Molecular Characterization of a Novel Cold-Active Hormone-Sensitive Lipase (HaHSL) from Halocynthiibacter Arcticus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Cloning and Purification
2.3. Biochemical Assays
2.4. Immobilization Methods
2.5. Bioinformatic Analysis
3. Results and Discussion
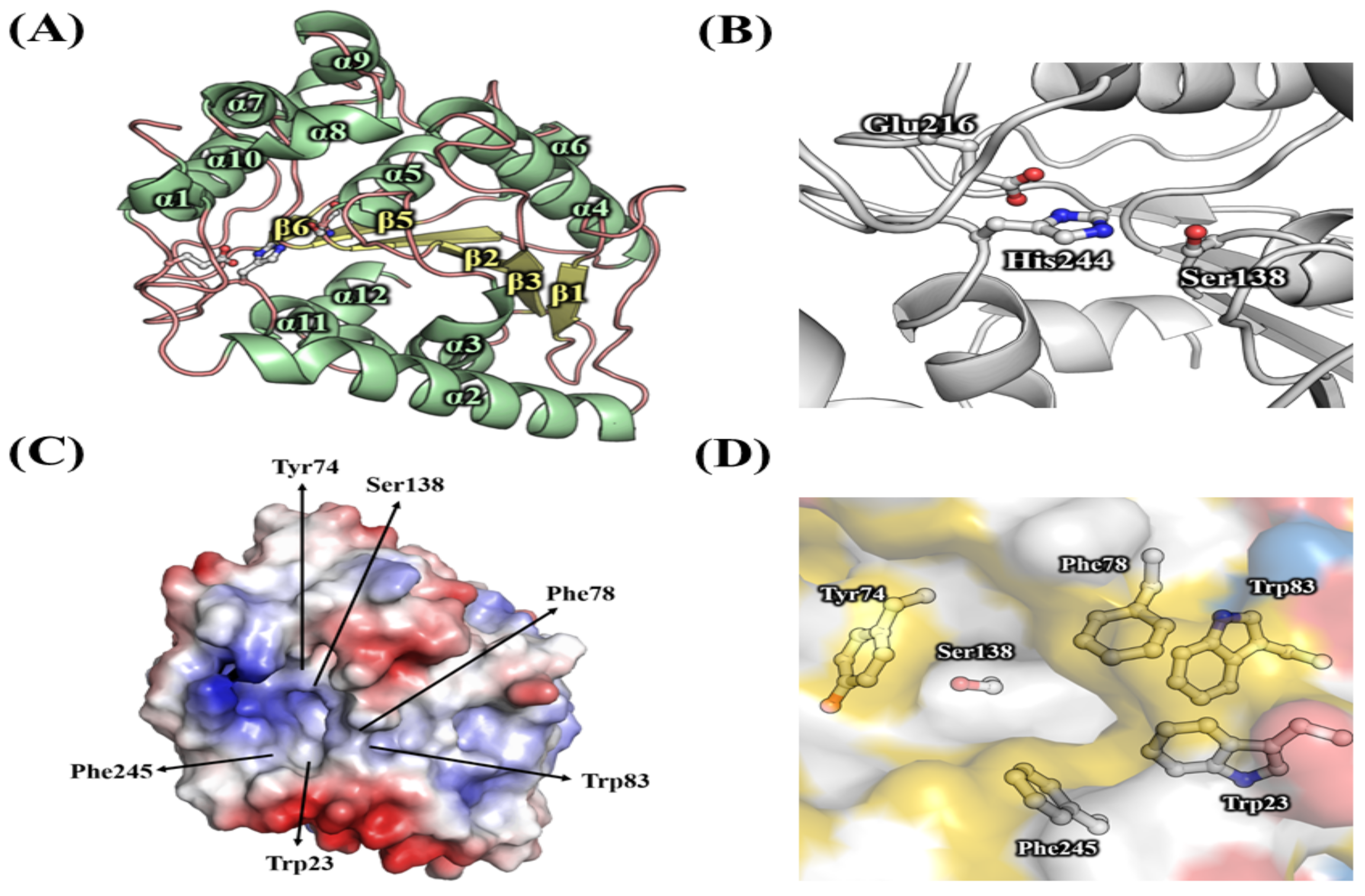
3.1. Bioinformatic Analysis of HaHSL
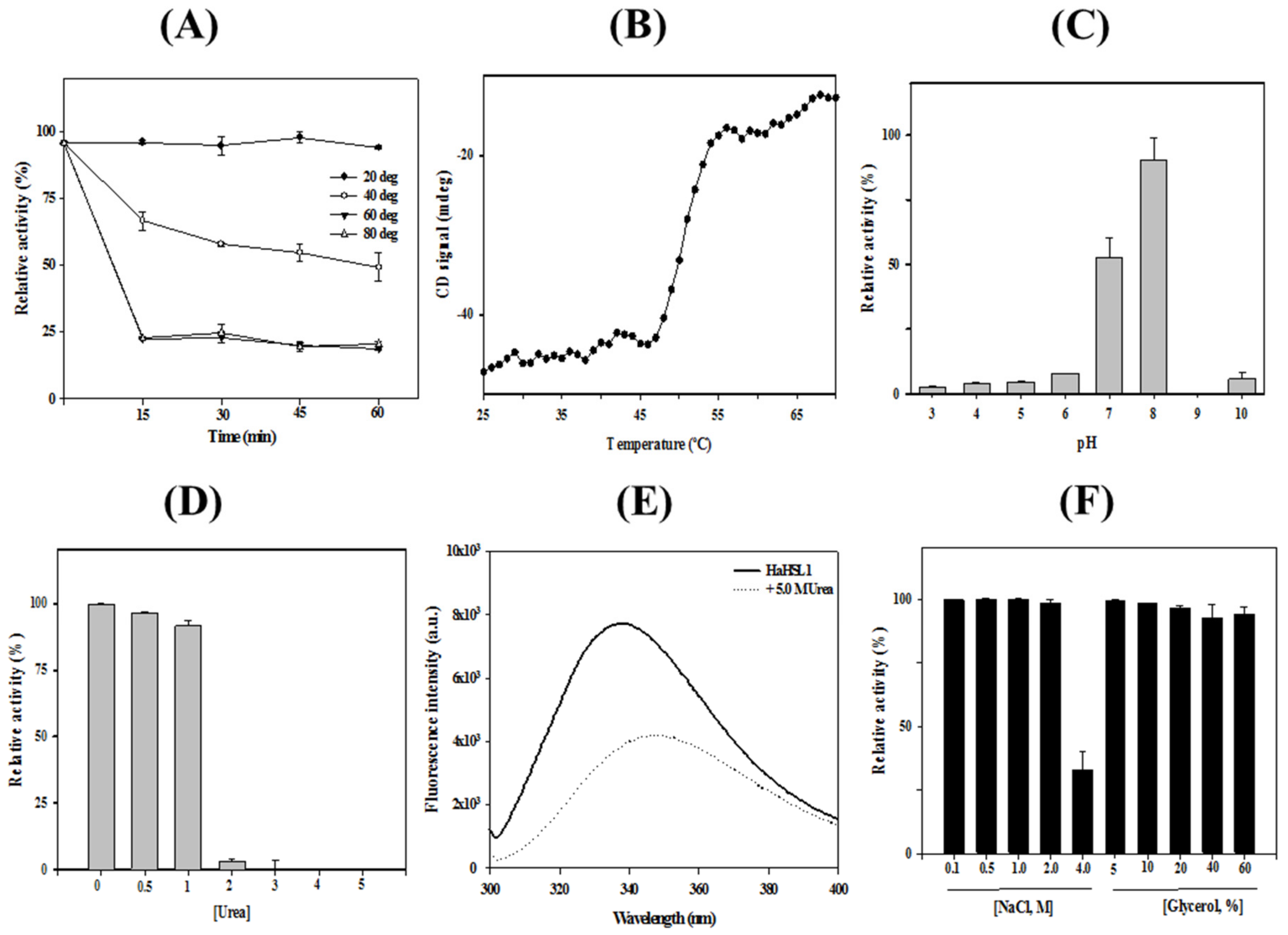
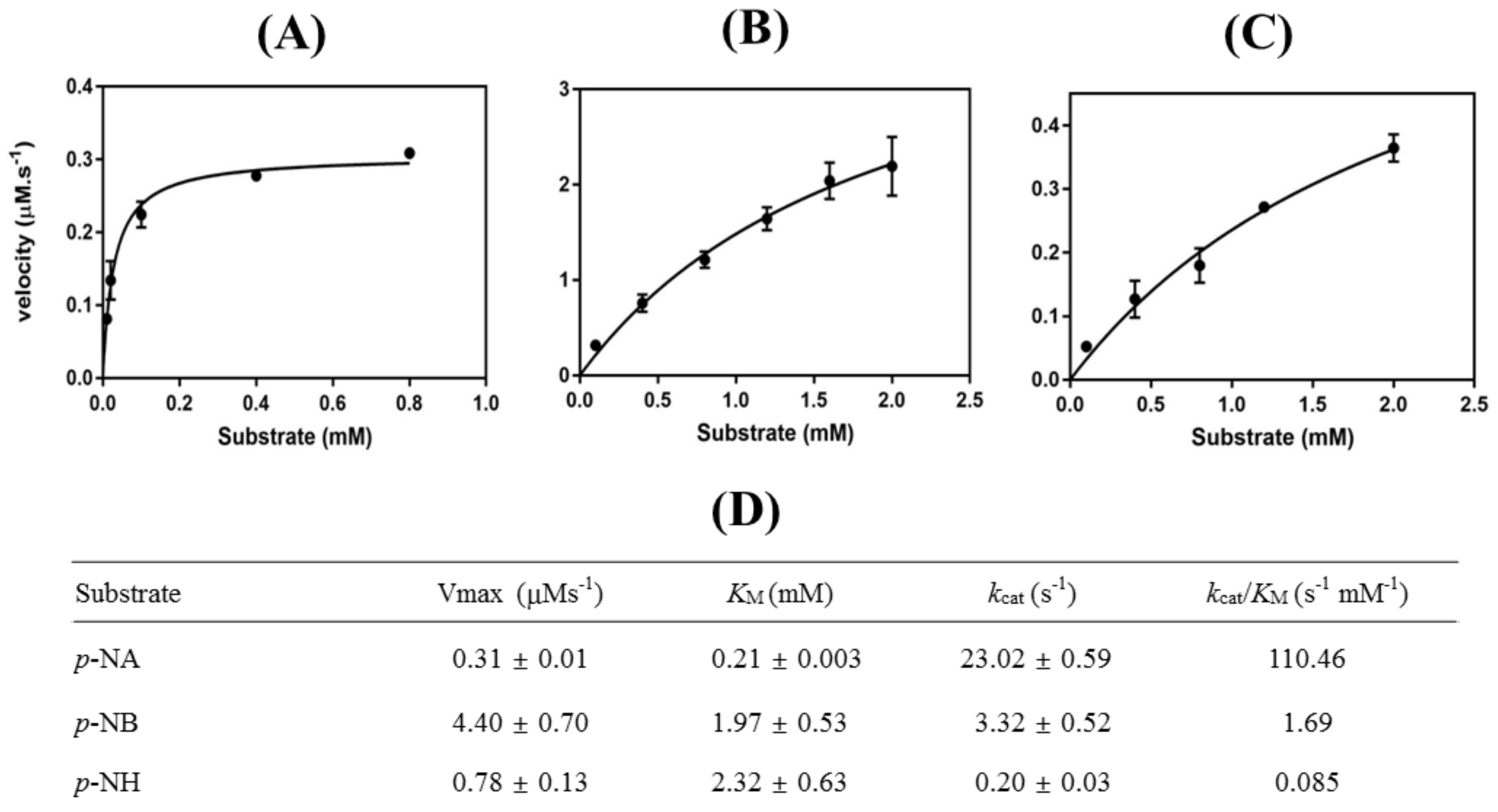
3.2. Characterizations of HaHSL
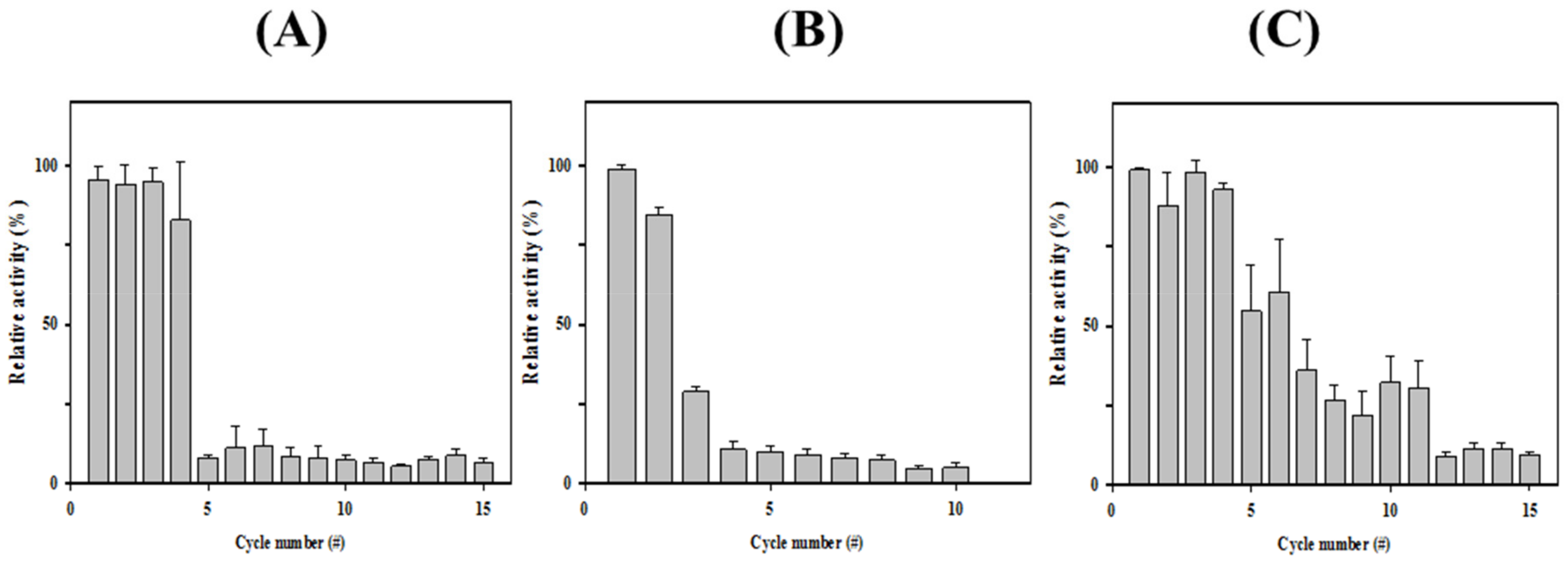
3.3. Immobilization of HaHSL
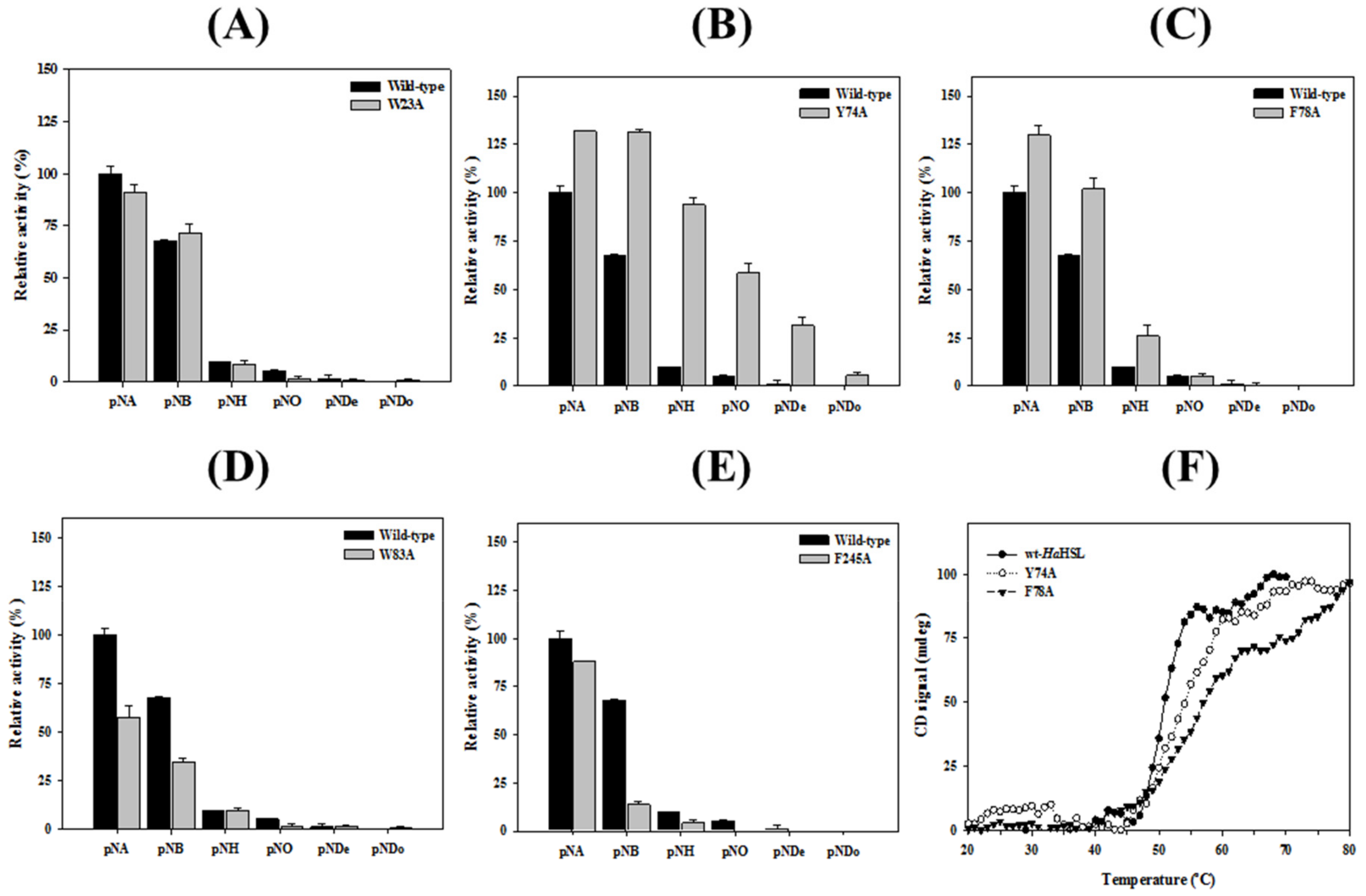
3.4. Effects of Mutations in the Substrate Binding Region
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lass, A.; Zimmermann, R.; Oberer, M.; Zechner, R. Lipolysis-a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog. Lipid. Res. 2011, 50, 14–27. [Google Scholar] [CrossRef]
- Casas-Godoy, L.; Duquesne, S.; Bordes, F.; Sandoval, G.; Marty, A. Lipases: An overview. Methods Mol. Biol. 2012, 861, 3–30. [Google Scholar]
- Sarmah, N.; Revathi, D.; Sheelu, G.; Yamuna Rani, K.; Sridhar, S.; Mehtab, V.; Sumana, C. Recent advances on sources and industrial applications of lipases. Biotechnol. Prog. 2018, 34, 5–28. [Google Scholar] [CrossRef]
