pH-Sensitive Co-Adsorption/Release of Doxorubicin and Paclitaxel by Carbon Nanotube, Fullerene, and Graphene Oxide in Combination with N-isopropylacrylamide: A Molecular Dynamics Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Molecular Dynamics
- Obtaining initial configuration of the particles including characteristics of atoms and the initial velocity as well as physical properties (i.e., mass, size, and type of atom)
- Calculation of neighbors list made for every atom of the system which contains all atoms within the force range of the targeted atom. This list alters at each step.
- Calculation of the force applied on each atom based on the configuration, primary conditions and acceleration of each particle as well as the new position and velocity of each particle through integral methods
2.2. Force Fields
2.3. System Preparation
3. Results and Discussion
3.1. Drug–Carrier Interactions
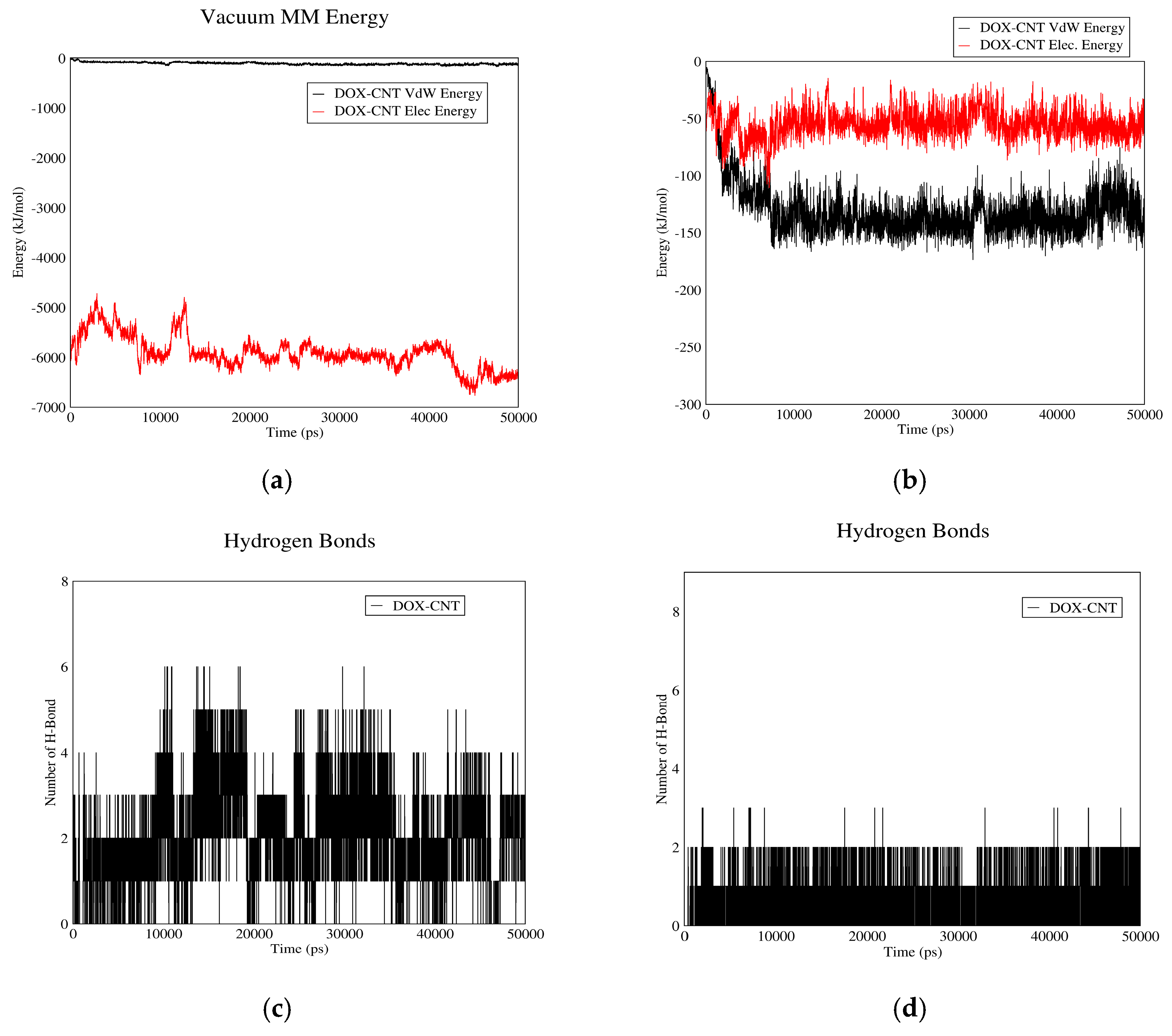
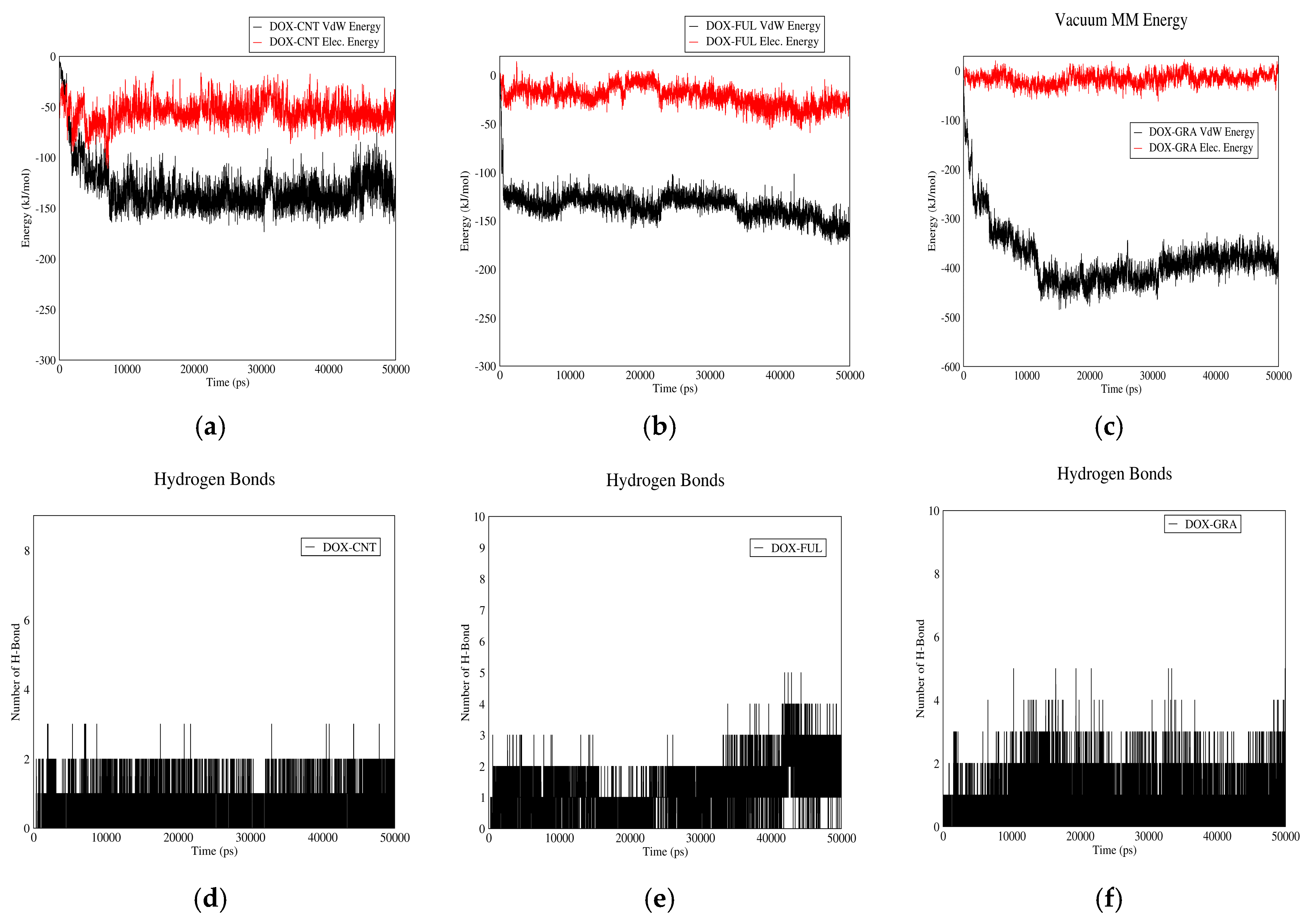
3.1.1. DOX-CNT Interactions
3.1.2. DOX–Fullerene Interactions
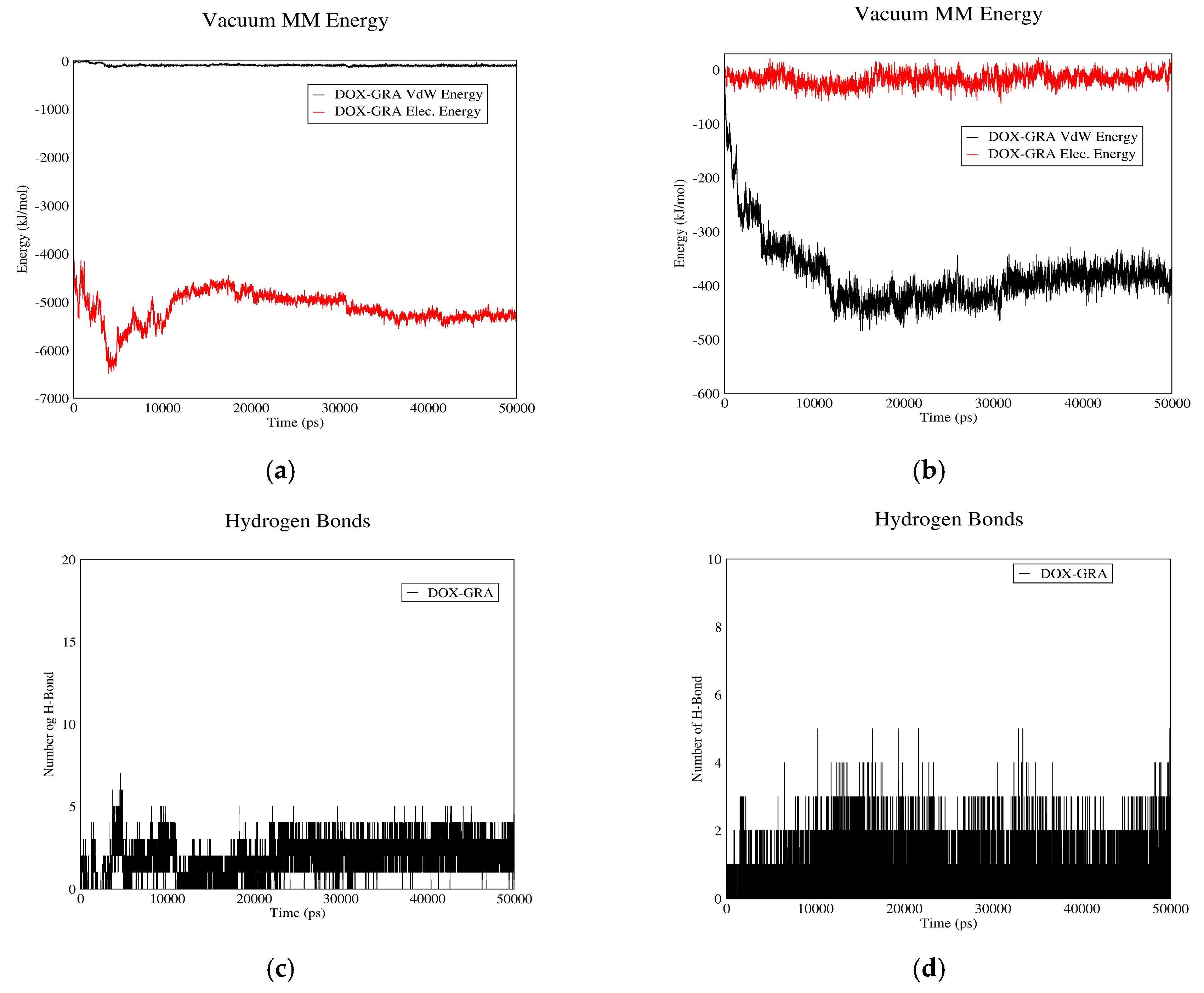
3.1.3. DOX–GO Interactions
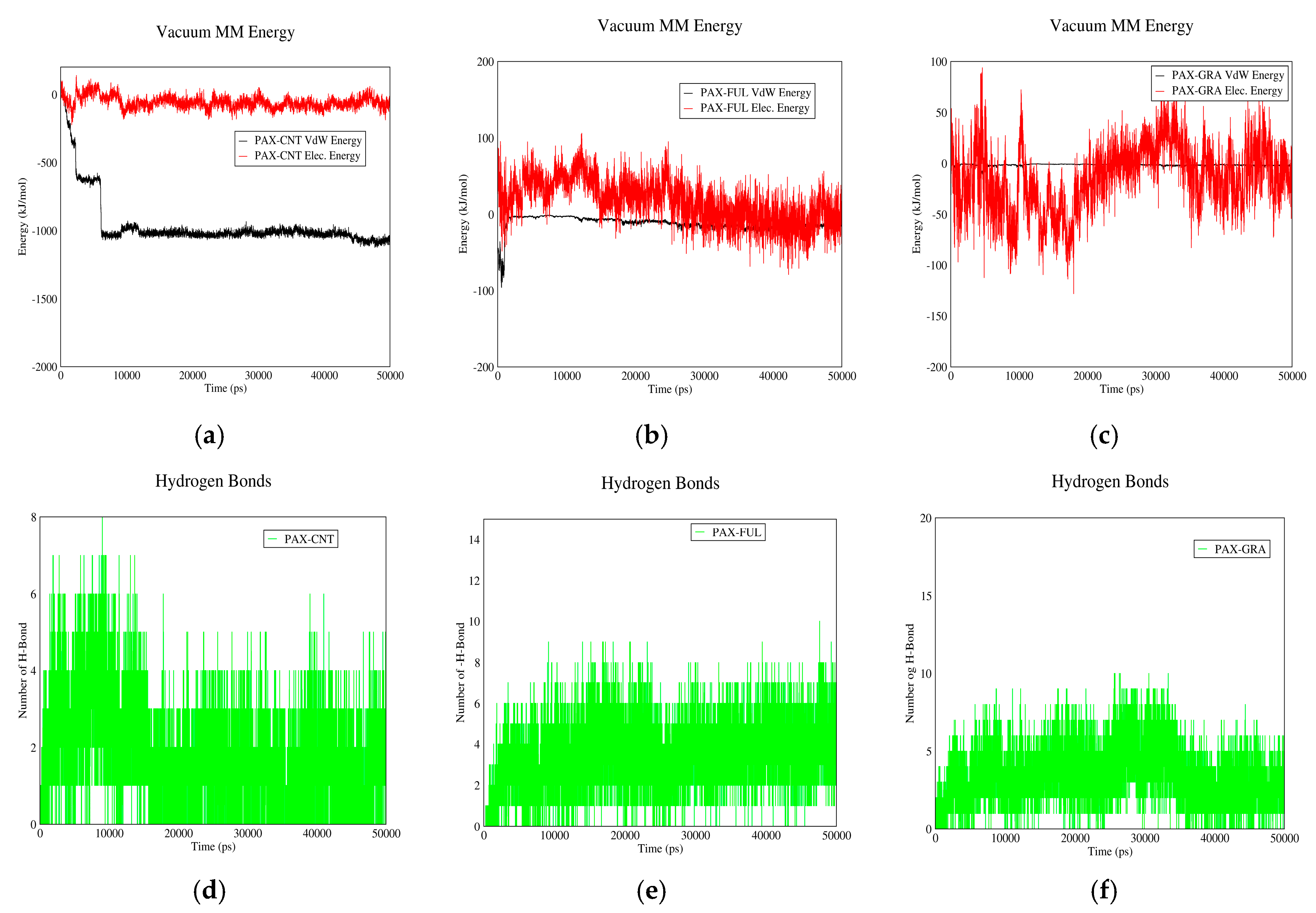
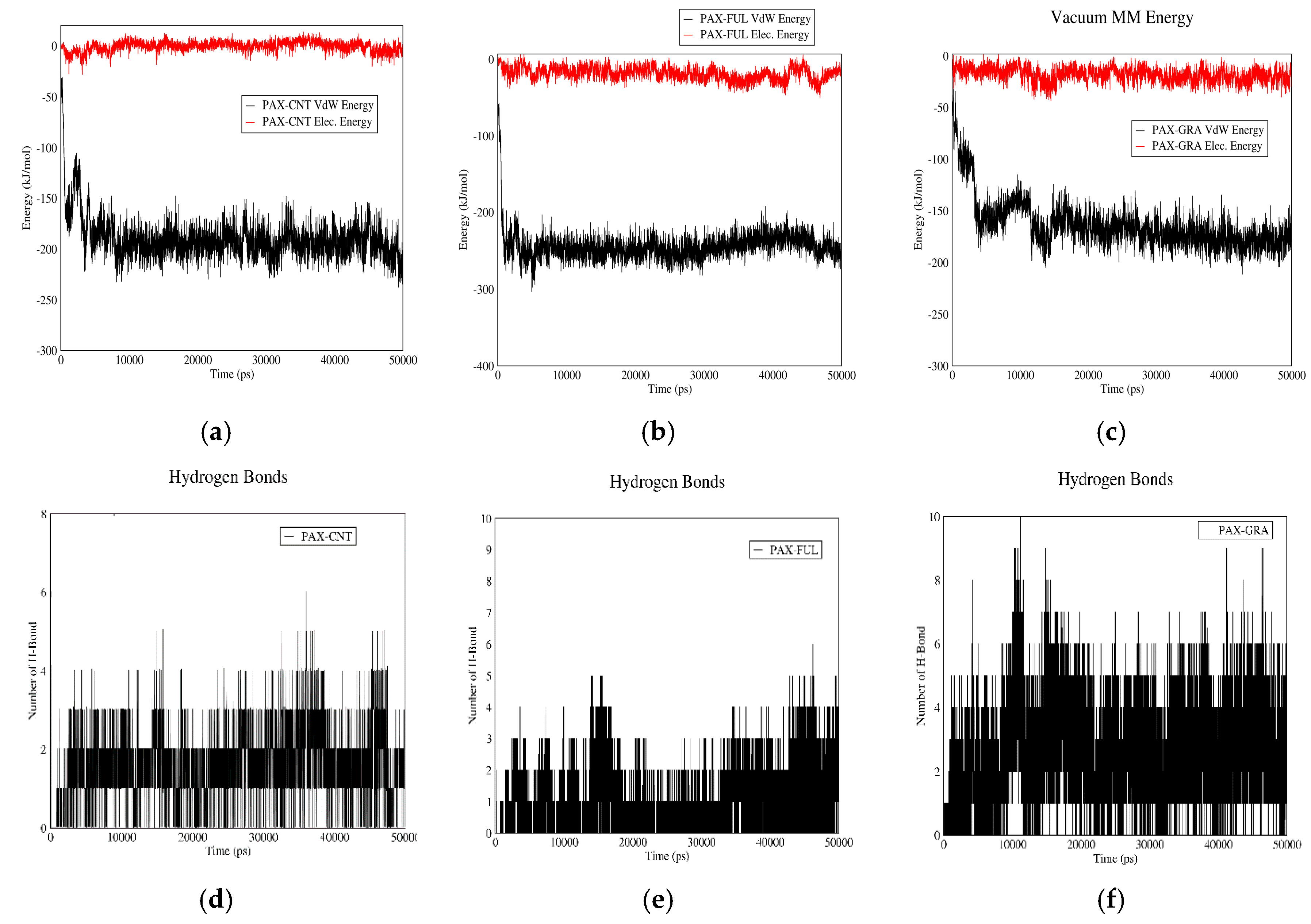
3.1.4. PAX–CNT Interactions
3.1.5. PAX–Fullerene Interactions
3.1.6. PAX–GO Interactions
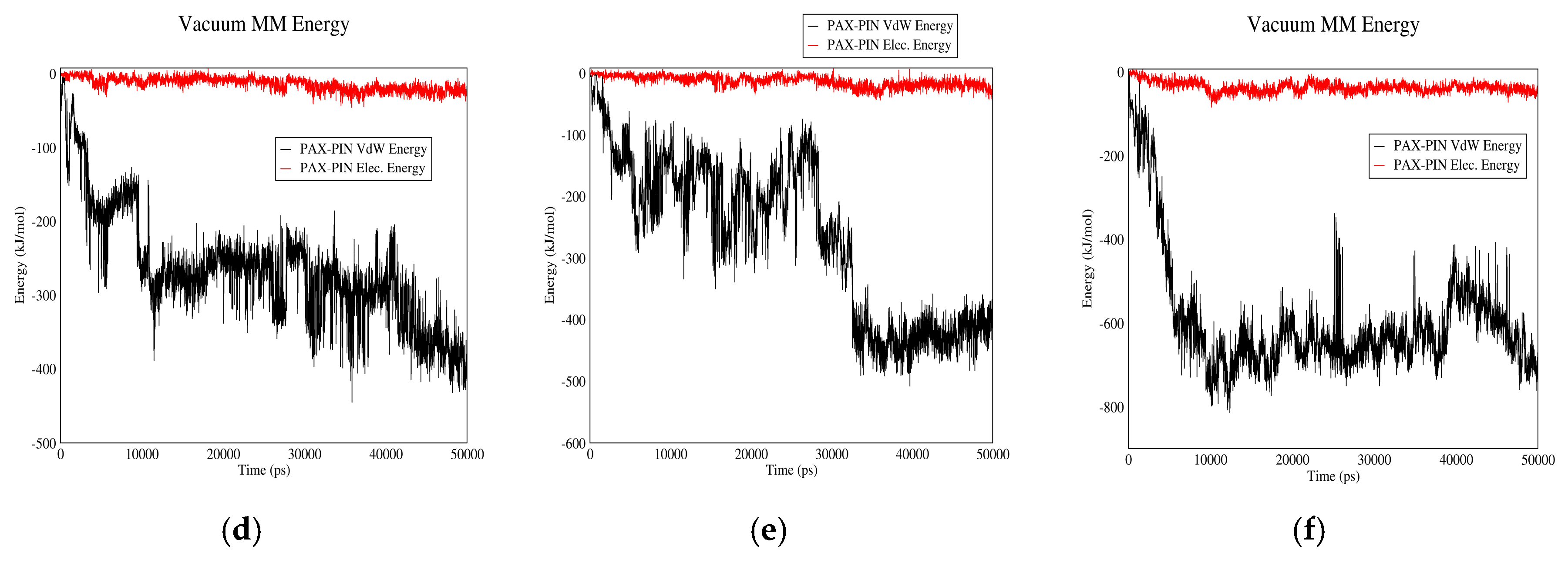
3.2. Drug–PIN Interactions
3.2.1. DOX–PIN Interactions
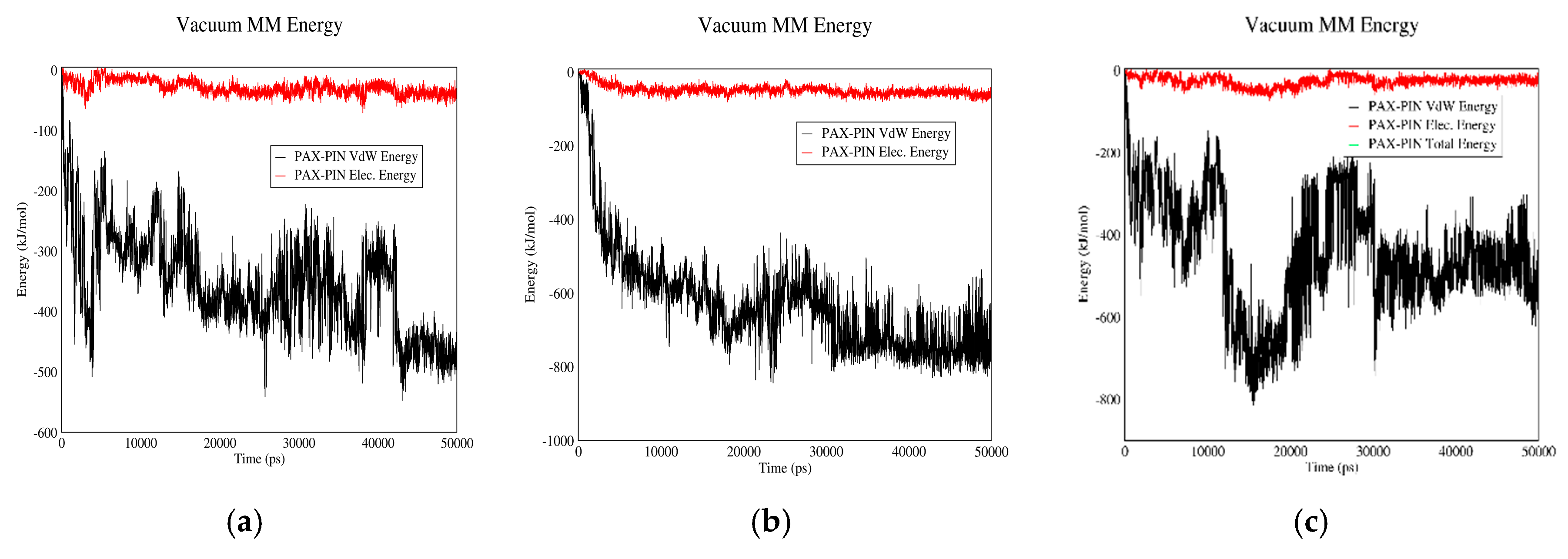
3.2.2. PAX–PIN Interactions
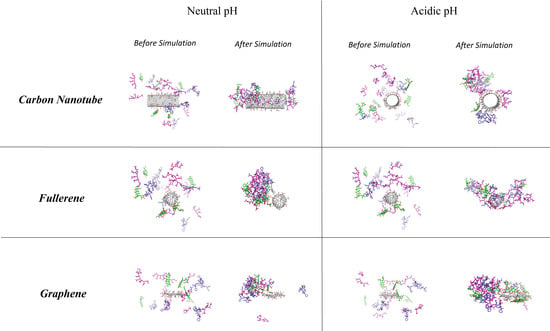
3.3. Comparison of Carriers
3.3.1. DOX–Carrier
3.3.2. PAX–Carrier
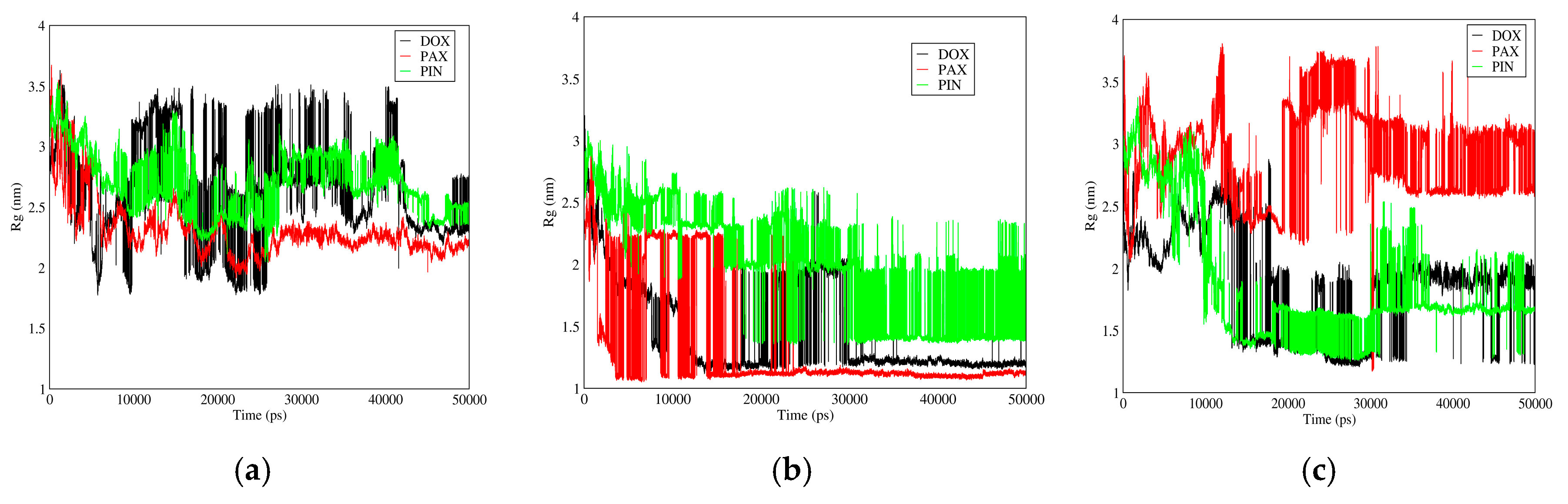
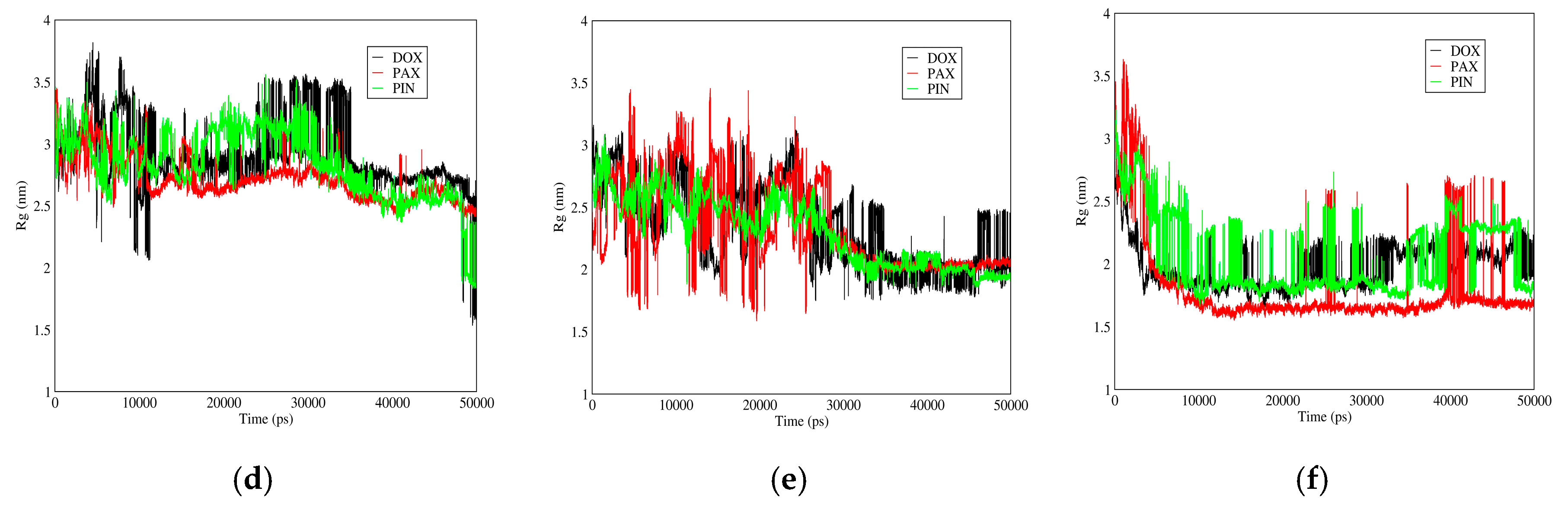
3.3.3. Radius of Gyration
3.3.4. MSD
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Aly, H.A. Cancer therapy and vaccination. J. Immunol. Methods 2012, 382, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Sumanasuriya, S.; De Bono, J. Treatment of advanced prostate cancer-a review of current therapies and future promise. Cold Spring Harb. Perspect. Med. 2018, 8, 030635. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Liang, X.; Mo, S.; Zhang, N.; Sun, D.; Dai, Z. Near-infrared cyanine-loaded liposome-like nanocapsules of camptothecin–floxuridine conjugate for enhanced chemophotothermal combination cancer therapy. ACS Appl. Mater. Interfaces 2018, 10, 3219–3228. [Google Scholar] [CrossRef] [PubMed]
