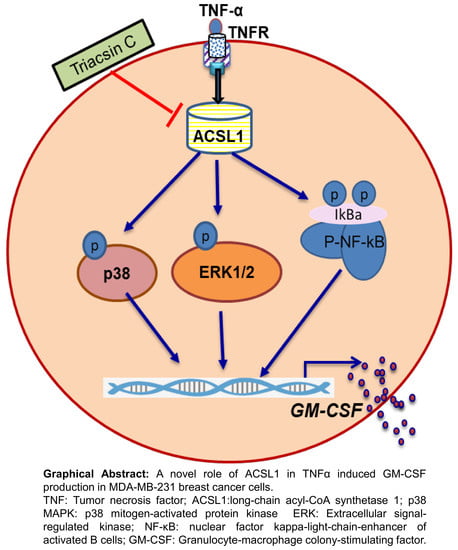
ACSL1 Regulates TNFα-Induced GM-CSF Production by Breast Cancer MDA-MB-231 Cells
Abstract
:1. Introduction
2. Results
2.1. TNFα Induces GM-CSF Gene Expression in Human MDA-MB-231 Cells
2.2. TNFα Induced GM-CSF Production is Suppressed by Inhibition of ACSL1
2.3. ACSL1 Deficiency Suppresses TNFα-Induced GM-CSF
2.4. ACSL1 is involved in TNFα-Activated MAPK and NF-κB Signaling Pathways
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Cell Stimulation
4.3. Real-Time Quantitative PCR
4.4. Small Interfering RNA (siRNA) Transfections
4.5. GM-CSF Determination
4.6. Western Blotting
4.7. Immunocytofluorescence
4.8. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Hong, I.S. Stimulatory versus suppressive effects of GM-CSF on tumor progression in multiple cancer types. Exp. Mol. Med. 2016, 48, e242. [Google Scholar] [CrossRef]
- Wicks, I.P.; Roberts, A.W. Targeting GM-CSF in inflammatory diseases. Nat. Rev. Rheumatol. 2016, 12, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Rasouli, J.; Ciric, B.; Imitola, J.; Gonnella, P.; Hwang, D.; Mahajan, K.; Mari, E.R.; Safavi, F.; Leist, T.P.; Zhang, G.X.; et al. Expression of GM-CSF in T Cells Is Increased in Multiple Sclerosis and Suppressed by IFN-beta Therapy. J. Immunol. 2015, 194, 5085–5093. [Google Scholar] [CrossRef] [PubMed]
- McInnes, I.B.; Schett, G. Pathogenetic insights from the treatment of rheumatoid arthritis. Lancet 2017, 389, 2328–2337. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Amezquita, R.A.; Kleinstein, S.H.; Stathopoulos, P.; Nowak, R.J.; O’Connor, K.C. Autoreactive T Cells from Patients with Myasthenia Gravis Are Characterized by Elevated IL-17, IFN-gamma, and GM-CSF and Diminished IL-10 Production. J. Immunol. 2016, 196, 2075–2084. [Google Scholar] [CrossRef]
- Borriello, F.; Galdiero, M.R.; Varricchi, G.; Loffredo, S.; Spadaro, G.; Marone, G. Innate Immune Modulation by GM-CSF and IL-3 in Health and Disease. Int. J. Mol. Sci. 2019, 20, 834. [Google Scholar] [CrossRef] [PubMed]
- Eyerich, K.; Dimartino, V.; Cavani, A. IL-17 and IL-22 in immunity: Driving protection and pathology. Eur. J. Immunol. 2017, 47, 607–614. [Google Scholar] [CrossRef] [Green Version]
- Lotfi, N.; Thome, R.; Rezaei, N.; Zhang, G.X.; Rezaei, A.; Rostami, A.; Esmaeil, N. Roles of GM-CSF in the Pathogenesis of Autoimmune Diseases: An Update. Front. Immunol. 2019, 10, 1265. [Google Scholar] [CrossRef]
- Van Overmeire, E.; Stijlemans, B.; Heymann, F.; Keirsse, J.; Morias, Y.; Elkrim, Y.; Brys, L.; Abels, C.; Lahmar, Q.; Ergen, C.; et al. M-CSF and GM-CSF Receptor Signaling Differentially Regulate Monocyte Maturation and Macrophage Polarization in the Tumor Microenvironment. Cancer Res. 2016, 76, 35–42. [Google Scholar] [CrossRef]
