Molecular Responses during Plant Grafting and Its Regulation by Auxins, Cytokinins, and Gibberellins
Abstract
:1. Introduction
2. Molecular Responses at the Grafting Site
2.1. Movement of Genetic Material at the Grafting Site
2.1.1. DNA (Organelle/Nuclear Genome) Movement across Graft Unions
2.1.2. RNA Movement across Graft Unions
2.2. Proteins at the Grafting Site
3. Plant Hormone Mediated Grafting and Its Underlying Molecular Mechanisms
3.1. Regulation of the Grafting by Auxins
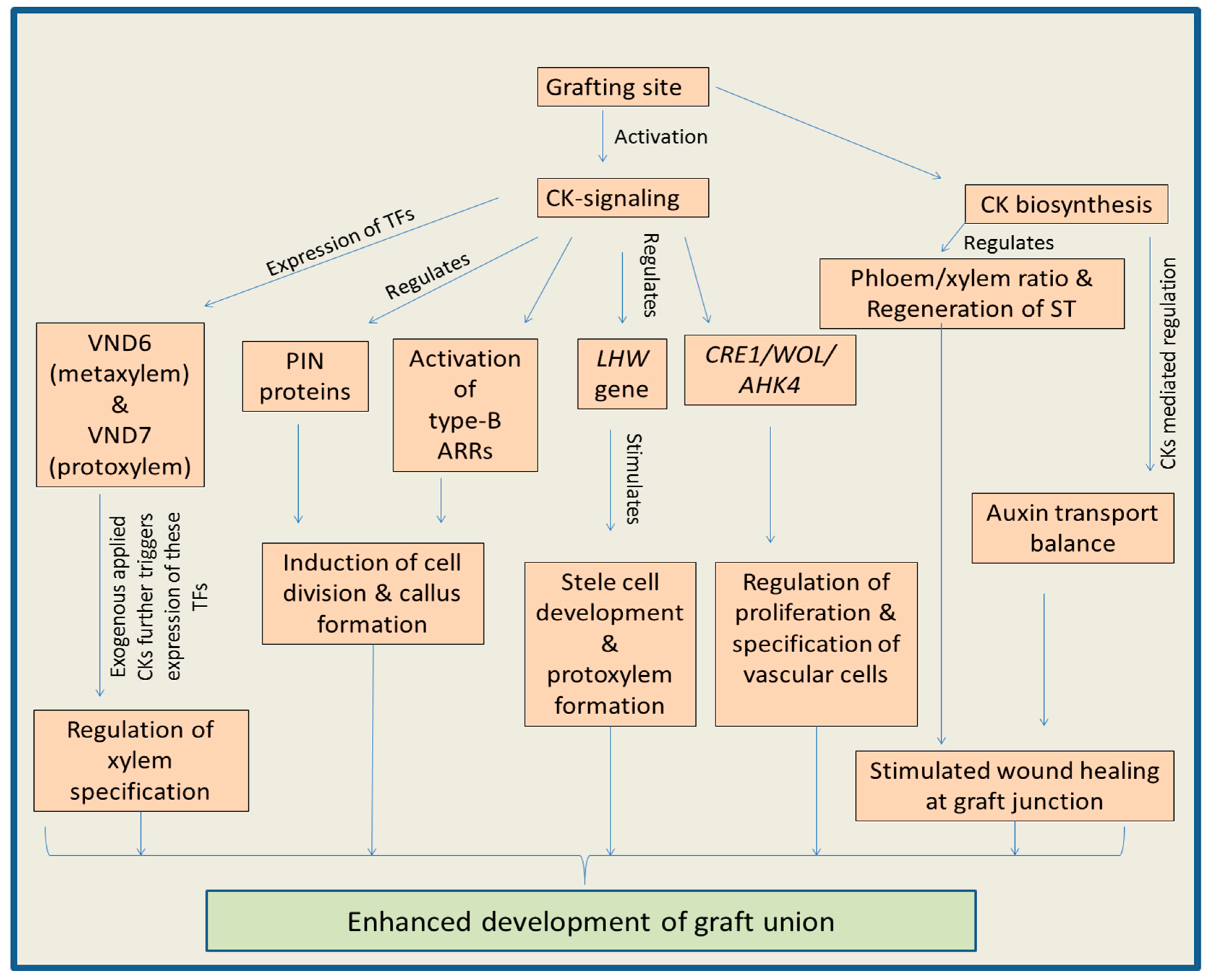
3.2. Regulation of the Grafting by Cytokinins
Cytokinins Regulate Cambium Activity and Secondary Growth to Favor Grafting
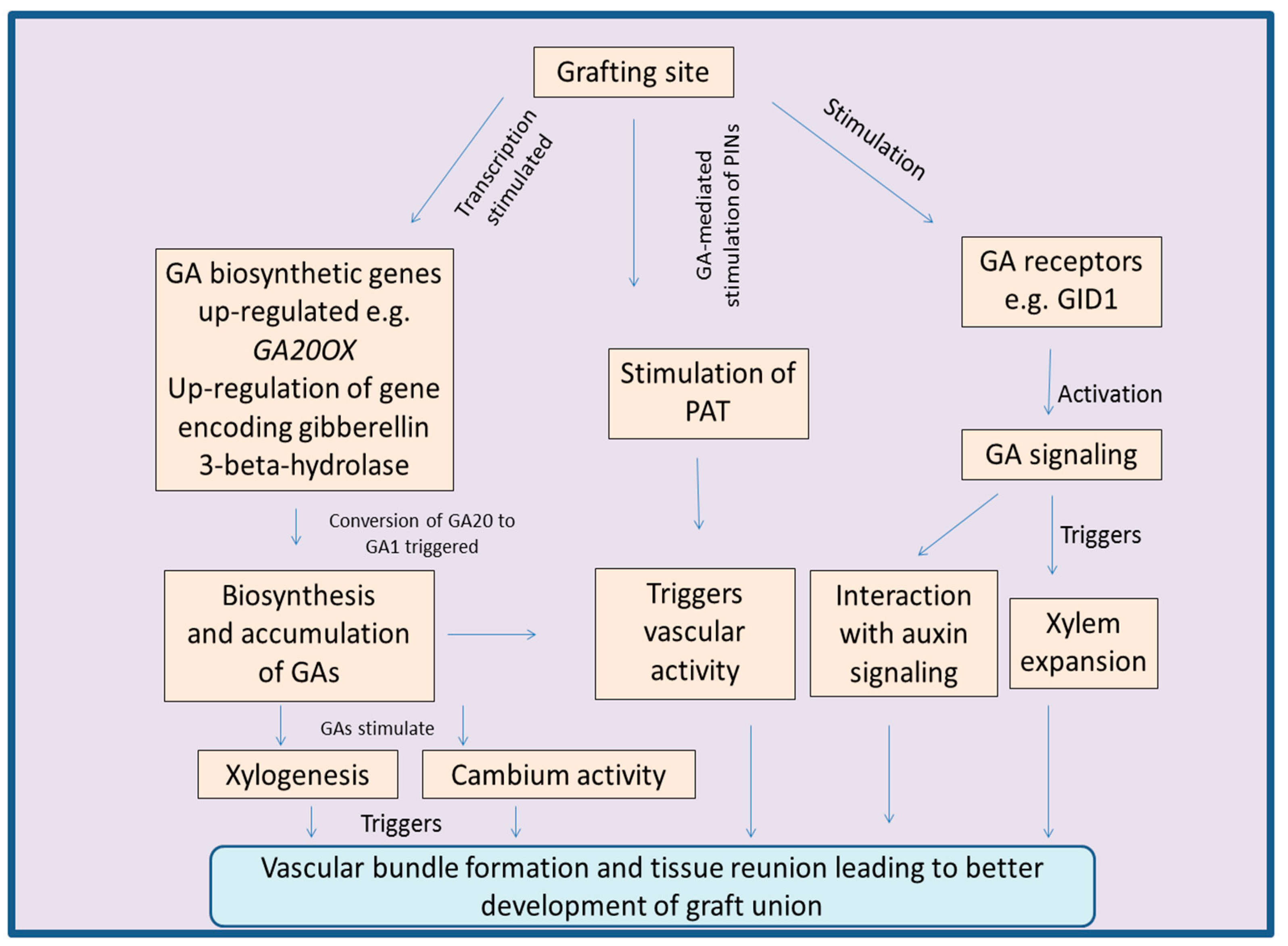
3.3. Regulation of Grafting by Gibberellins
4. Hormonal Crosstalk at Grafting Site
5. Conclusions and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Wang, J.; Jiang, L.; Wu, R. Plant grafting: How genetic exchange promotes vascular reconnection. New Phytol. 2017, 214, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Warschefsky, E.J.; Klein, L.L.; Frank, M.H.; Chitwood, D.H.; Londo, J.P.; von Wettberg, E.J.B.; Miller, A.J. Rootstocks: Diversity, Domestication, and Impacts on Shoot Phenotypes. Trends Plant Sci. 2016, 21, 418–437. [Google Scholar] [CrossRef] [PubMed]
- Goldschmidt, E.E. Plant grafting: New mechanisms, evolutionary implications. Front. Plant Sci. 2014, 5, 727. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-M.; Kubota, C.; Tsao, S.; Bie, Z.; Echevarria, P.H.; Morra, L.; Oda, M. Current status of vegetable grafting: Diffusion, grafting techniques, automation. Sci. Hortic. 2010, 127, 93–105. [Google Scholar] [CrossRef]
- Zhao, X.; Guo, Y.; Huber, D.J.; Lee, J. Grafting effects on postharvest ripening and quality of 1-methylcyclopropene-treated muskmelon fruit. Sci. Hortic. 2011, 130, 581–587. [Google Scholar] [CrossRef]
- Tsaballa, A.; Athanasiadis, C.; Pasentsis, K.; Ganopoulos, I.; Nianiou-Obeidat, I.; Tsaftaris, A. Molecular studies of inheritable grafting induced changes in pepper (Capsicum annuum) fruit shape. Sci. Hortic. 2013, 149, 2–8. [Google Scholar] [CrossRef]
- Santa-Cruz, A.; Martinez-Rodriguez, M.M.; Perez-Alfocea, F.; Romero-Aranda, R.; Bolarin, M.C. The rootstock effect on the tomato salinity response depends on the shoot genotype. Plant Sci. 2002, 162, 825–831. [Google Scholar] [CrossRef]
- Louws, F.J.; Rivard, C.L.; Kubota, C. Grafting fruiting vegetables to manage soilborne pathogens, foliar pathogens, arthropods and weeds. Sci. Hortic. 2010, 127, 127–146. [Google Scholar] [CrossRef]
- Schwarz, D.; Rouphael, Y.; Colla, G.; Venema, J.H. Grafting as a tool to improve tolerance of vegetables to abiotic stresses: Thermal stress, water stress and organic pollutants. Sci. Hortic. 2010, 127, 162–171. [Google Scholar] [CrossRef]
- Bhatt, R.M.; Upreti, K.K.; Divya, M.; Bhat, S.; Pavithra, C.; Sadashiva, A. Interspecific grafting to enhance physiological resilience to flooding stress in tomato (Solanum lycopersicum L.). Sci. Hortic. 2015, 182, 8–17. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Wang, Z.; Guo, X.; Wang, F.; Xia, X.J.; Zhou, J.; Shi, K.; Yu, J.Q.; Zhou, Y.H. Microarray and genetic analysis reveals that csa-miR159b plays a critical role in abscisic acid-mediated heat tolerance in grafted cucumber plants. Plant Cell Environ. 2016, 39, 1790–1804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penella, C.; Landi, M.; Guidi, L.; Nebauer, S.G.; Pellegrini, E.; San Bautista, A.; Remorini, D.; Nali, C.; Lopez-Galarza, S.; Calatayud, A. Salt-tolerant rootstock increases yield of pepper under salinity through maintenance of photosynthetic performance and sinks strength. J. Plant Physiol. 2016, 193, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dor, E.; Alperin, B.; Wininger, S.; Ben-Dor, B.; Somvanshi, V.S.; Koltai, H.; Kapulnik, Y.; Hershenhorn, J. Characterization of a novel tomato mutant resistant to the weedy parasites Orobanche and Phelipanche spp. Euphytica 2010, 171, 371–380. [Google Scholar] [CrossRef]
- Huang, Y.; Li, J.; Hua, B.; Liu, Z.; Fan, M.; Bie, Z. Grafting onto different rootstocks as a means to improve watermelon tolerance to low potassium stress. Sci. Hortic. 2013, 149, 80–85. [Google Scholar] [CrossRef]
- Albacete, A.; Martinez-Andujar, C.; Martinez-Perez, A.; Thompson, A.J.; Dodd, I.C.; Perez-Alfocea, F. Unravelling rootstockxscion interactions to improve food security. J. Exp. Bot. 2015, 66, 2211–2226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meister, R.; Rajani, M.S.; Ruzicka, D.; Schachtman, D.P. Challenges of modifying root traits in crops for agriculture. Trends Plant Sci. 2014, 19, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Wissuwa, M.; Kretzschmar, T.; Rose, T.J. From promise to application: Root traits for enhanced nutrient capture in rice breeding. J. Exp. Bot. 2016, 67, 3605–3615. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.J.; Atkinson, C.J.; Bengough, A.G.; Else, M.A.; Fernandez-Fernandez, F.; Harrison, R.J.; Schmidt, S. Contributions of roots and rootstocks to sustainable, intensified crop production. J. Exp. Bot. 2013, 64, 1209–1222. [Google Scholar] [CrossRef] [Green Version]