- Borrelli, G.M.; Trono, D. Recombinant Lipases and Phospholipases and Their Use as Biocatalysts for Industrial Applications. Int. J. Mol. Sci. 2015, 16, 20774–20840. [Google Scholar] [CrossRef] [Green Version]
- Arner, P.; Langin, D. The role of neutral lipases in human adipose tissue lipolysis. Curr. Opin. Lipidol. 2007, 18, 246–250. [Google Scholar] [CrossRef]
- Lampidonis, A.D.; Rogdakis, E.; Voutsinas, G.E.; Stravopodis, D.J. The resurgence of Hormone-Sensitive Lipase (HSL) in mammalian lipolysis. Gene 2011, 477, 1–11. [Google Scholar] [CrossRef]
- Krintel, C.; Klint, C.; Lindvall, H.; Mörgelin, M.; Holm, C. Quarternary structure and enzymological properties of the different hormone-sensitive lipase (HSL) isoforms. PLoS ONE. 2010, 5, e11193. [Google Scholar] [CrossRef]
- Kim, T.D. Bacterial Hormone-Sensitive Lipases (bHSLs): Emerging enzymes for biotechnological applications. J. Microbiol. Biotechnol. 2017, 27, 1907–1915. [Google Scholar] [CrossRef]
- Osterlund, T.; Contreras, J.A.; Holm, C. Identification of essential aspartic acid and histidine residues of hormone-sensitive lipase: Apparent residues of the catalytic triad. FEBS Lett. 1997, 403, 259–262. [Google Scholar] [CrossRef]