- Park, S.H.; Breitbach, C.J.; Lee, J.; Park, J.O.; Lim, H.Y.; Kang, W.K.; Moon, A.; Mun, J.H.; Sommermann, E.M.; Maruri Avidal, L.; et al. Phase 1b Trial of Biweekly Intravenous Pexa-Vec (JX-594), an Oncolytic and Immunotherapeutic Vaccinia Virus in Colorectal Cancer. Mol. Ther. J. Am. Soc. Gene Ther. 2015, 23, 1532–1540. [Google Scholar] [CrossRef]
- Sakakura, K.; Takahashi, H.; Kaira, K.; Toyoda, M.; Murata, T.; Ohnishi, H.; Oyama, T.; Chikamatsu, K. Relationship between tumor-associated macrophage subsets and CD47 expression in squamous cell carcinoma of the head and neck in the tumor microenvironment. Lab. Investig. J. Tech. Methods Pathol. 2016, 96, 994–1003. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, B.; Sonobe, Y.; Kawanokuchi, J.; Doi, Y.; Noda, M.; Takeuchi, H.; Mizuno, T.; Suzumura, A. GM-CSF increases LPS-induced production of proinflammatory mediators via upregulation of TLR4 and CD14 in murine microglia. J. Neuroinflammation 2012, 9, 268. [Google Scholar] [CrossRef] [PubMed]
- Salmiheimo, A.N.; Mustonen, H.K.; Vainionpaa, S.A.; Shen, Z.; Kemppainen, E.A.; Seppanen, H.E.; Puolakkainen, P.A. Increasing the Inflammatory Competence of Macrophages with IL-6 or with Combination of IL-4 and LPS Restrains the Invasiveness of Pancreatic Cancer Cells. J. Cancer 2016, 7, 42–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadeghi, K.; Wisgrill, L.; Wessely, I.; Diesner, S.C.; Schuller, S.; Durr, C.; Heinle, A.; Sachet, M.; Pollak, A.; Forster-Waldl, E.; et al. GM-CSF Down-Regulates TLR Expression via the Transcription Factor PU.1 in Human Monocytes. PLoS ONE 2016, 11, e0162667. [Google Scholar] [CrossRef] [PubMed]
- Komohara, Y.; Fujiwara, Y.; Ohnishi, K.; Takeya, M. Tumor-associated macrophages: Potential therapeutic targets for anti-cancer therapy. Adv. Drug Deliv. Rev. 2016, 99, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Al-Rashed, F.; Ahmad, Z.; Iskandar, M.A.; Tuomilehto, J.; Al-Mulla, F.; Ahmad, R. TNF-alpha Induces a Pro-Inflammatory Phenotypic Shift in Monocytes through ACSL1: Relevance to Metabolic Inflammation. Cell Physiol. Biochem. 2019, 52, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Choi, J.H.; Kim, J.B.; Nam, S.J.; Yang, J.H.; Kim, J.H.; Lee, J.E. Berberine suppresses TNF-alpha-induced MMP-9 and cell invasion through inhibition of AP-1 activity in MDA-MB-231 human breast cancer cells. Molecules 2008, 13, 2975–2985. [Google Scholar] [CrossRef] [PubMed]
- Bornfeldt, K.E. Growing evidence for a role for acyl-CoA synthetase 1 in immunometabolism. J. Leukoc. Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.M.; Li, L.O.; Wu, P.C.; Koves, T.R.; Ilkayeva, O.; Stevens, R.D.; Watkins, S.M.; Muoio, D.M.; Coleman, R.A. Adipose acyl-CoA synthetase-1 directs fatty acids toward beta-oxidation and is required for cold thermogenesis. Cell Metab. 