- Oliveri, V.; Viale, M.; Caron, G.; Aiello, C.; Gangemi, R.; Vecchio, G. Glycosylated copper(II) ionophores as prodrugs for β-glucosidase activation in targeted cancer therapy. Dalton Trans. 2013, 42, 2023–2034. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular vesicles in cancer—Implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef] [PubMed]
- Pastor, F. Aptamers: A new technological platform in cancer immunotherapy. Pharmaceuticals 2016, 9, 64. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, P.; Hameed, S.; Dai, Z. Recent advances in anti-angiogenic nanomedicines for cancer therapy. Nanoscale 2018, 10, 5393–5423. [Google Scholar] [CrossRef] [PubMed]
- Panagiotaki, K.N.; Sideratou, Z.; Vlahopoulos, S.A.; Paravatou-Petsotas, M.; Zachariadis, M.; Khoury, N.; Zoumpourlis, V.; Tsiourvas, D. A triphenylphosphonium-functionalized mitochondriotropic nanocarrier for efficient co-delivery of doxorubicin and chloroquine and enhanced antineoplastic activity. Pharmaceuticals 2017, 10, 91. [Google Scholar] [CrossRef] [PubMed]
- Paleos, C.M.; Sideratou, Z.; Theodossiou, T.A.; Tsiourvas, D. Carboxylated hydroxyethyl starch: A novel polysaccharide for the delivery of doxorubicin. Chem. Biol. Drug Des. 2015, 85, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Hameed, S.; Bhattarai, P.; Dai, Z. Cerasomes and bicelles: Hybrid bilayered nanostructures with silica-like surface in cancer theranostics. Front. Chem. 2018, 6, 127. [Google Scholar] [CrossRef] [PubMed]
- Viale, M.; Giglio, V.; Monticone, M.; Maric, I.; Lentini, G.; Rocco, M.; Vecchio, G. New doxorubicin nanocarriers based on cyclodextrins. Investig. New Drugs 2017, 35, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Oliveri, V.; Bellia, F.; Viale, M.; Maric, I.; Vecchio, G. Linear polymers of β and γ cyclodextrins with a polyglutamic acid backbone as carriers for doxorubicin. Carbohydr. Polym. 