- Melnyk, C.W.; Schuster, C.; Leyser, O.; Meyerowitz, E.M. A Developmental Framework for Graft Formation and Vascular Reconnection in Arabidopsis thaliana. Curr. Biol. 2015, 25, 1306–1318. [Google Scholar] [CrossRef]
- Ribeiro, L.M.; Nery, L.A.; Vieira, L.M.; Mercadante-Simões, M.O. Histological study of micrografting in passionfruit. Plant Cell Tissue Organ Cult. 2015, 123, 173–181. [Google Scholar] [CrossRef]
- Jeffree, C.; Yeoman, M. Development of intercellular connections between opposing cells in a graft union. New Phytol. 1983, 93, 491–509. [Google Scholar] [CrossRef]
- Aloni, B.; Cohen, R.; Karni, L.; Aktas, H.; Edelstein, M. Hormonal signaling in rootstock–scion interactions. Sci. Hortic. 2010, 127, 119–126. [Google Scholar] [CrossRef]
- Yin, H.; Yan, B.; Sun, J.; Jia, P.; Zhang, Z.; Yan, X.; Chai, J.; Ren, Z.; Zheng, G.; Liu, H. Graft-union development: A delicate process that involves cell-cell communication between scion and stock for local auxin accumulation. J. Exp. Bot. 2012, 63, 4219–4232. [Google Scholar] [CrossRef] [PubMed]
- Nanda, A.K.; Melnyk, C.W. The role of plant hormones during grafting. J. Plant Res. 2018, 131, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Taller, J.; Hirata, Y.; Yagishita, N.; Kita, M.; Ogata, S. Graft-induced genetic changes and the inheritance of several characteristics in pepper (Capsicum annuum L.). Theor. Appl. Genet. 1998, 97, 705–713. [Google Scholar] [CrossRef]
- Stegemann, S.; Bock, R. Exchange of genetic material between cells in plant tissue grafts. Science 2009, 324, 649–651. [Google Scholar] [CrossRef] [PubMed]
- Stegemann, S.; Keuthe, M.; Greiner, S.; Bock, R. Horizontal transfer of chloroplast genomes between plant species. Proc. Nat. Acad. Sci. USA 2012, 109, 2434–2438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, R.; Wang, X.; Lin, Y.; Ma, Y.; Liu, G.; Yu, X.; Zhong, S.; Liu, B. Inter-species grafting caused extensive and heritable alterations of DNA methylation in Solanaceae plants. PLoS ONE 2013, 8, e61995. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, I.; Stegemann, S.; Golczyk, H.; Karcher, D.; Bock, R. Horizontal genome transfer as an asexual path to the formation of new species. Nature 2014, 511, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Molnar, A.; Melnyk, C.W.; Bassett, A.; Hardcastle, T.J.; Dunn, R.; Baulcombe, D.C. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 2010, 328, 872–875. [Google Scholar] [CrossRef]
- Chitwood, D.H.; Timmermans, M.C. Small RNAs are on the move. Nature 2010, 467, 415. [Google Scholar] [CrossRef] [PubMed]
- Fragoso, V.; Goddard, H.; Baldwin, I.T.; Kim, S.G. A simple and efficient micrografting method for stably transformed Nicotiana attenuata plants to examine shoot-root signaling. Plant Met. 2011, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Melnyk, C.W.; Molnar, A.; Bassett, A.; Baulcombe, D.C. Mobile 24 nt small RNAs direct transcriptional gene silencing in the root meristems of Arabidopsis thaliana. Curr. Biol. 2011, 21, 1678–1683. [Google Scholar] [CrossRef] [PubMed]
- Sima, X.; Jiang, B.; Fang, J.; He, Y.; Fang, Z.; Km, S.K.; Ren, W.; Qiu, L.; Chen, X.; Zheng, B. Identification by deep sequencing and profiling of conserved and novel hickory microRNAs involved in the graft process. Plant Biotechnol. Rep. 2015, 9, 115–124. [Google Scholar] [CrossRef]
- Alscher, R.G.; Erturk, N.; Heath, L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef]
- Notaguchi, M.; Higashiyama, T.; Suzuki, T. Identification of mRNAs that move over long distances using an RNA-Seq analysis of Arabidopsis/Nicotiana benthamiana heterografts. Plant Cell Physiol. 2015, 56, 311–321. [Google Scholar] [CrossRef]
- Thieme, C.J.; Rojas-Triana, M.; Stecyk, E.; Schudoma, C.; Zhang, W.; Yang, L.; Minambres, M.; Walther, D.; Schulze, W.X.; Paz-Ares, J.; et al. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 2015, 1, 15025. [Google Scholar] [CrossRef]
- Yang, Y.; Mao, L.; Jittayasothorn, Y.; Kang, Y.; Jiao, C.; Fei, Z.; Zhong, G.Y. Messenger RNA exchange between scions and rootstocks in grafted grapevines. BMC Plant Biol. 2015, 15, 251. [Google Scholar] [CrossRef]
- Zhang, Z.; Zheng, Y.; Ham, B.K.; Chen, J.; Yoshida, A.; Kochian, L.V.; Fei, Z.; Lucas, W.J. Vascular-mediated signalling involved in early phosphate stress response in plants. Nat. Plants 2016, 2, 16033. [Google Scholar] [CrossRef]
- Zhang, W.; Thieme, C.J.; Kollwig, G.; Apelt, F.; Yang, L.; Winter, N.; Andresen, N.; Walther, D.; Kragler, F. tRNA-Related Sequences Trigger Systemic mRNA Transport in Plants. Plant Cell 2016, 28, 1237–1249. [Google Scholar] [CrossRef] [Green Version]
- Calderwood, A.; Kopriva, S.; Morris, R.J. Transcript Abundance Explains mRNA Mobility Data in Arabidopsis thaliana. Plant Cell 2016, 28, 610–615. [Google Scholar] [CrossRef] [PubMed]
