Genome-Wide Investigation of the NAC Gene Family and Its Potential Association with the Secondary Cell Wall in Moso Bamboo
Abstract
:1. Introduction
2. Results
2.1. Identification of NACs in Moso Bamboo
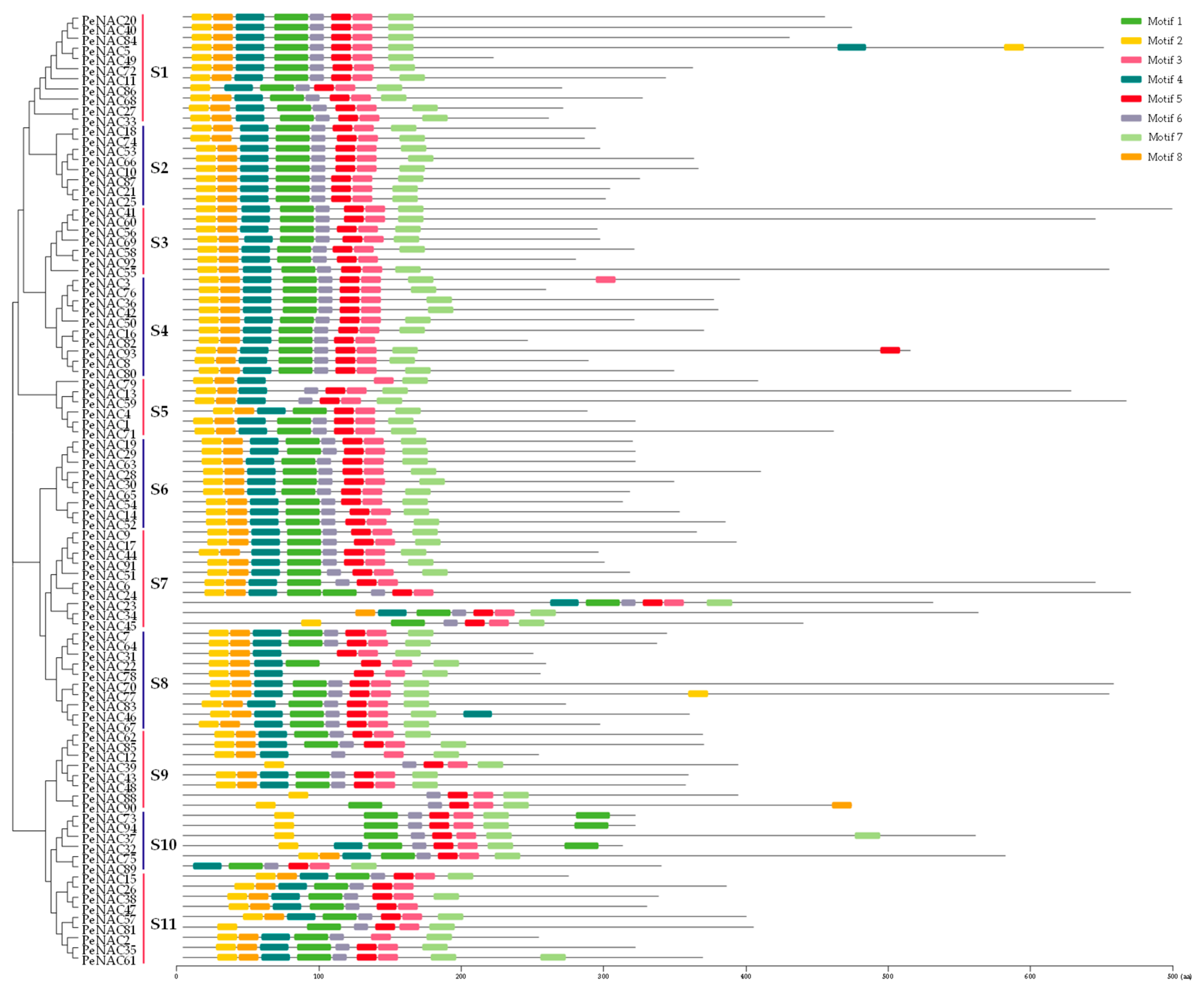
2.2. Gene Structure and Conserved Motif Analyses of PeNACs
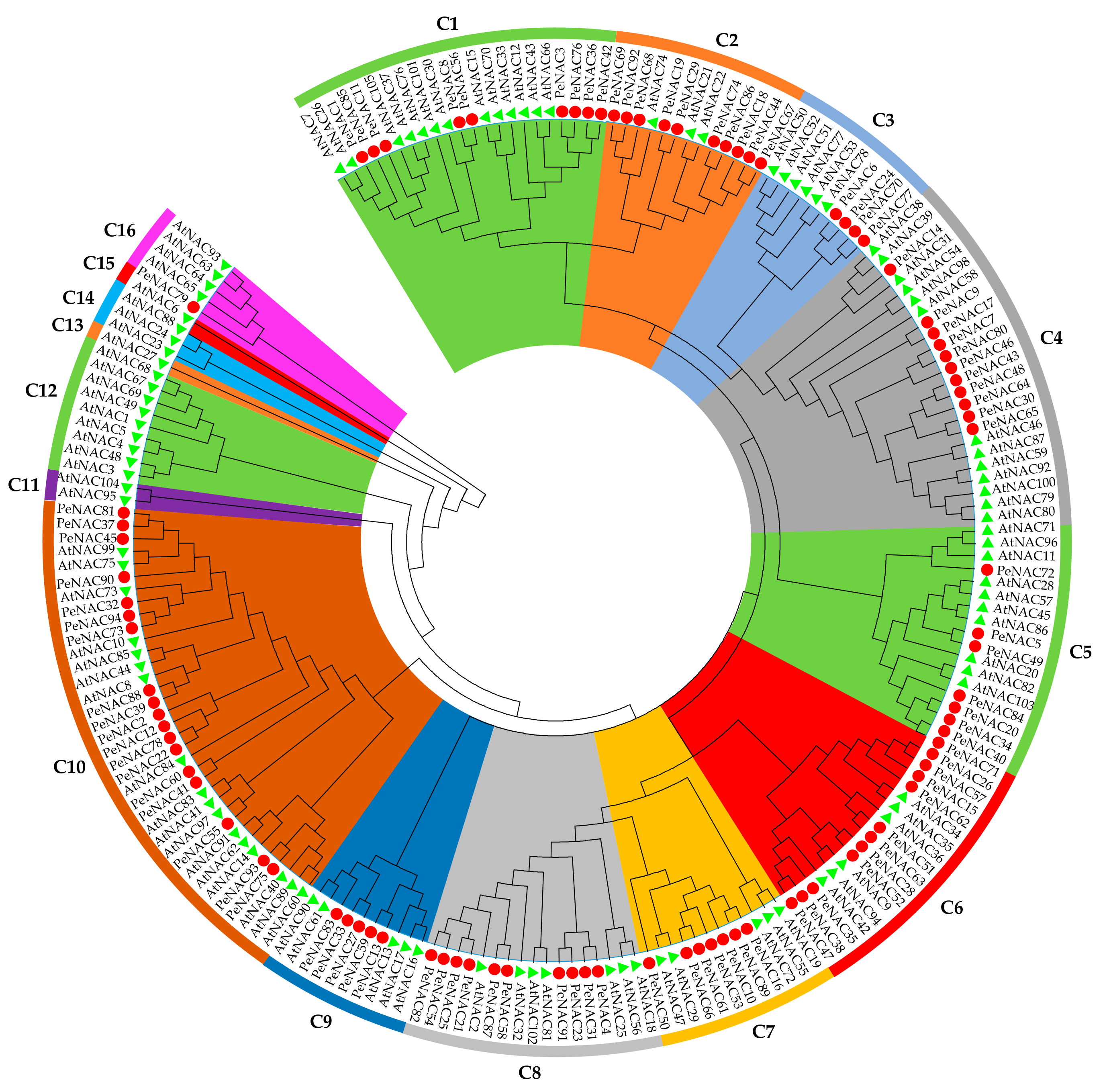
2.3. Phylogenetic Analysis and Classification of PeNACs
2.4. Putative Functions of PeNACs in Moso Bamboo
2.5. Expression Patterns of PeNACs Based on Transcriptome Data of Moso Bamboo
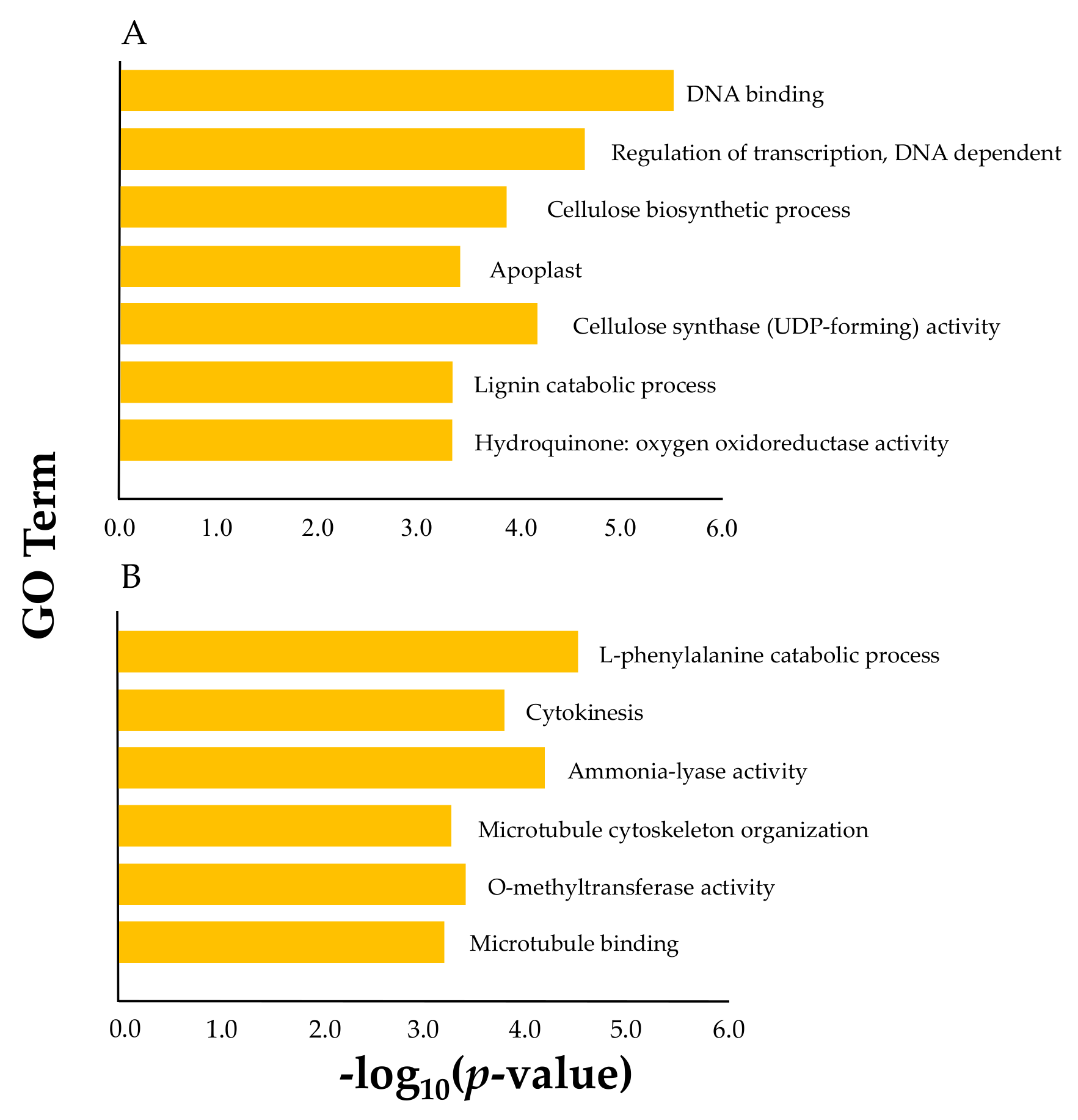
2.6. Expression Correlation and Co-Expression Networks
2.7. Prediction of miRNA Target Sites in PeNACs
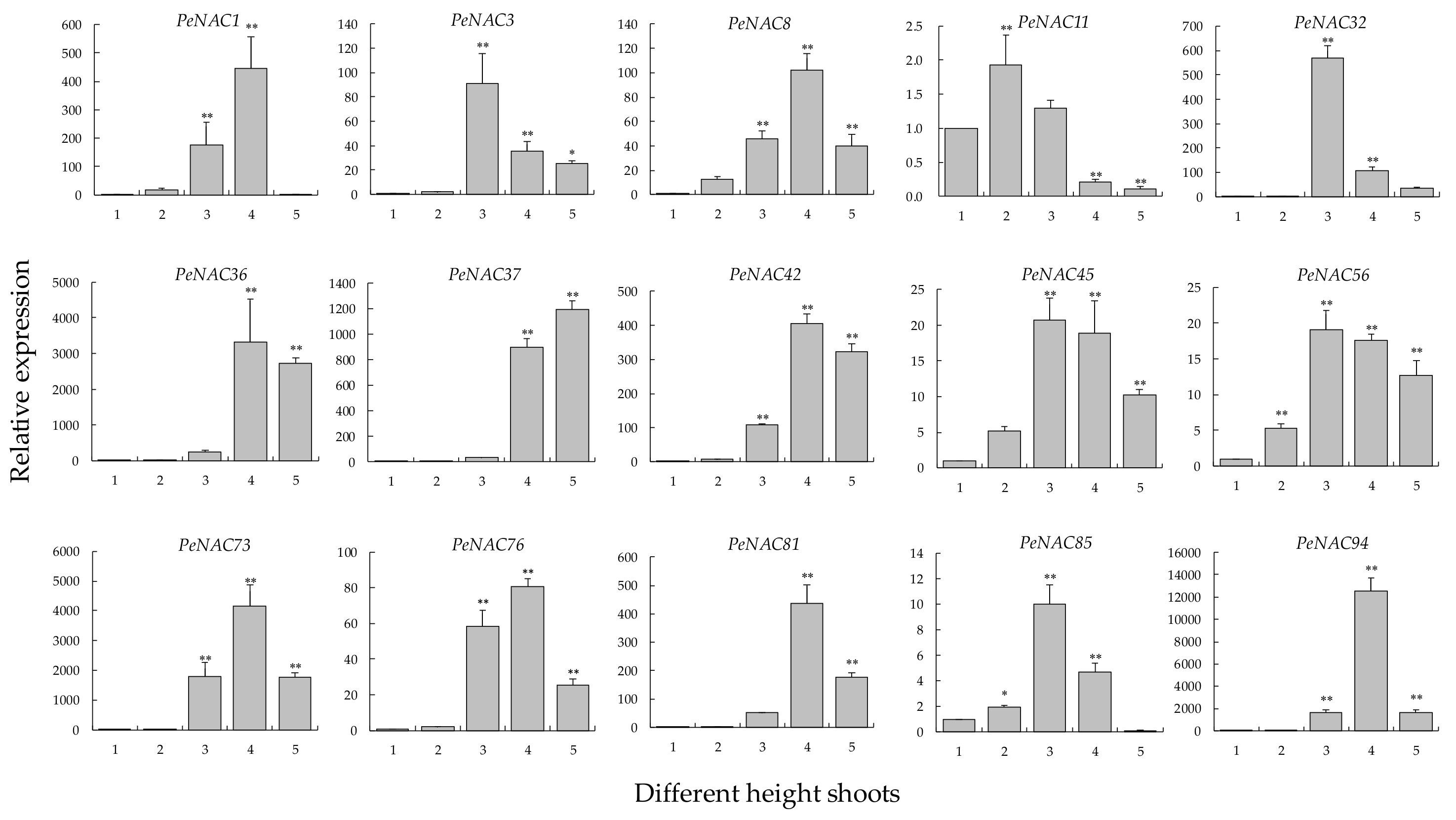
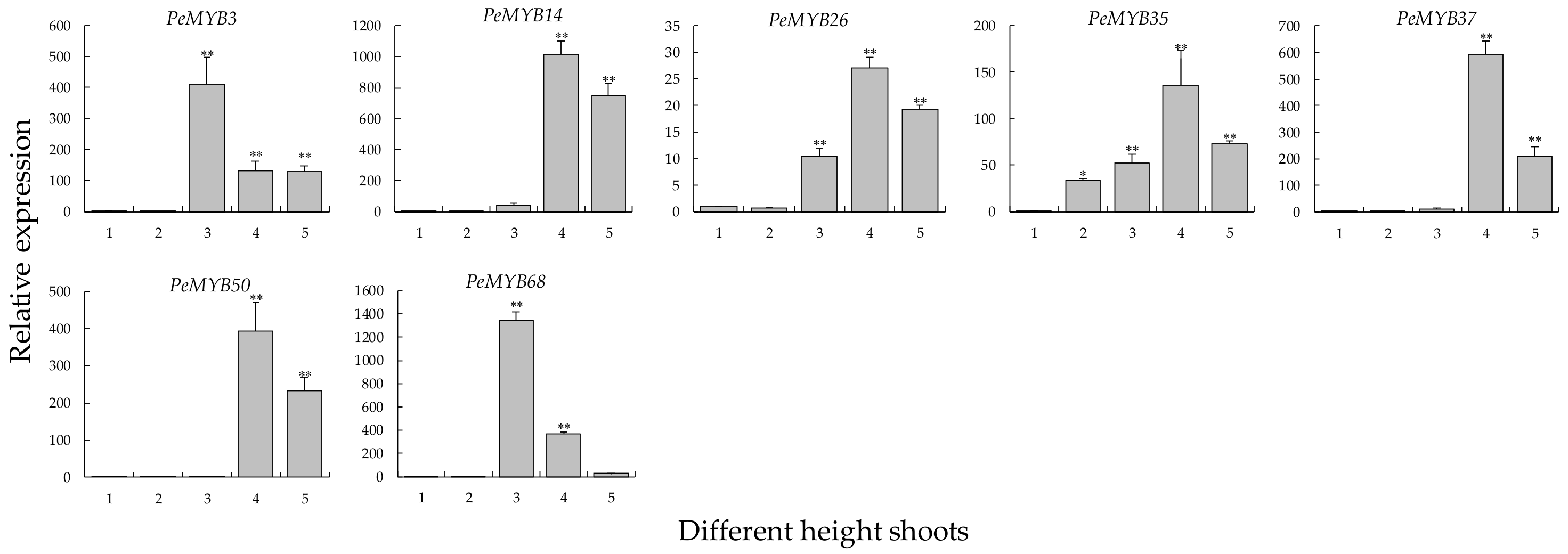
2.8. Expression Changes in Different-Height Shoots of Moso Bamboo
2.9. Transcriptional Activity Assay
3. Discussion
3.1. Conservation and Diversity of PeNACs
3.2. Homologous Genes Indicating Similar Functions
3.3. Diverse Expression Patterns of PeNACs
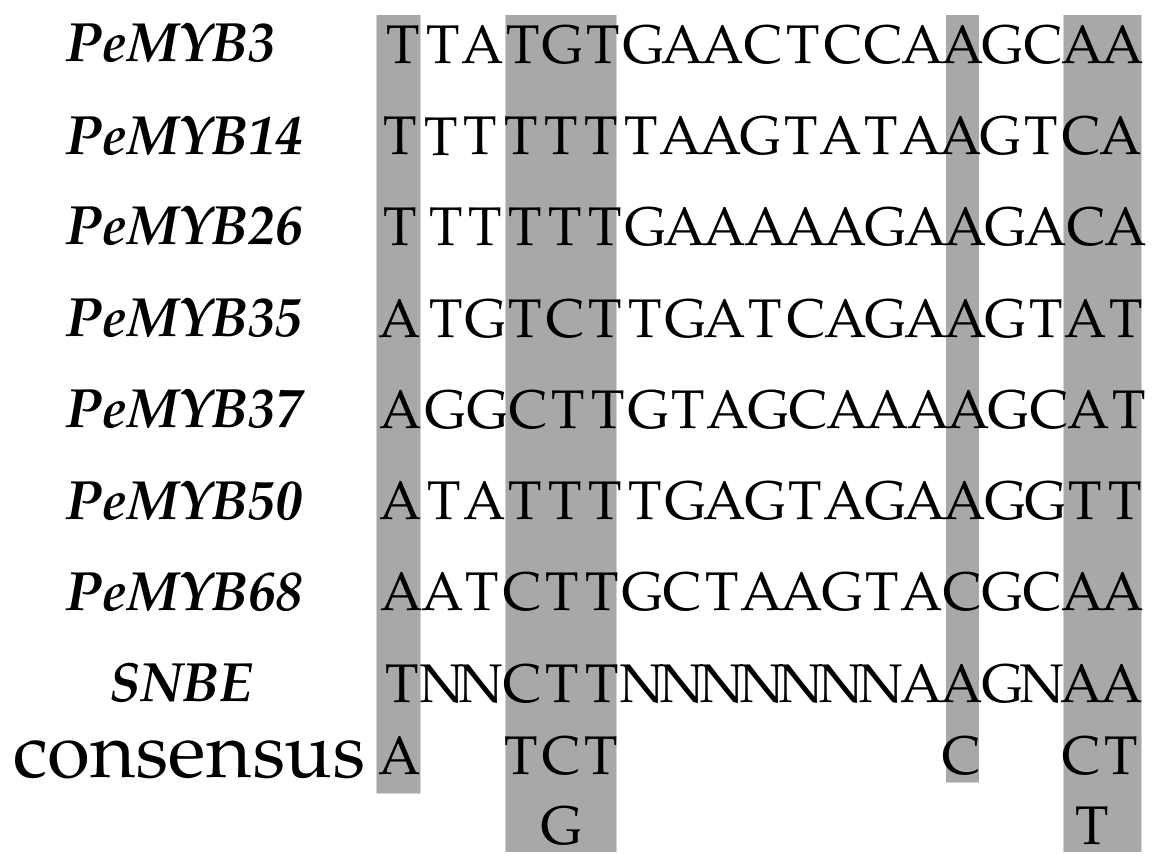
3.4. Complexity of SCW Biosynthesis
4. Materials and Methods
4.1. Identification and Nomenclature of PeNACs
4.2. Gene Structure, Protein Characterization, and Sequence Analyses
4.3. Phylogenetic Analysis and Functional Prediction
4.4. Tissue-Specific Expression Analysis of PeNACs Based on Transcriptome Data
4.5. Co-Expression Network of PeNACs and Subsequent GO Analysis
4.6. Plant Material Collection and cDNA Synthesis
4.7. Real-Time Quantitative PCR Analysis
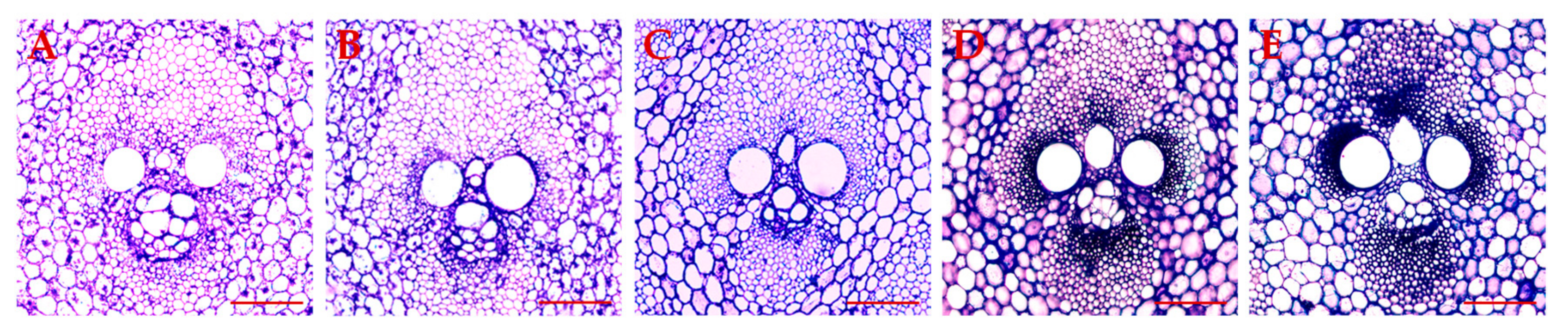
4.8. Histological Analysis Method
4.9. Transcriptional Activity Analysis of PeNACs
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yang, X.D.; Li, J.K. Consideration and suggestion on solving timber shortage problems in China. J. Beijing Fore Univ. 2009, 8, 105–108. [Google Scholar]
- Tang, Y.Y. The industrial development and utilization of bamboo timber resources in China. J. Bamboo Research 1997, 2, 26–33. [Google Scholar]