- Ngo, T.D.; Ryu, B.H.; Ju, H.; Jang, E.; Park, K.; Kim, K.K.; Kim, T.D. Structural and functional analyses of a bacterial homologue of hormone-sensitive lipase from a metagenomic library. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1726–1737. [Google Scholar] [CrossRef]
- Santiago, M.; Ramírez-Sarmiento, C.A.; Zamora, R.A.; Parra, L.P. Discovery, molecular mechanisms, and industrial applications of cold-active enzymes. Front. Microbiol. 2016, 7, 1408. [Google Scholar] [CrossRef]
- Littlechild, J.A. Enzymes from extreme environments and their industrial applications. Front. Bioeng. Biotechnol. 2015, 3, 161. [Google Scholar] [CrossRef]
- Raddadi, N.; Cherif, A.; Daffonchio, D.; Neifar, M.; Fava, F. Biotechnological applications of extremophiles, extremozymes and extremolytes. Appl. Microbiol. Biotechnol. 2015, 99, 7907–7913. [Google Scholar] [CrossRef] [Green Version]
- Baek, K.; Lee, Y.M.; Shin, S.C.; Hwang, K.; Hwang, C.Y.; Hong, S.G.; Lee, H.K. Halocynthiibacter arcticus sp. nov., isolated from Arctic marine sediment. Int. J. Syst. Evol. Microbiol. 2015, 65, 3861–3865. [Google Scholar] [CrossRef]