2017, 177, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.L.; Sill, M.W.; Coleman, R.L.; Sood, A.K.; Pearl, M.L.; Kehoe, S.M.; Carney, M.E.; Hanjani, P.; Van Le, L.; Zhou, X.C.; et al. Paclitaxel with and without pazopanib for persistent or recurrent ovarian cancer: A randomized clinical trial. JAMA Oncol. 2018, 4, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Tampaki, E.C.; Tampakis, A.; Alifieris, C.E.; Krikelis, D.; Pazaiti, A.; Kontos, M.; Trafalis, D.T. Efficacy and safety of neoadjuvant treatment with bevacizumab, liposomal doxorubicin, cyclophosphamide and paclitaxel combination in locally/regionally advanced, HER2-negative, grade III at premenopausal status breast cancer: A phase ii study. Clin. Drug Investig. 2018. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Zhu, F.; Li, B.; Zhao, L.; Liang, H.; Yan, Y.; Tan, H. Folate-conjugated and pH-triggered doxorubicin and paclitaxel co-delivery micellar system for targeted anticancer drug delivery. Mater. Chem. Front. 2018, 2, 1529–1538. [Google Scholar] [CrossRef]
- Rezazadeh, M.; Akbari, V.; Amuaghae, E.; Emami, J. Preparation and characterization of an injectable thermosensitive hydrogel for simultaneous delivery of paclitaxel and doxorubicin. Res. Pharm. Sci. 2018, 13, 181–191. [Google Scholar] [PubMed]
- Catuogno, S.; Esposito, C.L.; de Franciscis, V. Aptamer-mediated targeted delivery of therapeutics: An update. Pharmaceuticals 2016, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.K.; Jang, E.; Lee, K.; Haam, S.; Huh, Y.M. Delivery of cancer therapeutics using nanotechnology. Pharmaceutics 2013, 5, 294–317. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.R.; Vivas-Mejia, P.E. Nanoparticles as drug delivery systems in cancer medicine: Emphasis on RNAi-containing nanoliposomes. Pharmaceuticals 2013, 6, 1361–1380. [Google Scholar] [CrossRef] [PubMed]
- Karavolos, M.; Holban, A. Nanosized drug delivery systems in gastrointestinal targeting: Interactions with microbiota. Pharmaceuticals 2016, 9, 62. [Google Scholar] [CrossRef] [PubMed]