- Wu, H.; Lv, H.; Li, L.; Liu, J.; Mu, S.; Li, X.; Gao, J. Genome-Wide Analysis of the AP2/ERF Transcription Factors Family and the Expression Patterns of DREB Genes in Moso Bamboo (Phyllostachys edulis). PLoS ONE 2015, 10, e0126657. [Google Scholar] [CrossRef] [PubMed]
- Seth, M.K. Trees and Their Economic Importance. Bot. Rev. 2003, 69, 321–376. [Google Scholar] [CrossRef]
- Carroll, A.; Somerville, C. Cellulosic biofuels. Annu. Rev. Plant Biol. 2009, 60, 165–182. [Google Scholar] [CrossRef]
- Hirano, K.; Kondo, M.; Aya, K.; Miyao, A.; Sato, Y.; Antonio, B.A.; Namiki, N.; Nagamura, Y.; Matsuoka, M. Identification of Transcription Factors Involved in Rice Secondary Cell Wall Formation. Plant Cell Physiol. 2013, 54, 1791–1802. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Demura, T. Transcriptional regulation of secondary wall formation controlled by NAC domain proteins. Plant Biotechnol. 2010, 27, 237–242. [Google Scholar] [CrossRef] [Green Version]
- Zhong, R.; Lee, C.; Ye, Z.-H. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 2010, 15, 625–632. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, Q.; Chen, F.; Wang, M.; Dixon, R.A. NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J. 2011, 68, 1104–1114. [Google Scholar] [CrossRef]
- Hussey, S.G.; Mizrachi, E.; Creux, N.M.; Myburg, A.A. Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front. Plant Sci. 2013, 4, 325. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Ying, Y.Q.; Wang, J.; Zhao, X.H.; Zeng, W.; Beahan, C.; He, J.B.; Chen, X.Y.; Bacic, A.; Song, L.L.; et al. Transcriptome analysis provides insights into xylogenesis formation in moso bamboo (Phyllostachys edulis) shoot. Sci. Rep. 2018, 8, 3951. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.F.; Gan, S.S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Gil Nam, H.; Lim, P.O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 2016, 33, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Duval, M.; Hsieh, T.F.; Kim, S.Y.; Thomas, T.L. Molecular characterization of AtNAM: A member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 2002, 50, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Hussey, S.G.; Mizrachi, E.; Spokevicius, A.V.; Bossinger, G.; Berger, D.K.; Myburg, A.A. SND2, a NAC transcription actor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol. 2011, 11, 173. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.Q.; Ye, Z.H. Secondary cell walls: Biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol. 2015, 56, 195–214. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. The NAC Transcription Factors NST1 and NST2 of Arabidopsis Regulate Secondary Wall Thickenings and Are Required for Anther Dehiscence. Plant Cell 2005, 17, 2993–3006. [Google Scholar] [CrossRef]
- Kubo, M.; Udagawa, M.; Nishikubo, N.; Horiguchi, G.; Yamaguchi, M.; Ito, J.; Mimura, T.; Fukuda, H.; Demura, T. Transcription switches for protoxylem and metaxylem vessel formation. Genome Res. 2005, 19, 1855–1860. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, M.; Kubo, M.; Fukuda, H.; Demura, T. VASCULAR-RELATED NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008, 55, 652–664. [Google Scholar] [CrossRef]
- Zhou, J.; Zhong, R.; Ye, Z.H. Arabidopsis NAC Domain Proteins, VND1 to VND5, Are Transcriptional Regulators of Secondary Wall Biosynthesis in Vessels. PLoS ONE 2014, 9, e105726. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, G.Q.; Zou, D.; Yan, J.Q.; Li, Y.; Hu, S.; Li, X.B. The cotton (Gossypium hirsutum) NAC transcription factor (FSN1) as a positive regulator participates in controlling secondary cell wall biosynthesis and modification of fibers. New Phytol. 2018, 217, 625–640. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.H.; Wang, H. Molecular Mechanisms for Vascular Development and Secondary Cell Wall Formation. Front. Plant Sci. 2016, 7, 387. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Hu, M.; Li, J.; Chen, L.; Li, M.; Zhang, S.; Zhang, X.; Yang, X. Comprehensive analysis of NAC transcription factors uncovers their roles during fiber development and stress response in cotton. BMC Plant Biol. 2018, 18, 150. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Ohtani, M.; Yamaguchi, M.; Toyooka, K.; Wakazaki, M.; Sato, M.; Kubo, M.; Nakano, Y.; Sano, R.; Hiwatashi, Y.; et al. Contribution of NAC Transcription Factors to Plant Adaptation to Land. Science 2014, 343, 1505–1508. [Google Scholar] [CrossRef]
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288. [Google Scholar] [CrossRef]
- Littlewood, J.; Wang, L.; Turnbull, C.; Murphy, R.J. Techno-economic potential of bioethanol from bamboo in China. Biotechnol. Biofuels 2013, 6, 173. [Google Scholar] [CrossRef]
- Jiang, Z.H. Bamboo and Rattan in the World; Liaoning Science Technology Publishing House: Beijing, China, 2002. [Google Scholar]
- Peng, Z.H.; Lu, Y.; Li, L.B.; Zhao, Q.; Feng, Q.; Gao, Z.M.; Lu, H.Y.; Hu, T.; Yao, N.; Liu, K.Y.; et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat. Genet. 2013, 45, 456–461. [Google Scholar] [CrossRef]
- Yang, K.; Li, Y.; Wang, S.; Xu, X.; Sun, H.; Zhao, H.; Li, X.; Gao, Z. Genome-wide identification and expression analysis of the MYB transcription factor in moso bamboo (Phyllostachys edulis). PeerJ 2019, 6, e6242. [Google Scholar] [CrossRef]
- Yu, H.Q. Study on property of bamboo culms. World Bamboo Ratt. 2013, 1, 5–9. [Google Scholar]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. Curr. Neuropharmacol. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Puranik, S.; Sahu, P.P.; Mandal, S.N.; Suresh, B.V.; Parida, S.K.; Prasad, M. Comprehensive Genome-Wide Survey, Genomic Constitution and Expression Profiling of the NAC Transcription Factor Family in Foxtail Millet (Setaria italica L.). PLoS ONE 2013, 8, e64594. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Peng, Z.; Fei, B.; Li, L.; Hu, T.; Gao, Z.; Jiang, Z. BambooGDB: A bamboo genome database with functional annotation and an analysis platform. Database 2014, 2014, bau006. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhao, H.; Xu, W.; You, Q.; Yan, H.; Gao, Z.; Su, Z. Co-expression Gene Network Analysis and Functional Module Identification in Bamboo Growth and Development. Front. Genet. 2018, 9, 574. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Ye, Z.-H. Global Analysis of Direct Targets of Secondary Wall NAC Master Switches in Arabidopsis. Mol. Plant 2010, 3, 1087–1103. [Google Scholar] [CrossRef]
- Wang, C.Y.; Zhang, S.; Yü, Y.; Luo, Y.C.; Liu, Q.; Ju, C.; Zhang, Y.C.; Qu, L.H.; Lucas, W.J.; Wang, X.; et al. MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol. J. 2014, 12, 1132–1142. [Google Scholar] [CrossRef] [Green Version]
- Du, Q.; Wang, H. The role of HD-ZIP III transcription factors and miR165/166 in vascular development and secondary cell wall formation. Plant Signal. Behav. 2015, 10, e1078955. [Google Scholar] [CrossRef]
- Sun, X.; Wang, C.; Xiang, N.; Li, X.; Yang, S.; Du, J.; Yang, Y.; Yang, Y. Activation of secondary cell wall biosynthesis by miR319-targeted TCP4 transcription factor. Plant Biotechnol. J. 2017, 15, 1284–1294. [Google Scholar] [CrossRef]
- Wang, L.L.; Zhao, H.S.; Chen, D.L.; Li, L.C.; Sun, H.Y.; Lou, Y.F.; Gao, Z.M. Characterization and primary functional analysis of a bamboo NAC gene targeted by miR164b. Plant Cell Rep. 2016, 35, 1371–1383. [Google Scholar] [CrossRef]
- Fan, C.; Ma, J.; Guo, Q.; Li, X.; Wang, H.; Lu, M. Selection of Reference Genes for Quantitative Real-Time PCR in Bamboo (Phyllostachys edulis). PLoS ONE 2013, 8, e56573. [Google Scholar] [CrossRef]
