the putative
HoxA11AS encoding regions have been subjected to a different selective pressure during vertebrates’ evolution. Indeed, the regions that overlap with the complementary sequence of
HoxA11, in the first exon, seem to be significantly conserved among tetrapods. However, another
HoxA11AS encoding region, which seems to be complementary to the
HoxA11 enhancer/promoter region (ENST00000522674.1), is apparently less conserved [
74]. Within mammals, this particular
HoxA11 antisense is detectable in eutherians and is expressed during embryonic development, but also in the endometrium during pregnancy [
74]. The potential role of this
HoxA11AS in mammal reproduction may justify its generalized presence within eutherians’ genomes. In non-mammal organisms, such as in the frog, the degree of sequence conservation drops to less than 50% [
74]. Thus, this amphibian lncRNA, if functional, might be exclusively associated with the regulation of limb development and, as such, under lower selective pressure. Moreover, cross species alignment also revealed an absence of significant similarity between human
HOXA11AS gene and the syntenic zebrafish, stickleback and coelacanth regions. Moreover, when Bartel and colleagues characterized the lncRNAs during zebrafish development using chromatin marks, poly(A)-site mapping and RNA-Seq data, they did not find any potential homologues of
HOXA11AS, either by sequence conservation or genomic location [
83]. This might be due to the striking lack of conservation of the teleost HoxA intergenic regions, when compared to the ones from other gnathostome lineages [
84]. In fact, teleosts have duplicated and more compact HoxA clusters, with shorter intergenic regions [
84], which may suggest that they lack most of the genomic region able to transcribe
HoxA11AS. It remains to be uncovered if fish species that diverge prior to teleost radiation and where the length of the HoxA intergenic regions more closely resemble the ones from the tetrapods have the capacity to produce these lncRNAs (e.g., shark, paddlefish).
Given the possible inhibitory effect of
HoxA11AS on
HoxA11 and what seems to be a tetrapod-specific characteristic of
HoxA11AS, we present the hypothesis that the evolutionary origin of
HoxA11AS was one of the molecular events that might have contributed to the evolution of the fin/limb morphology. According to this hypothesis, the appearance of
HoxA11AS expression in the distal limb region inhibits
HoxA11 expression in that limb domain, thus uncoupling
HoxA11 and
HoxA13 limb expression domains and enabling the definition of a distal
HoxA13 single positive autopod domain. The apparent lack of
HoxA11AS homologues in the zebrafish, according to our hypothesis, may help to explain why the expression domains of
HoxA11/HoxA13 do not split during fin development. In contrast, the transient separation of these domains detected during shark and paddlefish fin development [
26,
35,
36] suggests that these organisms may produce
HoxA11AS. However, these
HoxA11AS are probably transcribed in the antisense direction within their own
HoxA11 loci; therefore, they may not be conserved among vertebrates. Taken as inspiration the work from Hsieh-Li and colleagues [
57], it would be interesting to analyze the expression pattern of sense and antisense
HoxA11 transcripts in these organisms.
An important step to gain insight into our preferential hypothesis, which relates the origin of
HoxA11AS with the evolution of vertebrate fins, is to functionally characterize these transcripts to better understand their biological relevance to limb development. To achieve this goal, a possible strategy would be to take advantage of the chick model, which seems to have conserved
HoxA11AS, and induce its overexpression, knockdown and knockout
in ovo, following the strategy used by McGlinn and colleagues [
76]. This would allow testing the impact of
HoxA11AS in the definition of the
HoxA11 expression domain during the formation of limbs. Another important step to explore our hypothesis is to assess the evolutionary importance of these lncRNAs during vertebrate fin/limb development. To this end, an innovative strategy would be the knock-in of
HoxA11AS in zebrafish and the characterization of its impact during fin development, assuming that the capacity to metabolize these lncRNAs was assembled prior to the divergence of tetrapods.
4. Cis-Regulation of 5’ HoxA Genes and the Fin-to-Limb Transition
According to the previously-presented hypothesis, the definition of the autopod domain is associated with the proximal restriction of
HoxA11 expression during the development of appendicular structures in stem groups of tetrapods [
26]. However, the autopod morphology, regarding digit size and number, acquired modifications during evolution probably due to an increase in the transcription of 5’
HoxA and
HoxD genes [
8,
85,
86]. Thus, recruitment of novel non-coding regulatory elements, which are able to modulate the transcription of these genes, seems to have been a crucial step for the evolution of the autopod [
85,
86]. The enhancer elements that govern the expression of 5’
HoxD have been characterized during the development of mouse limbs [
85,
86]. These reports highlighted that a particular combination of regulatory elements is required for the specification of the appropriate time, dosage and pattern of 5’
HoxD gene expression.