- Stockhofe, K.; Postema, J.M.; Schieferstein, H.; Ross, T.L. Radiolabeling of nanoparticles and polymers for pet imaging. Pharmaceuticals 2014, 7, 392–418. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Shum, K.T.; Burnett, J.C.; Rossi, J.J. Nanoparticle-based delivery of RNAi therapeutics: Progress and challenges. Pharmaceuticals 2013, 6, 85–107. [Google Scholar] [CrossRef] [PubMed]
- Debbage, P.; Thurner, G.C. Nanomedicine faces barriers. Pharmaceuticals 2010, 3, 3371–3416. [Google Scholar] [CrossRef]
- Ozalp, V.C.; Eyidogan, F.; Oktem, H.A. Aptamer-gated nanoparticles for smart drug delivery. Pharmaceuticals 2011, 4, 1137–1157. [Google Scholar] [CrossRef]
- Johnson, R.; Sabnis, N.; McConathy, W.J.; Lacko, A.G. The potential role of nanotechnology in therapeutic approaches for triple negative breast cancer. Pharmaceutics 2013, 5, 353–370. [Google Scholar] [CrossRef] [PubMed]
- Belleperche, M.; DeRosa, C.M. pH-control in aptamer-based diagnostics, therapeutics, and analytical applications. Pharmaceuticals 2018, 11, 80. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Sharma, G.; Kumar, R.; Singh, B.; Katare, O.P.; Raza, K. Lysine-based C60-fullerene nanoconjugates for monomethyl fumarate delivery: A novel nanomedicine for brain cancer cells. ACS Biomater. Sci. Eng. 2018, 4, 2134–2142. [Google Scholar] [CrossRef]
- Goodarzi, S.; Da Ros, T.; Conde, J.; Sefat, F.; Mozafari, M. Fullerene: Biomedical engineers get to revisit an old friend. Mater. Today 2017, 20, 460–480. [Google Scholar] [CrossRef]
- Lapin, N.A.; Vergara, L.A.; Mackeyev, Y.; Newton, J.M.; Dilliard, S.A.; Wilson, L.J.; Curley, S.A.; Serda, R.E. Biotransport kinetics and intratumoral biodistribution of malonodiserinolamide-derivatized [60]fullerene in a murine model of breast adenocarcinoma. Int. J. Nanomed. 2017, 12, 8289–8307. [Google Scholar] [CrossRef] [PubMed]
- Grebinyk, A.; Grebinyk, S.; Prylutska, S.; Ritter, U.; Matyshevska, O.; Dandekar, T.; Frohme, M. C60 fullerene accumulation in human leukemic cells and perspectives of led-mediated photodynamic therapy. Free Radic. Biol. Med. 2018, 124, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Thotakura, N.; Sharma, G.; Singh, B.; Kumar, V.; Raza, K. Aspartic acid derivatized hydroxylated fullerenes as drug delivery vehicles for docetaxel: An explorative study. Artif. Cells Nanomed. Biotechnol. 