- Mach, J. Ticket to Ride: tRNA-Related Sequences and Systemic Movement of mRNAs. Plant Cell 2016, 28, 1231–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, C.; Zheng, Y.; Huang, J.; Zhou, X.; Li, R.; Zha, M.; Wang, S.; Huang, Z.; Lan, H.; Turgeon, R.; et al. Elucidation of the Mechanisms of Long-Distance mRNA Movement in a Nicotiana benthamiana/Tomato Heterograft System. Plant Physiol. 2018, 177, 745–758. [Google Scholar] [CrossRef] [PubMed]
- Haywood, V.; Yu, T.S.; Huang, N.C.; Lucas, W.J. Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 2005, 42, 49–68. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.C.; Yu, T.S. The sequences of Arabidopsis GA-INSENSITIVE RNA constitute the motifs that are necessary and sufficient for RNA long-distance trafficking. Plant J. 2009, 59, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Xoconostle-Cazares, B.; Xiang, Y.; Ruiz-Medrano, R.; Wang, H.L.; Monzer, J.; Yoo, B.C.; McFarland, K.C.; Franceschi, V.R.; Lucas, W.J. Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 1999, 283, 94–98. [Google Scholar] [CrossRef]
- Duan, X.; Zhang, W.; Huang, J.; Zhao, L.; Ma, C.; Hao, L.; Yuan, H.; Harada, T.; Li, T. KNOTTED1 mRNA undergoes long-distance transport and interacts with movement protein binding protein 2C in pear (Pyrus betulaefolia). Plant Cell Tissue Organ Cult. 2015, 121, 109–119. [Google Scholar] [CrossRef]
- Duan, X.; Zhang, W.; Huang, J.; Hao, L.; Wang, S.; Wang, A.; Meng, D.; Zhang, Q.; Chen, Q.; Li, T. PbWoxT1 mRNA from pear (Pyrus betulaefolia) undergoes long-distance transport assisted by a polypyrimidine tract binding protein. New Phytol. 2016, 210, 511–524. [Google Scholar] [CrossRef]
- Spiegelman, Z.; Ham, B.K.; Zhang, Z.; Toal, T.W.; Brady, S.M.; Zheng, Y.; Fei, Z.; Lucas, W.J.; Wolf, S. A tomato phloem-mobile protein regulates the shoot-to-root ratio by mediating the auxin response in distant organs. Plant J. 2015, 83, 853–863. [Google Scholar] [CrossRef]
- Paultre, D.S.G.; Gustin, M.P.; Molnar, A.; Oparka, K.J. Lost in Transit: Long-Distance Trafficking and Phloem Unloading of Protein Signals in Arabidopsis Homografts. Plant Cell 2016, 28, 2016–2025. [Google Scholar] [CrossRef]
- Wu, Y.; Cosgrove, D.J. Adaptation of roots to low water potentials by changes in cell wall extensibility and cell wall proteins. J. Exp. Bot. 2000, 51, 1543–1553. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Choi, D.; Kende, H. Expansins: Ever-expanding numbers and functions. Curr. Opin. Plant Biol. 2001, 4, 527–532. [Google Scholar] [CrossRef]
- Malz, S.; Sauter, M. Expression of two PIP genes in rapidly growing internodes of rice is not primarily controlled by meristem activity or cell expansion. Plant Mol. Biol. 1999, 40, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Javot, H.; Maurel, C. The role of aquaporins in root water uptake. Ann. Bot. 2002, 90, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Liu, L.; Huang, J.; Cheng, X.; Zhu, Y.; Xu, H. Analysis on physiological and biochemical traits of survival of Carya cathayensis grafted seedling. J. Fujian Coll. For. 2002, 22, 320–324. [Google Scholar]
- Zheng, B.S.; Chu, H.L.; Jin, S.H.; Huang, Y.J.; Wang, Z.J.; Chen, M.; Huang, J.Q. cDNA-AFLP analysis of gene expression in hickory (Carya cathayensis) during graft process. Tree Physiol. 2010, 30, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Saravana Kumar, R.M.; Gao, L.X.; Yuan, H.W.; Xu, D.B.; Liang, Z.; Tao, S.C.; Guo, W.B.; Yan, D.L.; Zheng, B.S.; Edqvist, J. Auxin enhances grafting success in Carya cathayensis (Chinese hickory). Planta 2018, 247, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Yuan, H.; Tong, Y.; Zhao, L.; Qiu, L.; Guo, W.; Shen, C.; Liu, H.; Yan, D.; Zheng, B. Comparative Proteomic Analysis of the Graft Unions in Hickory (Carya cathayensis) Provides Insights into Response Mechanisms to Grafting Process. Front. Plant Sci. 2017, 8, 676. [Google Scholar] [CrossRef]
- Zhao, Y. Essential Roles of Local Auxin Biosynthesis in Plant Development and in Adaptation to Environmental Changes. Ann. Rev. Plant Biol. 2018, 69, 417–435. [Google Scholar] [CrossRef]
- Lv, B.; Yan, Z.; Tian, H.; Zhang, X.; Ding, Z. Local Auxin Biosynthesis Mediates Plant Growth and Development. Trends Plant Sci. 2019, 24, 6–9. [Google Scholar] [CrossRef]
- Ljung, K. Auxin metabolism and homeostasis during plant development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef] [Green Version]
- Yao, X.; Tian, L.; Yang, J.; Zhao, Y.N.; Zhu, Y.X.; Dai, X.; Zhao, Y.; Yang, Z.N. Auxin production in diploid microsporocytes is necessary and sufficient for early stages of pollen development. PLoS Genet. 2018, 14, e1007397. [Google Scholar] [CrossRef]
- Chen, Q.; Dai, X.; De-Paoli, H.; Cheng, Y.; Takebayashi, Y.; Kasahara, H.; Kamiya, Y.; Zhao, Y. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 2014, 55, 1072–1079. [Google Scholar] [CrossRef]
- Ruzicka, K.; Ursache, R.; Hejatko, J.; Helariutta, Y. Xylem development-from the cradle to the grave. New Phytol. 2015, 207, 519–535. [Google Scholar] [CrossRef]
- Matsuoka, K.; Sugawara, E.; Aoki, R.; Takuma, K.; Terao-Morita, M.; Satoh, S.; Asahina, M. Differential Cellular Control by Cotyledon-Derived Phytohormones Involved in Graft Reunion of Arabidopsis Hypocotyls. Plant Cell Physiol. 2016, 57, 2620–2631. [Google Scholar] [CrossRef]