Regarding the
HoxA11 and
HoxA13 genes, despite their crucial role for the correct development and proximo-distal patterning of the limbs, information on their
cis-regulatory network is still scarce, although the research teams led by Innis [
47], Kmita [
66], Gómez-Skarmeta and Shubin [
43] have shed some light into the enhancers that drive
HoxA13 expression in the developing limbs and their possible impact in the fin-to-limb transition. From the works published to date on mice, the enhancer network that regulates distal
HoxA13 expression in the developing limbs is comprised of at least nineteen different elements located upstream of, and excluding, the
HoxA cluster (
Figure 4) [
66]. Some of those enhancers are present within genes located upstream of the
HoxA cluster, namely the
Hibadh,
Tax1bp1 and
Jazf1 genes [
66]. A curious observation is that the enhancers driving
HoxA13 distal expression in mouse limb form several sub-megabase topological domains (sub-TADs) that contact each other in a fashion that, in some cases, seems to be independent of the enhancer’s transcriptional activity. This suggests that the sub-TADs organization is governed by a developmental structure/function interplay instead of a mere clustering of active enhancers [
66].
When the conservation of the mouse
HoxA13 enhancer network was assayed, using sequence homology and ATAC-Seq (assay for transposase-accessible chromatin using sequencing) in zebrafish and gar, only the gar genome showed conservation of three (e10, e13 and e16) of the 19 enhancers that regulate
HoxA13 transcription in mouse limb, and at least the gar e16 region induces, in zebrafish and mouse transgene reporter assays, an expression pattern that is very similar to the one driven by mouse e16 [
43]. Taking into account that lack of sequence conservation is not necessarily sufficient to exclude the presence of enhancers in fish, these data nevertheless suggest two possible circumstances. Firstly, the zebrafish
hoxa13a and
hoxa13b enhancer network, if present, represents a derived state, as no sequence homology was detected between zebrafish, gar and mouse
HoxA enhancers [
43]. This observation may result from extensive remodeling of
HoxA regulatory elements caused by the occurrence of a third whole genome duplication in teleosts followed by gene sub-functionalization, thus leading to the inability of the zebrafish
hoxa13 paralogs to respond to a “distal limb program” during development [
43]. Secondly, the enhancer network that regulates
HoxA13’s late phase expression suffered extensive elaboration/remodeling over the course of the vertebrates’ evolution, although some of its elements might have already been present in the last common ancestor of bony vertebrates [
43]. The extensive elaboration of
HoxA13 enhancer elements throughout vertebrate evolution suggests that the fish
HoxA13 transcriptional level during fin development is too low to elicit an adequate “distal limb identity”, thus suggesting that
HoxA13 gene dosage increase was a requirement for the fin-to-limb transition. This hypothesis finds support in the work published, in 1997, by Zákány and colleagues [
8], where by analyzing a set of distal
HoxA and
HoxD compound mutants, this team found that overall autopod morphology, and in particular digit number and length, greatly varies in response to
Hox dosage [
8] and to a minor extent to the qualitative nature of the expressed HoxA and HoxD proteins (
Hox code) [
85]. Moreover, overexpression of zebrafish
hoxd13a was shown to cause changes in the morphogenesis of the fins towards a limb-like identity [
22]. Testing out the hypothesis that an increase of
HoxA13 dosage was an evolutionary requirement for the fin-to-limb transition could be easily achieved in zebrafish following a strategy previously described by Freitas and colleagues [
22]. Performing such a proof-of-principle study not only upholds the potential to disclose how increasing levels of HoxA13 proteins may have contributed to the morphological transition between fish fins and tetrapod limbs, but may also shed light on how regulation and function of
Hox13 paralogs genes may have set the grounds for autopod evolution.
5. Conclusions and Future Directions
In this review, we attempted to survey the evolution of HoxA11 and HoxA13 transcription factors and their regulatory control in vertebrates. Evaluating the current state of the art and performing complementary analyses, using information currently available for distinct vertebrate lineages, we support the hypotheses implicating changes in the alanine content, capacity to generate long-non-coding RNAs and incorporation of a novel
cis-regulatory region in the developmental mechanisms involved in the fin-to-limb transition in vertebrates (
Figure 4).