2017, 1–10. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, M.; Visani de Luna, L.A.; Fonseca, L.C.; Giorgio, S.; Alves, O.L. Folic-acid-functionalized graphene oxide nanocarrier: Synthetic approaches, characterization, drug delivery study, and antitumor screening. ACS Appl. Nano Mater. 2018, 1, 922–932. [Google Scholar] [CrossRef]
- Cheng, S.J.; Chiu, H.Y.; Kumar, P.V.; Hsieh, K.Y.; Yang, J.W.; Lin, Y.R.; Shen, Y.C.; Chen, G.Y. Simultaneous drug delivery and cellular imaging using graphene oxide. Biomater. Sci. 2018, 6, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi, M.; Rahmani, F.; Nouranian, S. Molecular simulation of pH-dependent diffusion, loading, and release of doxorubicin in graphene and graphene oxide drug delivery systems. J. Mater. Chem. B 2016, 4, 7441–7451. [Google Scholar] [CrossRef]
- Motlagh, N.S.H.; Parvin, P.; Refahizadeh, M.; Bavali, A. Fluorescence properties of doxorubicin coupled carbon nanocarriers. Appl. Opt. 2017, 56, 7498–7503. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-C.; Zhao, J.-J.; Zhang, R.; Li, H.; Chen, B.; Zhang, L.-L.; Yang, H. Multifunctionalization of graphene and graphene oxide for controlled release and targeted delivery of anticancer drugs. Am. J. Transl. Res. 2017, 9, 5197–5219. [Google Scholar] [PubMed]
- Khang, D.W.; Kang, S.S.; Choi, J.; Nam, T.H. Carbon Nanotube-Based Anti-Cancer Agent Capable of Suppressing Drug Resistance. U.S. Patent 9981042, 29 May 2018. [Google Scholar]
- Comparetti, E.J.; Pedrosa, V.d.A.; Kaneno, R. Carbon nanotube as a tool for fighting cancer. Bioconj. Chem. 2017, 29, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Shen, J.W.; Wang, Q. Molecular dynamics study on DNA nanotubes as drug delivery vehicle for anticancer drugs. Colloids Surf. B Biointerfaces 2017, 153, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Kim, K.; Zhang, J.; Escalada, A.; Tunuguntla, R.; Comolli, L.R.; Allen, F.I.; Shnyrova, A.V.; Cho, K.R.; Munoz, D. Stochastic transport through carbon nanotubes in lipid bilayers and live cell membranes. Nature 2014, 514, 612–615. [Google Scholar] [CrossRef] [PubMed]