- Armengot, L.; Marques-Bueno, M.M.; Jaillais, Y. Regulation of polar auxin transport by protein and lipid kinases. J. Exp. Bot. 2016, 67, 4015–4037. [Google Scholar] [CrossRef] [Green Version]
- Melnyk, C.W.; Gabel, A.; Hardcastle, T.J.; Robinson, S.; Miyashima, S.; Grosse, I.; Meyerowitz, E.M. Transcriptome dynamics at Arabidopsis graft junctions reveal an intertissue recognition mechanism that activates vascular regeneration. Proc. Nat. Acad. Sci. USA 2018, 115, E2447–E2456. [Google Scholar] [CrossRef]
- Kumar, R.M.S.; Ji, G.; Guo, H.; Zhao, L.; Zheng, B. Over-expression of a grafting-responsive gene from hickory increases abiotic stress tolerance in Arabidopsis. Plant Cell Rep. 2018, 37, 541–552. [Google Scholar] [CrossRef]
- Sauer, M.; Balla, J.; Luschnig, C.; Wisniewska, J.; Reinohl, V.; Friml, J.; Benkova, E. Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 2006, 20, 2902–2911. [Google Scholar] [CrossRef] [Green Version]
- Mazur, E.; Benkova, E.; Friml, J. Vascular cambium regeneration and vessel formation in wounded inflorescence stems of Arabidopsis. Sci. Rep. 2016, 6, 33754. [Google Scholar] [CrossRef]
- Yu, C.; Dong, W.; Zhan, Y.; Huang, Z.A.; Li, Z.; Kim, I.S.; Zhang, C. Genome-wide identification and expression analysis of ClLAX, ClPIN and ClABCB genes families in Citrullus lanatus under various abiotic stresses and grafting. BMC Genet. 2017, 18, 33. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zhao, L.; Qiu, L.; Xu, D.; Tong, Y.; Guo, W.; Yang, X.; Shen, C.; Yan, D.; Zheng, B. Transcriptome and hormonal analysis of grafting process by investigating the homeostasis of a series of metabolic pathways in Torreya grandis cv. Merrillii. Ind. Crops Prod. 2017, 108, 814–823. [Google Scholar] [CrossRef]
- Wisniewska, J.; Xu, J.; Seifertova, D.; Brewer, P.B.; Ruzicka, K.; Blilou, I.; Rouquie, D.; Benkova, E.; Scheres, B.; Friml, J. Polar PIN localization directs auxin flow in plants. Science 2006, 312, 883. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, N.; Taylor, C.; Leyser, O. Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol. 2013, 11, e1001474. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Ma, J.; Tan, M.; Mao, J.; An, N.; Sha, G.; Zhang, D.; Zhao, C.; Han, M. Transcriptome analysis reveals the effects of sugar metabolism and auxin and cytokinin signaling pathways on root growth and development of grafted apple. BMC Genom. 2016, 17, 150. [Google Scholar] [CrossRef] [PubMed]
- Dubrovsky, J.G.; Sauer, M.; Napsucialy-Mendivil, S.; Ivanchenko, M.G.; Friml, J.; Shishkova, S.; Celenza, J.; Benkova, E. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Nat. Acad. Sci. USA 2008, 105, 8790–8794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marhavy, P.; Vanstraelen, M.; De Rybel, B.; Zhaojun, D.; Bennett, M.J.; Beeckman, T.; Benkova, E. Auxin reflux between the endodermis and pericycle promotes lateral root initiation. EMBO J. 2013, 32, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Vermeer, J.E.; von Wangenheim, D.; Barberon, M.; Lee, Y.; Stelzer, E.H.; Maizel, A.; Geldner, N. A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 2014, 343, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jin, Z.; Yin, H.; Yan, B.; Ren, Z.; Xu, J.; Mu, C.; Zhang, Y.; Wang, M.; Liu, H. Auxin redistribution and shifts in PIN gene expression during Arabidopsis grafting. Russ. J. Plant Physiol. 2014, 61, 688–696. [Google Scholar] [CrossRef]
- Li, W.; Fang, C.; Krishnan, S.; Chen, J.; Yu, H.; Murphy, A.S.; Merewitz, E.; Katin-Grazzini, L.; McAvoy, R.J.; Deng, Z.; et al. Elevated auxin and reduced cytokinin contents in rootstocks improve their performance and grafting success. Plant Biotechnol. J. 2017, 15, 1556–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kramer, E.M. PIN and AUX/LAX proteins: Their role in auxin accumulation. Trends Plant Sci. 2004, 9, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Swarup, R.; Peret, B. AUX/LAX family of auxin influx carriers-an overview. Front. Plant Sci. 2012, 3, 225. [Google Scholar] [CrossRef] [PubMed]
- Peret, B.; Swarup, K.; Ferguson, A.; Seth, M.; Yang, Y.; Dhondt, S.; James, N.; Casimiro, I.; Perry, P.; Syed, A.; et al. AUX/LAX genes encode a family of auxin influx transporters that perform distinct functions during Arabidopsis development. Plant Cell 2012, 24, 2874–2885. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zhao, L.; Chen, J.; Yang, Y.; Xu, D.; Tao, S.; Zheng, S.; Shen, Y.; He, Y.; Shen, C.; et al. Identification and expression profiling of the Aux/IAA gene family in Chinese hickory (Carya cathayensis Sarg.) during the grafting process. Plant Physiol. Biochem. 2018, 127, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Lechner, E.; Yamada, M.; Hobbie, L.; Ehrismann, J.S.; Jurgens, G.; Estelle, M. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 2005, 9, 109–119. [Google Scholar] [CrossRef]
- Fukaki, H.; Okushima, Y.; Tasaka, M. Auxin-mediated lateral root formation in higher plants. Int. Rev. Cytol. 2007, 256, 111–137. [Google Scholar] [CrossRef]