Conserved polyalanine tracts with a minimum of four residues are common within the HoxA11 and HoxA13 proteins of tetrapods (
Figure 2,
Figure 3 and
Figure 4). However, these types of sequences are undetectable in the orthologs of fish. In the HoxA11 proteins, these enriched alanine domains co-localize with the hydrophilic region, being higher the percentages of alanines detectable in amniotes. Among the fish analyzed, the hydrophilic region of paddlefish seems to be the one that has a higher content of alanines. These observations point us to two hypotheses: (1) an increased alanine content within HoxA11 proteins may have impacted the formation of specific amniote limb structures during evolution, such as the hand plates that precede digit differentiation; (2) incorporation of extra alanine residues into the hydrophilic regions of HoxA11 protein probably started prior to the tetrapod divergence, in the common ancestor of actinopterygians and sarcopterygians. In the HoxA13 proteins, most polyalanine tracts (I–VII) are conserved in mammals (
Figure 3 and
Figure 4). However, the polyalanine Tract III seems to be present in the chick HoxA13 proteins, which may suggest a particular role of these tracts in the formation of amniote-specific limb structures, the hand plates.
Taking into consideration the functional potential found for this type of amino acid repeats [
40,
47,
49], their integration into the HoxA11 and Hoxa13 proteins during evolution might have modified the way they interact with their downstream targets, contributing to the phenotypic variation found in the evolutionary transition from fish fins to tetrapod limbs. For example, the polyalanine repeats found in the HoxA11 and HoxA13 proteins might have enhanced their repressor role upon other molecules, helping to establish novel domains of cell identity, leading to the differentiation of the hand plate in amniotes. The repressor potential of HoxA13 in particular might have inhibited the expression of
HoxA11 in the most distal mesenchyme of the appendages, generating a novel field of cell identity, which might have potentiated the formation of additional skeletal elements. However, the proof of principle of these hypotheses is lacking. Here, we suggest, as a future working perspective, the use of CRISPR-Cas9 technology [
70] to harbor polyalanine repeats within the zebrafish
HoxA11 or
HoxA13 in order to evaluate the phenotypic consequences of such sequences during appendage development. These experiments would certainly add to our knowledge of how changes in the alanine content of transcription factors impacted the diversification of vertebrate structures.
In addition, we show here that the capacity to generate
HoxA11AS, which are long-non-coding RNAs, seems to be exclusive to tetrapods (
Figure 4) [
42,
57,
74,
81]. Moreover the expression of particular
HoxA11 antisense RNAs was found to be complementary to
HoxA11, suggesting that while
HoxA11 is progressively repressed distally,
HoxA11AS expression expands proximally. This observation points to a repressor role of
HoxA11AS on
HoxA11 that might have contributed to split the
HoxA11/HoxA13 expression domains, potentiating the formation of the zeugopod and the autopod. Given that during catshark and paddlefish fin development, the expression domains of
hoxa11 and
hoxa13 also split transiently into two non-overlapping territories, it would be important to search for
HoxA11AS homologues in these organisms and evaluate the potential repressor role on
HoxA11. These ideas also lack functional validation, and a first step would be to gain more insight into the expression of HoxA11 antisense in fish representing distinct phylogenetic positions. Moreover, it would be important to better characterize the role of
HoxA11AS during tetrapod limb development, to test their developmental and evolutionary potential harboring of
HoxA11AS in zebrafish and to characterize the impact during appendage development.
Finally, we suggest that the transcriptional levels of
HoxA11 and
HoxA13 might have also influenced appendage evolution, as has been proposed for
HoxD13 [
22]. Indeed, the autopod morphology in mouse, in particular digit number and length, is influenced by the
Hox dosage [
8], which often depends on the enhancer capacity of
cis-regulatory regions within their genomic landscape [
87]. Marked differences in the putative regulatory regions of the 5’
HoxA genes were identified in fish species representing distinct phylogenetic positions, such as the spotted gar and the zebrafish (
Figure 4). Thus,
cis-regulatory changes may have interfered with the differential fin morphology of distinct fish groups (
Figure 4). Within tetrapods, subtle changes were also identified in the putative regulatory regions of these genes that may have contributed to the distinct autopod anatomy found in mammals and birds, for example [
43]. Additionally, fish
versus tetrapod comparisons suggest a significant increase of
cis-regulatory modules in tetrapods [
43], a phenomena that might have been crucial to achieve the appropriate levels of Hox expression required for the origin of the autopod. To further explore the consequences of increased levels of 5’ HoxA proteins and the addition of novel regulatory modules for the developmental processes resulting in the evolution of limbs, we emphasize the advantages of the zebrafish model to perform the appropriate proof of principle studies due to its current technical repertoire, allowing a variety of functional assays [
22].