- Guven, A.; Villares, G.J.; Hilsenbeck, S.G.; Lewis, A.; Landua, J.D.; Dobrolecki, L.E.; Wilson, L.J.; Lewis, M.T. Carbon nanotube capsules enhance the in vivo efficacy of cisplatin. Acta Biomater. 2017, 58, 466–478. [Google Scholar] [CrossRef] [PubMed]
- Karthik, R.; Sasikumar, R.; Chen, S.-M.; Kumar, J.V.; Elangovan, A.; Muthuraj, V.; Muthukrishnan, P.; Al-Hemaid, F.M.; Ali, M.A.; Elshikh, M.S. A highly sensitive and selective electrochemical determination of non-steroidal prostate anti-cancer drug nilutamide based on f-MWCNT in tablet and human blood serum sample. J. Colloid Interface Sci. 2017, 487, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Chen, E.-Y.; Shen, J.-W.; Wang, Q. Molecular modelling of translocation of biomolecules in carbon nanotubes: Method, mechanism and application. Mol. Simul. 2016, 42, 827–835. [Google Scholar] [CrossRef]
- Augustine, S.; Singh, J.; Srivastava, M.; Sharma, M.; Das, A.; Malhotra, B.D. Recent advances in carbon based nanosystems for cancer theranostics. Biomater. Sci. 2017, 5, 901–952. [Google Scholar] [CrossRef] [PubMed]
- Paleos, C.M.; Tsiourvas, D.; Sideratou, Z.; Tziveleka, L.A. Drug delivery using multifunctional dendrimers and hyperbranched polymers. Expert Opin. Drug Deliv. 2010, 7, 1387–1398. [Google Scholar] [CrossRef] [PubMed]
- Kontoyianni, C.; Sideratou, Z.; Theodossiou, T.; Tziveleka, L.A.; Tsiourvas, D.; Paleos, C.M. A novel micellar pegylated hyperbranched polyester as a prospective drug delivery system for paclitaxel. Macromol. Biosci. 2008, 8, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Bartelmess, J.; Quinn, S.; Giordani, S. Carbon nanomaterials: Multi-functional agents for biomedical fluorescence and Raman imaging. Chem. Soc. Rev. 2015, 44, 4672–4698. [Google Scholar] [CrossRef] [PubMed]
- Paleos, C.M.; Tsiourvas, D.; Sideratou, Z.; Tziveleka, L. Multifunctional dendritic drug delivery systems: Design, synthesis, controlled and triggered release. Curr. Top. Med. Chem. 2008, 8, 1204–1224. [Google Scholar] [CrossRef] [PubMed]