- Zhao, Y. Auxin biosynthesis and its role in plant development. Ann. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef]
- Ivanchenko, M.G.; Coffeen, W.C.; Lomax, T.L.; Dubrovsky, J.G. Mutations in the Diageotropica (Dgt) gene uncouple patterned cell division during lateral root initiation from proliferative cell division in the pericycle. Plant J. 2006, 46, 436–447. [Google Scholar] [CrossRef]
- Ivanchenko, M.G.; Zhu, J.; Wang, B.; Medvecka, E.; Du, Y.; Azzarello, E.; Mancuso, S.; Megraw, M.; Filichkin, S.; Dubrovsky, J.G.; et al. The cyclophilin A DIAGEOTROPICA gene affects auxin transport in both root and shoot to control lateral root formation. Development 2015, 142, 712–721. [Google Scholar] [CrossRef] [Green Version]
- Xie, Q.; Frugis, G.; Colgan, D.; Chua, N.H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000, 14, 3024–3036. [Google Scholar] [CrossRef]
- Willemsen, V.; Bauch, M.; Bennett, T.; Campilho, A.; Wolkenfelt, H.; Xu, J.; Haseloff, J.; Scheres, B. The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev. Cell 2008, 15, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Avci, U.; Grant, E.H.; Haigler, C.H.; Beers, E.P. XND1, a member of the NAC domain family in Arabidopsis thaliana, negatively regulates lignocellulose synthesis and programmed cell death in xylem. Plant J. 2008, 53, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Celenza, J.L., Jr.; Grisafi, P.L.; Fink, G.R. A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 1995, 9, 2131–2142. [Google Scholar] [CrossRef] [PubMed]
- DiDonato, R.J.; Arbuckle, E.; Buker, S.; Sheets, J.; Tobar, J.; Totong, R.; Grisafi, P.; Fink, G.R.; Celenza, J.L. Arabidopsis ALF4 encodes a nuclear-localized protein required for lateral root formation. Plant J. 2004, 37, 340–353. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Jiao, Y.; Meyerowitz, E.M. Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 2010, 18, 463–471. [Google Scholar] [CrossRef]
- Notaguchi, M.; Wolf, S.; Lucas, W.J. Phloem-Mobile Aux/IAA Transcripts Target to the Root Tip and Modify Root Architecture F. J. Integr. Plant Biol. 2012, 54, 760–772. [Google Scholar] [CrossRef] [PubMed]
- Wybouw, B.; De Rybel, B. Cytokinin–A developing story. Trends Plant Sci. 2018, 24, 177–185. [Google Scholar] [CrossRef]
- Magyar-Tábori, K.; Dobránszki, J.; da Silva, J.A.T.; Bulley, S.M.; Hudák, I. The role of cytokinins in shoot organogenesis in apple. Plant Cell Tissue Organ Cult. 2010, 101, 251–267. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Nat. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef] [Green Version]
- Cortleven, A.; Leuendorf, J.E.; Frank, M.; Pezzetta, D.; Bolt, S.; Schmulling, T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 2019, 42, 998–1018. [Google Scholar] [CrossRef]
- Schaller, G.E.; Street, I.H.; Kieber, J.J. Cytokinin and the cell cycle. Curr. Opin. Plant Biol. 2014, 21, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.; Vankova, R.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Cytokinins: Metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 2012, 17, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Köse, C.; Güleryüz, M. Effects of auxins and cytokinins on graft union of grapevine (Vitis vinifera). N. Z. J. Crop Hortic. Sci. 2006, 34, 145–150. [Google Scholar] [CrossRef]
- Mitchell, J.; Van Staden, J. Cytokinins and the wounding response in potato tissue. Z. Pflanzenphysiol. 1983, 109, 1–5. [Google Scholar] [CrossRef]
- Mo, Z.; Feng, G.; Su, W.; Liu, Z.; Peng, F. Transcriptomic Analysis Provides Insights into Grafting Union Development in Pecan (Carya illinoinensis). Genes 2018, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Aloni, R. The role of cytokinin in organised differentiation of vascular tissues. Funct. Plant Biol. 1993, 20, 601–608. [Google Scholar] [CrossRef]
- Shehata, S.; El-Shraiy, A.M. Regulating cucumber grafting by interactions of cytokinins in xylem exudates of rootstock and basipetol polar auxin transport of scion at graft union. Aus. J. Basic Appl. Sci. 2010, 4, 6179–6184. [Google Scholar]
- Valdes, A.E.; Fernandez, B.; Centeno, M.L. Hormonal changes throughout maturation and ageing in Pinus pinea. Plant Physiol. Biochem. 2004, 42, 335–340. [Google Scholar] [CrossRef]
- Autio, A.M.; Day, M.E. Cytokinin Phytohormonal Effects on Crown Structure. Arboricult. Urban Forest. 2016, 42, 1–20. [Google Scholar]
- Meier, A.R.; Saunders, M.R.; Michler, C.H. Epicormic buds in trees: A review of bud establishment, development and dormancy release. Tree Physiol. 2012, 32, 565–584. [Google Scholar] [CrossRef]
- Dello Ioio, R.; Linhares, F.S.; Scacchi, E.; Casamitjana-Martinez, E.; Heidstra, R.; Costantino, P.; Sabatini, S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 2007, 17, 678–682. [Google Scholar] [CrossRef] [PubMed]