- Hibara, K.I.; Takada, S.; Hibara, K.; Tasaka, M. CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J. 2003, 36, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Maruyama, K.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Tran, L.-S.P.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Ohme-Takagi, M.; et al. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 2004, 39, 863–876. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Kim, S.G.; Park, J.E.; Park, H.Y.; Lim, M.H.; Chua, N.H.; Park, C.M. A Membrane-Bound NAC Transcription Factor Regulates Cell Division in Arabidopsis. Plant Cell 2006, 18, 3132–3144. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lin, Y.C.; Sun, Y.H.; Song, J.; Chen, H.; Zhang, X.H.; Sederoff, R.R.; Chiang, V.L. Splice variant of the SND1 transcription factor is a dominant negative of SND1 members and their regulation in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 2012, 109, 14699–14704. [Google Scholar] [CrossRef]
- Han, Q.Q.; Qiao, P.; Song, Y.Z.; Zhang, J.Y. Structural analysis and tissue-specific expression patterns of a novel salt-inducible NAC transcription factor gene from Nicotiana tabacum cv. Xanthi. J. Hortic. Sci. Biotechnol. 2014, 89, 700–706. [Google Scholar] [CrossRef]
- Ahmad, M.; Yan, X.; Li, J.; Yang, Q.; Jamil, W.; Teng, Y.; Bai, S. Genome wide identification and predicted functional analyses of NAC transcription factors in Asian pears. BMC Plant Biol. 2018, 18, 214. [Google Scholar] [CrossRef]
- Gong, X.; Zhao, L.; Song, X.; Lin, Z.; Gu, B.; Yan, J.; Zhang, S.; Tao, S.; Huang, X. Genome-wide analyses and expression patterns under abiotic stress of NAC transcription factors in white pear (Pyrus bretschneideri). BMC Plant Biol. 2019, 19, 161. [Google Scholar] [CrossRef]
- Staiger, D.; Brown, J.W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef]
- Li, J.; Meng, X.; Zong, Y.; Chen, K.; Zhang, H.; Liu, J.; Li, J.; Gao, C. Gene replacements and insertions in rice by intron targeting using CRISPR–Cas9. Nat. Plants 2016, 2, 16139. [Google Scholar] [CrossRef]