- Lee, Y.M.; Baek, K.; Lee, J.; Lee, H.K.; Park, H.; Shin, S.C. Complete genome sequence of Halocynthiibacter arcticus PAMC 20958(T) from an Arctic marine sediment sample. J. Biotechnol. 2016, 224, 12–13. [Google Scholar] [CrossRef]
- Oh, C.; Ryu, B.H.; Yoo, W.; Nguyen, D.D.; Kim, T.; Ha, S.; Kim, T.D.; Kim, K.K. Identification and crystallographic analysis of a new carbohydrate acetylesterase (SmAcE1) from Sinorhizobium meliloti. Crystals 2018, 8, 12. [Google Scholar] [CrossRef]
- Lee, C.W.; Kwon, S.; Park, S.H.; Kim, B.Y.; Yoo, W.; Ryu, B.H.; Kim, H.W.; Shin, S.C.; Kim, S.; Park, H.; et al. Crystal structure and functional characterization of an Esterase (EaEST) from Exiguobacterium antarcticum. PLoS ONE 2017, 12, e0169540. [Google Scholar] [CrossRef]
- Ryu, B.H.; Oh, C.; Yoo, W.; Kim, K.K.; Kim, T.D. Identification and crystallization of penicillin-binding protein/β-lactamase homolog (Rp46) from Ruegeria pomeroyi. Crystals 2017, 7, 6. [Google Scholar] [CrossRef]
- Wang, Y.; Ryu, B.H.; Yoo, W.; Lee, C.W.; Kim, K.K.; Lee, J.H.; Kim, T.D. Identification, characterization, immobilization, and mutational analysis of a novel acetylesterase with industrial potential (LaAcE) from Lactobacillus acidophilus. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 197–210. [Google Scholar] [CrossRef]