2010, 12, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, J.R.; Wang, X.; Lewandowski, E.D. Acyl CoA synthetase-1 links facilitated long chain fatty acid uptake to intracellular metabolic trafficking differently in hearts of male versus female mice. J. Mol. Cell. Cardiol. 2016, 94, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Hayden, M.S.; Ghosh, S. Regulation of NF-kappaB by TNF family cytokines. Semin. Immunol. 2014, 26, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, G.; Midiri, A.; Beninati, C.; Piraino, G.; Valenti, A.; Nicocia, G.; Teti, D.; Cook, J.; Teti, G. Mitogen-activated protein kinases and NF-kappa B are involved in TNF-alpha responses to group B streptococci. J. Immunol. 2002, 169, 1401–1409. [Google Scholar] [CrossRef] [PubMed]
- Lowenberg, B.; Salem, M.; Delwel, R. Effects of recombinant multi-CSF, GM-CSF, G-CSF and M-CSF on the proliferation and maturation of human AML in vitro. Blood Cells 1988, 14, 539–549. [Google Scholar] [PubMed]
- Reggiani, F.; Labanca, V.; Mancuso, P.; Rabascio, C.; Talarico, G.; Orecchioni, S.; Manconi, A.; Bertolini, F. Adipose Progenitor Cell Secretion of GM-CSF and MMP9 Promotes a Stromal and Immunological Microenvironment That Supports Breast Cancer Progression. Cancer Res. 2017, 77, 5169–5182. [Google Scholar] [CrossRef] [PubMed]
- Ghirelli, C.; Reyal, F.; Jeanmougin, M.; Zollinger, R.; Sirven, P.; Michea, P.; Caux, C.; Bendriss-Vermare, N.; Donnadieu, M.H.; Caly, M.; et al. Breast Cancer Cell-Derived GM-CSF Licenses Regulatory Th2 Induction by Plasmacytoid Predendritic Cells in Aggressive Disease Subtypes. Cancer Res. 2015, 75, 2775–2787. [Google Scholar] [CrossRef] [PubMed]
- Aliper, A.M.; Frieden-Korovkina, V.P.; Buzdin, A.; Roumiantsev, S.A.; Zhavoronkov, A. A role for G-CSF and GM-CSF in nonmyeloid cancers. Cancer Med. 2014, 3, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Braun, B.; Lange, M.; Oeckler, R.; Mueller, M.M. Expression of G-CSF and GM-CSF in human meningiomas correlates with increased tumor proliferation and vascularization. J. Neuro-Oncol. 2004, 68, 131–140. [Google Scholar] [CrossRef]
- Mueller, M.M.; Fusenig, N.E. Constitutive expression of G-CSF and GM-CSF in human skin carcinoma cells with functional consequence for tumor progression. Int. J. Cancer 1999, 83, 780–789. [Google Scholar] [CrossRef]
- Kohanbash, G.; McKaveney, K.; Sakaki, M.; Ueda, R.; Mintz, A.H.; Amankulor, N.; Fujita, M.; Ohlfest, J.R.; Okada, H. GM-CSF promotes the immunosuppressive activity of glioma-infiltrating myeloid cells through interleukin-4 receptor-alpha. Cancer Res. 2013, 73, 6413–6423. [Google Scholar] [CrossRef]
- Gutschalk, C.M.; Yanamandra, A.K.; Linde, N.; Meides, A.; Depner, S.; Mueller, M.M. GM-CSF enhances tumor invasion by elevated MMP-2, -9, and -26 expression. Cancer Med. 2013, 2, 117–129. [Google Scholar] [CrossRef]
- Zhou, X.L.; Fan, W.; Yang, G.; Yu, M.X. The clinical significance of PR, ER, NF- kappa B, and TNF- alpha in breast cancer. Dis Markers 2014, 2014, 494581. [Google Scholar] [CrossRef] [PubMed]