- Yu, G.; Luo, W.; Fu, G.; Wang, J. Interspecies gene function prediction using semantic similarity. BMC Syst. Biol. 2016, 10, 121–507. [Google Scholar] [CrossRef] [Green Version]
- Vroemen, C.W.; Mordhorst, A.P.; Albrecht, C.; Kwaaitaal, M.A.C.J.; De Vries, S.C. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. F1000 Post Publ. Peer Rev. Biomed. Lit. 2003, 15, 1563–1577. [Google Scholar] [CrossRef] [PubMed]
- Balazadeh, S.; Siddiqui, H.; Allu, A.D.; Matallana-Ramirez, L.P.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Koehler, B.; Mueller-Roeber, B. A gene regulatory network controlled by the NAC transcription factor AtNAC92/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 2010, 62, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Li, Y.; Wu, X.; Qi, G.; Li, N.; Zhang, K.; Wang, D.; Sun, X.S. Utilization of sorghum lignin to improve adhesion strength of soy protein adhesives on wood veneer. Ind. Crop. Prod. 2013, 50, 501–509. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Ye, Z.H. MYB46 and MYB83 bind to the SMRE sites and directly activate a suit of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 2012, 53, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, M.; Demura, T. The quest for transcriptional hubs of lignin biosynthesis: Beyond the NAC-MYB-gene regulatory network model. Curr. Opin. Biotechnol. 2019, 56, 82–87. [Google Scholar] [CrossRef]
- Ohashi-Ito, K.; Oda, Y.; Fukuda, H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 Directly Regulates the Genes That Govern Programmed Cell Death and Secondary Wall Formation during Xylem Differentiation. Plant Cell 2010, 22, 3461–3473. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Mitsuda, N.; Ohtani, M.; Ohme-Takagi, M.; Kato, K.; Demura, T.; Ohme-Takagi, M. VASCULAR-RELATED NAC-DOMAIN 7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J. 2011, 66, 579–590. [Google Scholar] [CrossRef]