- Kim, B.Y.; Yoo, W.; Huong Luu Le, L.T.; Kim, K.K.; Kim, H.W.; Lee, J.H.; Kim, Y.O.; Kim, T.D. Characterization and mutation anaylsis of a cold-active bacterial hormone-sensitive lipase from Salinisphaera sp. P7-4. Arch. Biochem. Biophys. 2019, 663, 132–142. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Gouet, P.; Robert, X.; Courcelle, E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003, 31, 3320–3323. [Google Scholar] [CrossRef] [PubMed]
- Maiangwa, J.; Ali, M.S.; Salleh, A.B.; Rahman, R.N.; Shariff, F.M.; Leow, T.C. Adaptational properties and applications of cold-active lipases from psychrophilic bacteria. Extremophiles 2015, 19, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Perfumo, A.; Banat, I.M.; Marchant, R. Going Green and Cold: Biosurfactants from low-temperature environments to biotechnology applications. Trends Biotechnol. 2018, 36, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Li, P.Y.; Ji, P.; Li, C.Y.; Zhang, Y.; Wang, G.L.; Zhang, X.Y.; Xie, B.B.; Qin, Q.L.; Chen, X.L.; Zhou, B.C.; et al. Structural basis for dimerization and catalysis of a novel esterase from the GTSAG motif subfamily of the bacterial hormone-sensitive lipase family. J. Biol. Chem. 2014, 289, 19031–19041. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Huo, Y.Y.; Ji, R.; Kuang, S.; Ji, C.; Xu, X.W.; Li, J. Structural insights of a hormone sensitive lipase homologue Est22. Sci. Rep. 2016, 6, 28550. [Google Scholar] [CrossRef] [PubMed]
- Lineweaver, H.; Burk, D. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 1934, 56, 658–666. [Google Scholar] [CrossRef]
- Sheldon, R.A. Characteristic features and biotechnological applications of cross-linked enzyme aggregates (CLEAs). Appl. Microbiol. Biotechnol. 2011, 92, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, J.; Majumder, A.B.; Gupta, M.N. Adding an appropriate amino acid during crosslinking results in more stable crosslinked enzyme aggregates. Anal. Biochem. 2016, 507, 27–32. [Google Scholar] [CrossRef]
- Ge, J.; Lei, J.; Zare, R.N. Protein-inorganic hybrid nanoflowers. Nat. Nanotechnol. 2012, 7, 428–432. [Google Scholar] [CrossRef]
- Dal Magro, L.; Silveira, V.C.C.; de Menezes, E.W.; Benvenutti, E.V.; Nicolodi, S.; Hertz, P.F.; Klein, M.P.; Rodrigues, R.C. Magnetic biocatalysts of pectinase and cellulase: Synthesis and characterization of two preparations for application in grape juice clarification. Int. J. Biol. Macromol. 