- Laplaze, L.; Benkova, E.; Casimiro, I.; Maes, L.; Vanneste, S.; Swarup, R.; Weijers, D.; Calvo, V.; Parizot, B.; Herrera-Rodriguez, M.B.; et al. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 2007, 19, 3889–3900. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto-Kitano, M.; Kusumoto, T.; Tarkowski, P.; Kinoshita-Tsujimura, K.; Vaclavikova, K.; Miyawaki, K.; Kakimoto, T. Cytokinins are central regulators of cambial activity. Proc. Nat. Acad. Sci. USA 2008, 105, 20027–20031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishopp, A.; Help, H.; El-Showk, S.; Weijers, D.; Scheres, B.; Friml, J.; Benkova, E.; Mahonen, A.P.; Helariutta, Y. A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr. Biol. 2011, 21, 917–926. [Google Scholar] [CrossRef] [PubMed]
- Bishopp, A.; Lehesranta, S.; Vaten, A.; Help, H.; El-Showk, S.; Scheres, B.; Helariutta, K.; Mahonen, A.P.; Sakakibara, H.; Helariutta, Y. Phloem-transported cytokinin regulates polar auxin transport and maintains vascular pattern in the root meristem. Curr. Biol. 2011, 21, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Melnyk, C.W. Plant grafting: Insights into tissue regeneration. Regeneration 2017, 4, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Nishitani, C.; Demura, T.; Fukuda, H. Analysis of early processes in wound-induced vascular regeneration using TED3 and ZeHB3 as molecular markers. Plant Cell Physiol. 2002, 43, 79–90. [Google Scholar] [CrossRef]
- Garner, R.J. The Grafter’s Handbook; Chelsea Green Publishing: White River Junction, VT, USA, 2013. [Google Scholar]
- Pina, A.; Errea, P.; Martens, H.J. Graft union formation and cell-to-cell communication via plasmodesmata in compatible and incompatible stem unions of Prunus spp. Sci. Hortic. 2012, 143, 144–150. [Google Scholar] [CrossRef]
- Mo, Z.; He, H.; Su, W.; Peng, F. Analysis of differentially accumulated proteins associated with graft union formation in pecan (Carya illinoensis). Sci. Hortic. 2017, 224, 126–134. [Google Scholar] [CrossRef]
- Melnyk, C.W. Connecting the plant vasculature to friend or foe. New Phytol. 2017, 213, 1611–1617. [Google Scholar] [CrossRef]
- Elo, A.; Immanen, J.; Nieminen, K.; Helariutta, Y. Stem cell function during plant vascular development. Sem. Cell Dev. Biol. 2009, 20, 1097–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahonen, A.P.; Bonke, M.; Kauppinen, L.; Riikonen, M.; Benfey, P.N.; Helariutta, Y. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 2000, 14, 2938–2943. [Google Scholar] [CrossRef] [PubMed]
- Mahonen, A.P.; Bishopp, A.; Higuchi, M.; Nieminen, K.M.; Kinoshita, K.; Tormakangas, K.; Ikeda, Y.; Oka, A.; Kakimoto, T.; Helariutta, Y. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science 2006, 311, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Nieminen, K.; Immanen, J.; Laxell, M.; Kauppinen, L.; Tarkowski, P.; Dolezal, K.; Tahtiharju, S.; Elo, A.; Decourteix, M.; Ljung, K.; et al. Cytokinin signaling regulates cambial development in poplar. Proc. Nat. Acad. Sci. USA 2008, 105, 20032–20037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Craig, J.C.; Petzold, H.E.; Dickerman, A.W.; Beers, E.P. The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol. 2005, 138, 803–818. [Google Scholar] [CrossRef]
- Hirose, N.; Takei, K.; Kuroha, T.; Kamada-Nobusada, T.; Hayashi, H.; Sakakibara, H. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 2008, 59, 75–83. [Google Scholar] [CrossRef]
- Aloni, R.; Aloni, E.; Langhans, M.; Ullrich, C.I. Role of cytokinin and auxin in shaping root architecture: Regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. 2006, 97, 883–893. [Google Scholar] [CrossRef]
- Miyawaki, K.; Tarkowski, P.; Matsumoto-Kitano, M.; Kato, T.; Sato, S.; Tarkowska, D.; Tabata, S.; Sandberg, G.; Kakimoto, T. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Nat. Acad. Sci. USA 2006, 103, 16598–16603. [Google Scholar] [CrossRef]
- Ye, Z.H. Vascular tissue differentiation and pattern formation in plants. Ann. Rev. Plant Biol. 2002, 53, 183–202. [Google Scholar] [CrossRef]
- Fukuda, H. Signals that control plant vascular cell differentiation. Nat. Rev. Mol. Cell Biol. 2004, 5, 379–391. [Google Scholar] [CrossRef]
- Sakai, H.; Honma, T.; Aoyama, T.; Sato, S.; Kato, T.; Tabata, S.; Oka, A. ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 2001, 294, 1519–1521. [Google Scholar] [CrossRef] [PubMed]
- Mason, M.G.; Mathews, D.E.; Argyros, D.A.; Maxwell, B.B.; Kieber, J.J.; Alonso, J.M.; Ecker, J.R.; Schaller, G.E. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 2005, 17, 3007–3018. [Google Scholar] [CrossRef] [PubMed]
- Pils, B.; Heyl, A. Unraveling the evolution of cytokinin signaling. Plant Physiol. 2009, 151, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Huh, S.U.; Kojima, M.; Sakakibara, H.; Paek, K.H.; Hwang, I. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell 2010, 19, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Proietti, S.; Caarls, L.; Coolen, S.; Van Pelt, J.A.; Van Wees, S.C.M.; Pieterse, C.M.J. Genome-wide association study reveals novel players in defense hormone crosstalk in Arabidopsis. Plant Cell Environ. 2018, 41, 2342–2356. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, J.; Elo, A.; Helariutta, Y. Hormone interactions during vascular development. Plant Mol. Biol. 2009, 69, 347. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Jiang, B.; Fang, J.; Shen, Y.; Fang, Z.; Rm, S.K.; Yi, K.; Shen, C.; Yan, D.; Zheng, B. Analysis of transcriptome in hickory (Carya cathayensis), and uncover the dynamics in the hormonal signaling pathway during graft process. BMC Genom. 2016, 17, 935. [Google Scholar] [CrossRef] [PubMed]