- Aihua, L.; Itaru, H.; Masaki, I.; Haoshen, Z. Poly(acrylic acid)-wrapped multi-walled carbon nanotubes composite solubilization in water: Definitive spectroscopic properties. Nanotechnology 2006, 17, 2845. [Google Scholar]
- Dong, X.; Wei, C.; Liang, J.; Liu, T.; Kong, D.; Lv, F. Thermosensitive hydrogel loaded with chitosan-carbon nanotubes for near infrared light triggered drug delivery. Colloids Surf. B Biointerfaces 2017, 154, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Etika, K.C.; Jochum, F.D.; Cox, M.A.; Schattling, P.; Theato, P.; Grunlan, J.C. Nanotube friendly polyN-isopropylacrylamide). Macromol. Rapid Commun. 2010, 31, 1368–1372. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, D.; Bosi, S.; Melchionna, M.; Da Ros, T.; Marchesan, S. The glitter of carbon nanostructures in hybrid/composite hydrogels for medicinal use. Curr. Top. Med. Chem. 2016, 16, 1976–1989. [Google Scholar] [CrossRef] [PubMed]
- Sideratou, Z.; Agathokleous, M.; Theodossiou, T.A.; Tsiourvas, D. Functionalized hyperbranched polyethylenimines as thermosensitive drug delivery nanocarriers with controlled transition temperatures. Biomacromolecules 2018, 19, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Borhani, D.W.; Shaw, D.E. The future of molecular dynamics simulations in drug discovery. J. Comput.-Aided Mol. Des. 2012, 26, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Discher, D.E.; Ortiz, V.; Srinivas, G.; Klein, M.L.; Kim, Y.; Christian, D.; Cai, S.; Photos, P.; Ahmed, F. Emerging applications of polymersomes in delivery: From molecular dynamics to shrinkage of tumors. Prog. Polym. Sci. 2007, 32, 838–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheong, D.W.; Boon, Y.D. Comparative study of force fields for molecular dynamics simulations of α-glycine crystal growth from solution. Cryst. Growth Des. 2010, 10, 5146–5158. [Google Scholar] [CrossRef]
- Rosano, C.; Viale, M.; Cosimelli, B.; Severi, E.; Gangemi, R.; Ciogli, A.; De Totero, D.; Spinelli, D. ABCB1 structural models, molecular docking, and synthesis of new oxadiazolothiazin-3-one inhibitors. ACS Med. Chem. Lett. 2013, 4, 694–698. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Peng, G.; Li, J.; Liang, L.; Kong, Z.; Wang, H.; Jia, L.; Wang, X.; Zhang, W.; Shen, J.-W. Molecular dynamics study on the configuration and arrangement of doxorubicin in carbon nanotubes. J. Mol. Liq. 2018, 262, 295–301. [Google Scholar] [CrossRef]