- Ko, J.-H.; Kim, W.-C.; Han, K.-H. Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J. 2009, 60, 649–665. [Google Scholar] [CrossRef]
- McCarthy, R.L.; Zhong, R.; Ye, Z.H. MYB83 Is a Direct Target of SND1 and Acts Redundantly with MYB46 in the Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis. Plant Cell Physiol. 2009, 50, 1950–1964. [Google Scholar] [CrossRef] [Green Version]
- Zhong, R.; Richardson, E.A.; Ye, Z.-H. The MYB46 Transcription Factor Is a Direct Target of SND1 and Regulates Secondary Wall Biosynthesis in Arabidopsis. Plant Cell 2007, 19, 2776–2792. [Google Scholar] [CrossRef]
- Jin, J.P.; Zhang, H.; Kong, L.; Gao, G.; Luo, J.C. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.Y. GSDS: A gene structure display server. Hereditas Beijing 2007, 29, 1023. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Boil. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Windham, G.L.; Williams, W.P.; Mylroie, J.E.; Reid, C.X.; Womack, E.D. A Histological Study of Aspergillus flavus Colonization of Wound Inoculated Maize Kernels of Resistant and Susceptible Maize Hybrids in the Field. Front. Microbiol. 2018, 9, 799. [Google Scholar] [CrossRef] [Green Version]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shan, X.; Yang, K.; Xu, X.; Zhu, C.; Gao, Z. Genome-Wide Investigation of the NAC Gene Family and Its Potential Association with the Secondary Cell Wall in Moso Bamboo. Biomolecules 2019, 9, 609. https://doi.org/10.3390/biom9100609
Shan X, Yang K, Xu X, Zhu C, Gao Z. Genome-Wide Investigation of the NAC Gene Family and Its Potential Association with the Secondary Cell Wall in Moso Bamboo. Biomolecules. 2019; 9(10):609. https://doi.org/10.3390/biom9100609
Chicago/Turabian StyleShan, Xuemeng, Kebin Yang, Xiurong Xu, Chenglei Zhu, and Zhimin Gao. 2019. "Genome-Wide Investigation of the NAC Gene Family and Its Potential Association with the Secondary Cell Wall in Moso Bamboo" Biomolecules 9, no. 10: 609. https://doi.org/10.3390/biom9100609