- Ohashi-Ito, K.; Bergmann, D.C. Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development 2007, 134, 2959–2968. [Google Scholar] [CrossRef]
- Kubo, M.; Udagawa, M.; Nishikubo, N.; Horiguchi, G.; Yamaguchi, M.; Ito, J.; Mimura, T.; Fukuda, H.; Demura, T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005, 19, 1855–1860. [Google Scholar] [CrossRef] [Green Version]
- Motose, H.; Sugiyama, M.; Fukuda, H. A proteoglycan mediates inductive interaction during plant vascular development. Nature 2004, 429, 873–878. [Google Scholar] [CrossRef]
- Kakimoto, T. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 1996, 274, 982–985. [Google Scholar] [CrossRef] [PubMed]
- Perilli, S.; Moubayidin, L.; Sabatini, S. The molecular basis of cytokinin function. Curr. Opin. Plant Biol. 2010, 13, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Hejatko, J.; Ryu, H.; Kim, G.T.; Dobesova, R.; Choi, S.; Choi, S.M.; Soucek, P.; Horak, J.; Pekarova, B.; Palme, K.; et al. The histidine kinases CYTOKININ-INDEPENDENT1 and ARABIDOPSIS HISTIDINE KINASE2 and 3 regulate vascular tissue development in Arabidopsis shoots. Plant Cell 2009, 21, 2008–2021. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Schmülling, T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009, 12, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef] [PubMed]
- Dayan, J.; Voronin, N.; Gong, F.; Sun, T.P.; Hedden, P.; Fromm, H.; Aloni, R. Leaf-induced gibberellin signaling is essential for internode elongation, cambial activity, and fiber differentiation in tobacco stems. Plant Cell 2012, 24, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhou, L.; Huang, M.; He, X.; Yang, Y.; Liu, X.; Li, Y.; Hou, X. Gibberellins play an essential role in late embryogenesis of Arabidopsis. Nat. Plants 2018, 4, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Martínez, C.; Espinosa-Ruiz, A.; Prat, S. Gibberellins and plant vegetative growth. Ann. Plant Rev. Online 2018, 49, 285–322. [Google Scholar]
- Li, H.; Wu, H.; Qi, Q.; Li, H.; Li, Z.; Chen, S.; Ding, Q.; Wang, Q.; Yan, Z.; Gai, Y.; et al. Gibberellins Play a Role in Regulating Tomato Fruit Ripening. Plant Cell Physiol. 2019, 60, 1619–1629. [Google Scholar] [CrossRef]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef] [Green Version]
- Ragni, L.; Nieminen, K.; Pacheco-Villalobos, D.; Sibout, R.; Schwechheimer, C.; Hardtke, C.S. Mobile gibberellin directly stimulates Arabidopsis hypocotyl xylem expansion. Plant Cell 2011, 23, 1322–1336. [Google Scholar] [CrossRef] [PubMed]
- Anushma, P.; Swamy, G.; Gangadhara, K. Effect of colored shade nets on softwood grafting success in jamun (Syzigium cuminiiskeels). Plant Arch. 2014, 14, 293–295. [Google Scholar]
- Eriksson, S.; Bohlenius, H.; Moritz, T.; Nilsson, O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 2006, 18, 2172–2181. [Google Scholar] [CrossRef] [PubMed]
- Regnault, T.; Daviere, J.M.; Wild, M.; Sakvarelidze-Achard, L.; Heintz, D.; Carrera Bergua, E.; Lopez Diaz, I.; Gong, F.; Hedden, P.; Achard, P. The gibberellin precursor GA12 acts as a long-distance growth signal in Arabidopsis. Nat. Plants 2015, 1, 15073. [Google Scholar] [CrossRef] [PubMed]
- Asahina, M.; Gocho, Y.; Kamada, H.; Satoh, S. Involvement of inorganic elements in tissue reunion in the hypocotyl cortex of Cucumis sativus. J. Plant Res. 2006, 119, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Asahina, M.; Iwai, H.; Kikuchi, A.; Yamaguchi, S.; Kamiya, Y.; Kamada, H.; Satoh, S. Gibberellin produced in the cotyledon is required for cell division during tissue reunion in the cortex of cut cucumber and tomato hypocotyls. Plant Physiol. 2002, 129, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Asahina, M.; Yamauchi, Y.; Hanada, A.; Kamiya, Y.; Kamada, H.; Satoh, S.; Yamaguchi, S. Effects of the removal of cotyledons on endogenous gibberellin levels in hypocotyls of young cucumber and tomato seedlings. Plant Biotechnol. 2007, 24, 99–106. [Google Scholar] [CrossRef] [Green Version]
- Mauriat, M.; Moritz, T. Analyses of GA20ox- and GID1-over-expressing aspen suggest that gibberellins play two distinct roles in wood formation. Plant J. 2009, 58, 989–1003. [Google Scholar] [CrossRef]
- Dayan, J.; Schwarzkopf, M.; Avni, A.; Aloni, R. Enhancing plant growth and fiber production by silencing GA 2-oxidase. Plant Biotechnol. J. 2010, 8, 425–435. [Google Scholar] [CrossRef]
- Eriksson, M.E.; Israelsson, M.; Olsson, O.; Moritz, T. Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 2000, 18, 784–788. [Google Scholar] [CrossRef]
- Biemelt, S.; Tschiersch, H.; Sonnewald, U. Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis, and photosynthesis in transgenic tobacco plants. Plant Physiol. 2004, 135, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Proebsting, W.M.; Hedden, P.; Lewis, M.J.; Croker, S.J.; Proebsting, L.N. Gibberellin concentration and transport in genetic lines of pea: Effects of grafting. Plant Physiol. 1992, 100, 1354–1360. [Google Scholar] [CrossRef] [PubMed]