2018, 115, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Meng, X.; Xu, X.; Liu, W.; Li, S. The molecular basis for lipase stereoselectivity. Appl. Microbiol. Biotechnol. 2018, 102, 3487–3495. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.S. Some like it hot, some like it cold: Temperature dependent biotechnological applications and improvements in extremophilic enzymes. Biotechnol. Adv. 2015, 33, 1912–1922. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.Y.; Yeom, S.J.; Jang, S.C.; Lee, C.S.; Roh, C.; Jeong, H.H. Simple analysis of lipid inhibition activity on an adipocyte micro-cell pattern chip. Biomolecules 2018, 8, 37. [Google Scholar] [CrossRef]


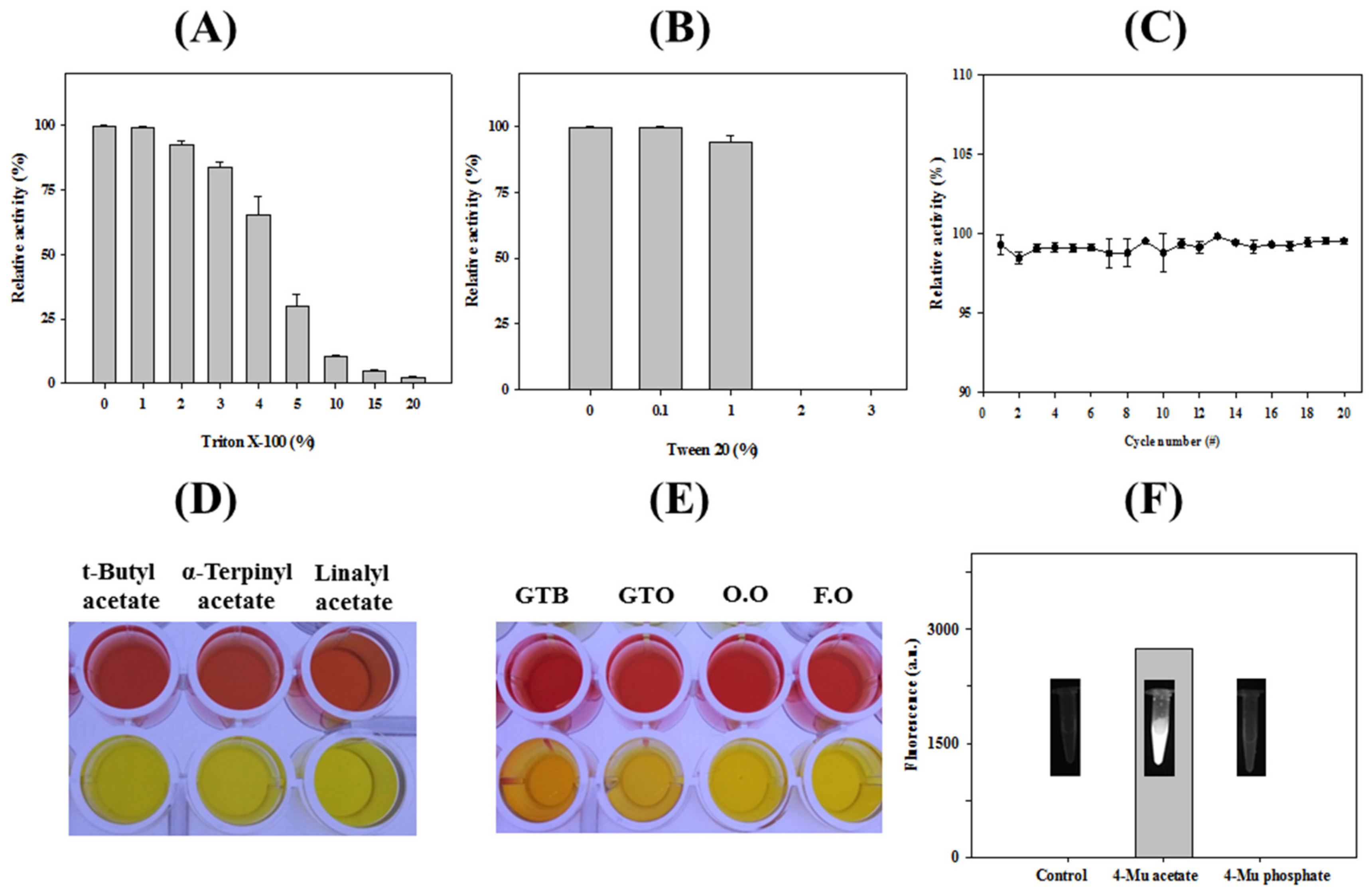


| Substrates | Concentration | Acetic Acid (g/L) | |||
|---|---|---|---|---|---|
| Tertiary alcohol esters | |||||
| tert-Butyl acetate | 0.25 M | 0.75 ± 0.0 | |||
| α-Terpinyl acetate | 0.25 M | 0.58 ± 0.0 | |||
| Linalyl acetate | 0.25 M | 0.54 ± 0.0 | |||
| Lipids | |||||
| Glyceryl tributyrate | 1.0% | 0.51 ± 0.0 | |||
| Glyceryl trioleate | 1.0% | 0.64 ± 0.0 | |||
| Olive oil | 1.0% | 0.68 ± 0.0 | |||