- Riniker, S. Fixed-charge atomistic force fields for molecular dynamics simulations in the condensed phase: An overview. J. Chem. Inf. Model. 2018, 58, 565–578. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Roy, S.; Nair, N. Coarse-grained molecular dynamics force-field for polyacrylamide in infinite dilution derived from iterative Boltzmann inversion and martini force-field. J. Phys. Chem. B 2018, 122, 1516–1524. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Lemkul, J.A.; Eastman, P.K.; MacKerell, A.D. Molecular dynamics simulations using the drude polarizable force field on GOUs with openMM: Implementation, validation, and benchmarks. J. Comput. Chem. 2018, 39, 1682–1689. [Google Scholar] [CrossRef] [PubMed]
- Molecular dynamics simulation of self- and mutual diffusion coefficients for confined mixtures. J. Chem. Phys. 2005, 123, 144701. [CrossRef] [PubMed]







© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rezaian, M.; Maleki, R.; Dahri Dahroud, M.; Alamdari, A.; Alimohammadi, M. pH-Sensitive Co-Adsorption/Release of Doxorubicin and Paclitaxel by Carbon Nanotube, Fullerene, and Graphene Oxide in Combination with N-isopropylacrylamide: A Molecular Dynamics Study. Biomolecules 2018, 8, 127. https://doi.org/10.3390/biom8040127
Rezaian M, Maleki R, Dahri Dahroud M, Alamdari A, Alimohammadi M. pH-Sensitive Co-Adsorption/Release of Doxorubicin and Paclitaxel by Carbon Nanotube, Fullerene, and Graphene Oxide in Combination with N-isopropylacrylamide: A Molecular Dynamics Study. Biomolecules. 2018; 8(4):127. https://doi.org/10.3390/biom8040127
Chicago/Turabian StyleRezaian, Milad, Reza Maleki, Mohammad Dahri Dahroud, Abdolmohammad Alamdari, and Milad Alimohammadi. 2018. "pH-Sensitive Co-Adsorption/Release of Doxorubicin and Paclitaxel by Carbon Nanotube, Fullerene, and Graphene Oxide in Combination with N-isopropylacrylamide: A Molecular Dynamics Study" Biomolecules 8, no. 4: 127. https://doi.org/10.3390/biom8040127