- Rubinow, K.B.; Wall, V.Z.; Nelson, J.; Mar, D.; Bomsztyk, K.; Askari, B.; Lai, M.A.; Smith, K.D.; Han, M.S.; Vivekanandan-Giri, A.; et al. Acyl-CoA synthetase 1 is induced by Gram-negative bacteria and lipopolysaccharide and is required for phospholipid turnover in stimulated macrophages. J. Biol. Chem. 2013, 288, 9957–9970. [Google Scholar] [CrossRef] [PubMed]
- Koga, Y.; Hisada, T.; Ishizuka, T.; Utsugi, M.; Ono, A.; Yatomi, M.; Kamide, Y.; Aoki-Saito, H.; Tsurumaki, H.; Dobashi, K.; et al. CREB regulates TNF-alpha-induced GM-CSF secretion via p38 MAPK in human lung fibroblasts. Allergol. Int. Off. J. Jpn. Soc. Allergol. 2016, 65, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, S.; Matsumoto, K.; Gon, Y.; Maruoka, S.; Kujime, K.; Hayashi, S.; Takeshita, I.; Horie, T. p38 MAP kinase regulates TNF alpha-, IL-1 alpha- and PAF-induced RANTES and GM-CSF production by human bronchial epithelial cells. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2000, 30, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Wray, G.M.; Foster, S.J.; Hinds, C.J.; Thiemermann, C. A cell wall component from pathogenic and non-pathogenic gram-positive bacteria (peptidoglycan) synergises with endotoxin to cause the release of tumour necrosis factor-alpha, nitric oxide production, shock, and multiple organ injury/dysfunction in the rat. Shock 2001, 15, 135–142. [Google Scholar] [CrossRef]
- Al-Rashed, F.; Kochumon, S.; Usmani, S.; Sindhu, S.; Ahmad, R. Pam3CSK4 Induces MMP-9 Expression in Human Monocytic THP-1 Cells. Cell Physiol. Biochem. 2017, 41, 1993–2003. [Google Scholar] [CrossRef] [PubMed]
- Sindhu, S.; Al-Roub, A.; Koshy, M.; Thomas, R.; Ahmad, R. Palmitate-Induced MMP-9 Expression in the Human Monocytic Cells is Mediated through the TLR4-MyD88 Dependent Mechanism. Cell Physiol. Biochem. 2016, 39, 889–900. [Google Scholar] [CrossRef]
- Ahmad, R.; Shihab, P.K.; Jasem, S.; Behbehani, K. FSL-1 induces MMP-9 production through TLR-2 and NF-κB /AP-1 signaling pathways in monocytic THP-1 cells. Cell Physiol. Biochem. 2014, 34, 929–942. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, R.; Al-Rashed, F.; Akhter, N.; Al-Mulla, F.; Ahmad, R. ACSL1 Regulates TNFα-Induced GM-CSF Production by Breast Cancer MDA-MB-231 Cells. Biomolecules 2019, 9, 555. https://doi.org/10.3390/biom9100555
Thomas R, Al-Rashed F, Akhter N, Al-Mulla F, Ahmad R. ACSL1 Regulates TNFα-Induced GM-CSF Production by Breast Cancer MDA-MB-231 Cells. Biomolecules. 2019; 9(10):555. https://doi.org/10.3390/biom9100555
Chicago/Turabian StyleThomas, Reeby, Fatema Al-Rashed, Nadeem Akhter, Fahd Al-Mulla, and Rasheed Ahmad. 2019. "ACSL1 Regulates TNFα-Induced GM-CSF Production by Breast Cancer MDA-MB-231 Cells" Biomolecules 9, no. 10: 555. https://doi.org/10.3390/biom9100555