- Bjorklund, S.; Antti, H.; Uddestrand, I.; Moritz, T.; Sundberg, B. Cross-talk between gibberellin and auxin in development of Populus wood: Gibberellin stimulates polar auxin transport and has a common transcriptome with auxin. Plant J. 2007, 52, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Chawla, R.; DeMason, D.A. Molecular expression of PsPIN1, a putative auxin efflux carrier gene from pea (Pisum sativum L.). Plant Growth Regul. 2004, 44, 1–14. [Google Scholar] [CrossRef]
- Asahina, M.; Satoh, S. Molecular and physiological mechanisms regulating tissue reunion in incised plant tissues. J. Plant Res. 2015, 128, 381–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooijdonk, V.; Woolley, D.; Warrington, I.; Tustin, D. Initial alteration of scion architecture by dwarfing apple rootstocks may involve shoot-root-shoot signalling by auxin, gibberellin, and cytokinin. J. Hortic. Sci. Biotechnol. 2010, 85, 59–65. [Google Scholar] [CrossRef]
- Liu, N.; Yang, J.; Fu, X.; Zhang, L.; Tang, K.; Guy, K.M.; Hu, Z.; Guo, S.; Xu, Y.; Zhang, M. Genome-wide identification and comparative analysis of grafting-responsive mRNA in watermelon grafted onto bottle gourd and squash rootstocks by high-throughput sequencing. Mol. Genet. Genom. 2016, 291, 621–633. [Google Scholar] [CrossRef]
- Ni, J.; Gao, C.; Chen, M.S.; Pan, B.Z.; Ye, K.; Xu, Z.F. Gibberellin Promotes Shoot Branching in the Perennial Woody Plant Jatropha curcas. Plant Cell Physiol. 2015, 56, 1655–1666. [Google Scholar] [CrossRef]
- Iwase, A.; Mitsuda, N.; Koyama, T.; Hiratsu, K.; Kojima, M.; Arai, T.; Inoue, Y.; Seki, M.; Sakakibara, H.; Sugimoto, K.; et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. 2011, 21, 508–514. [Google Scholar] [CrossRef]
- Asahina, M.; Azuma, K.; Pitaksaringkarn, W.; Yamazaki, T.; Mitsuda, N.; Ohme-Takagi, M.; Yamaguchi, S.; Kamiya, Y.; Okada, K.; Nishimura, T.; et al. Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proc. Nat. Acad. Sci. USA 2011, 108, 16128–16132. [Google Scholar] [CrossRef]
- Chandler, J.W.; Werr, W. Cytokinin-auxin crosstalk in cell type specification. Trends Plant Sci. 2015, 20, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Elfving, D.C.; Visser, D.B. Timing cyclanilide and cytokinin applications in the nursery to obtain desired lateral branch height in apple and sweet cherry trees. HortScience 2006, 41, 1238–1242. [Google Scholar] [CrossRef]
- Pitaksaringkarn, W.; Ishiguro, S.; Asahina, M.; Satoh, S. ARF6 and ARF8 contribute to tissue reunion in incised Arabidopsis inflorescence stems. Plant Biotechnol. 2014, 31, 49–53. [Google Scholar] [CrossRef]
- Else, M.A.; Taylor, J.M.; Young, S.; Atkinson, C.J. The effect of the graft union on hormonal and ionic signalling between rootstocks and scions of grafted apple (Malus pumila L. Mill.). Environ. Exp. Bot. 2018, 156, 325–336. [Google Scholar] [CrossRef]
- Benschop, J.J.; Jackson, M.B.; Gühl, K.; Vreeburg, R.A.; Croker, S.J.; Peeters, A.J.; Voesenek, L.A. Contrasting interactions between ethylene and abscisic acid in Rumex species differing in submergence tolerance. Plant J. 2005, 44, 756–768. [Google Scholar] [CrossRef] [PubMed]
- Cookson, S.J.; Clemente Moreno, M.J.; Hevin, C.; Nyamba Mendome, L.Z.; Delrot, S.; Trossat-Magnin, C.; Ollat, N. Graft union formation in grapevine induces transcriptional changes related to cell wall modification, wounding, hormone signalling, and secondary metabolism. J. Exp. Bot. 2013, 64, 2997–3008. [Google Scholar] [CrossRef] [PubMed]
- Rufato, L.; Marchioretto, L.D.R.; Orlandi, J.C.; Michelon, M.F.; De Rossi, A.; Sander, G.F.; de Macedo, T.A. Lateral branch induction at nursery with growth regulators in ‘maxi gala’ apple trees grafted on four rootstocks. Sci. Hortic. 2019, 253, 349–357. [Google Scholar] [CrossRef]
- Sorce, C.; Massai, R.; Picciarelli, P.; Lorenzi, R. Hormonal relationships in xylem sap of grafted and ungrafted Prunus rootstocks. Sci. Hortic. 2002, 93, 333–342. [Google Scholar] [CrossRef]
- Aloni, B.; Karni, L.; Deventurero, G.; Levin, Z.; Cohen, R.; Katzir, N.; Lotan-Pompan, M.; Edelstein, M.; Aktas, H.; Turhan, E. Physiological and biochemical changes at the rootstock-scion interface in graft combinations between Cucurbita rootstocks and a melon scion. J. Hortic. Sci. Biotechnol. 2008, 83, 777–783. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, A.; Zheng, B. Molecular Responses during Plant Grafting and Its Regulation by Auxins, Cytokinins, and Gibberellins. Biomolecules 2019, 9, 397. https://doi.org/10.3390/biom9090397
Sharma A, Zheng B. Molecular Responses during Plant Grafting and Its Regulation by Auxins, Cytokinins, and Gibberellins. Biomolecules. 2019; 9(9):397. https://doi.org/10.3390/biom9090397
Chicago/Turabian StyleSharma, Anket, and Bingsong Zheng. 2019. "Molecular Responses during Plant Grafting and Its Regulation by Auxins, Cytokinins, and Gibberellins" Biomolecules 9, no. 9: 397. https://doi.org/10.3390/biom9090397