| Fish oil | 1.0% | 0.70 ± 0.0 | |||
| Acetylated carbohydrates | |||||
| α-D-Mannose pentaacetate | 10 mM | 0.09 ± 0.0 | |||
| β-D-Galactose pentaacetate | 10 mM | 0.91 ± 0.2 | |||
| Propionic acids | |||||
| (Rac)-Naproxol acetate | 0.30 M | 0.19 ± 0.0 | |||
| (S)-Naproxol acetate | 0.15 M | 0.52 ± 0.0 | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, L.T.H.L.; Yoo, W.; Lee, C.; Wang, Y.; Jeon, S.; Kim, K.K.; Lee, J.H.; Kim, T.D. Molecular Characterization of a Novel Cold-Active Hormone-Sensitive Lipase (HaHSL) from Halocynthiibacter Arcticus. Biomolecules 2019, 9, 704. https://doi.org/10.3390/biom9110704
Le LTHL, Yoo W, Lee C, Wang Y, Jeon S, Kim KK, Lee JH, Kim TD. Molecular Characterization of a Novel Cold-Active Hormone-Sensitive Lipase (HaHSL) from Halocynthiibacter Arcticus. Biomolecules. 2019; 9(11):704. https://doi.org/10.3390/biom9110704
Chicago/Turabian StyleLe, Ly Thi Huong Luu, Wanki Yoo, Changwoo Lee, Ying Wang, Sangeun Jeon, Kyeong Kyu Kim, Jun Hyuck Lee, and T. Doohun Kim. 2019. "Molecular Characterization of a Novel Cold-Active Hormone-Sensitive Lipase (HaHSL) from Halocynthiibacter Arcticus" Biomolecules 9, no. 11: 704. https://doi.org/10.3390/biom9110704





