Factors That Shape Eukaryotic tRNAomes: Processing, Modification and Anticodon–Codon Use
Abstract
:1. Introduction
2. Eukaryotic tRNAome Expansions
3. Eukaryization of tRNA Genes: Monocistrony and Control by a Separate RNA Polymerase
4. The pre-tRNA Chaperone, La Protein, Is Directly Targeted to RNAP III Transcripts
5. Nuclear pre-tRNA Modification Enzymes Can Also Avert pre-tRNA Misfolding
6. Factors That Would Support Eukaryal tRNAome Expansions
7. Eukaryotic tRNA Anticodon-Sparing
8. Adenosine 34 Anticodon-Sparing
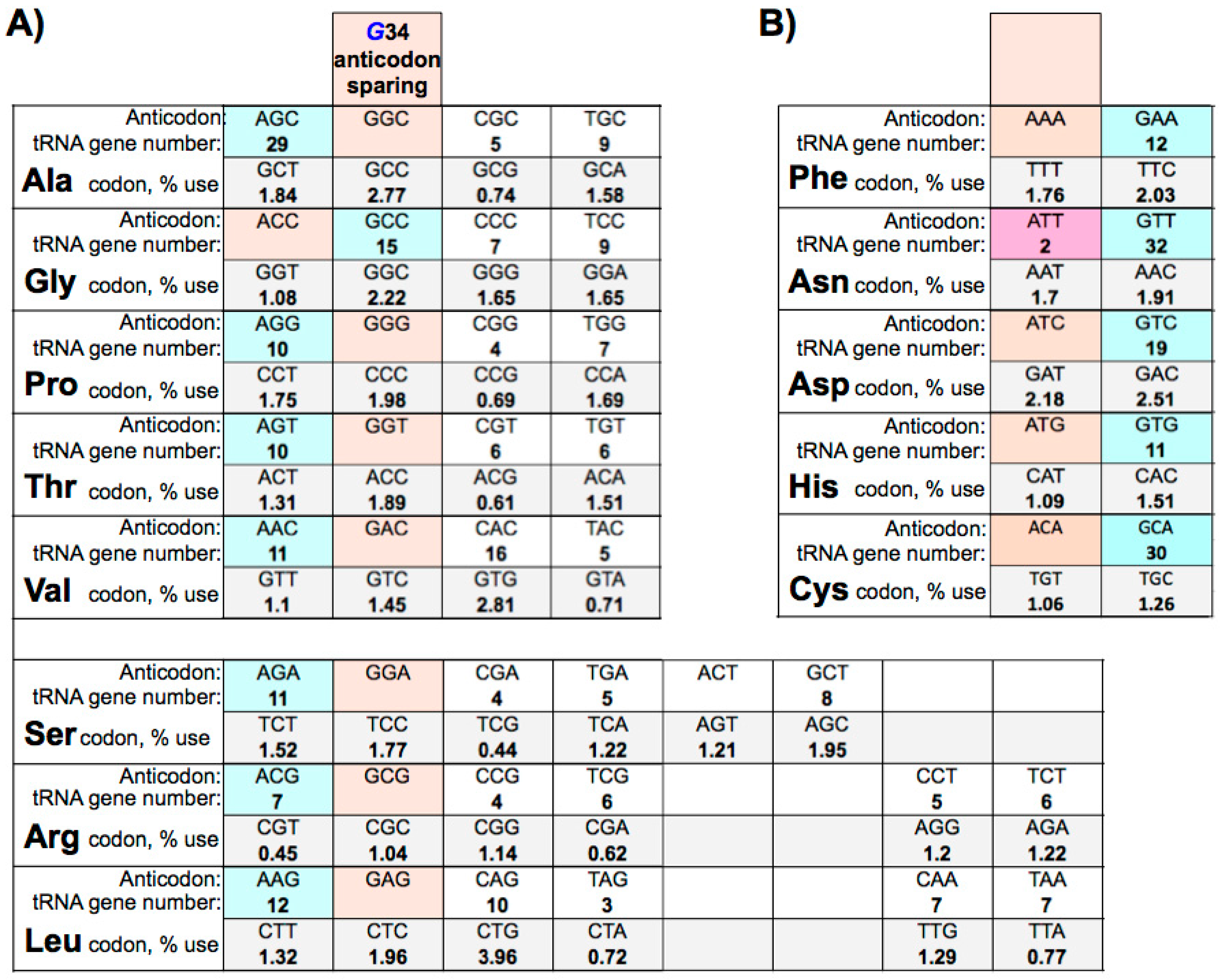
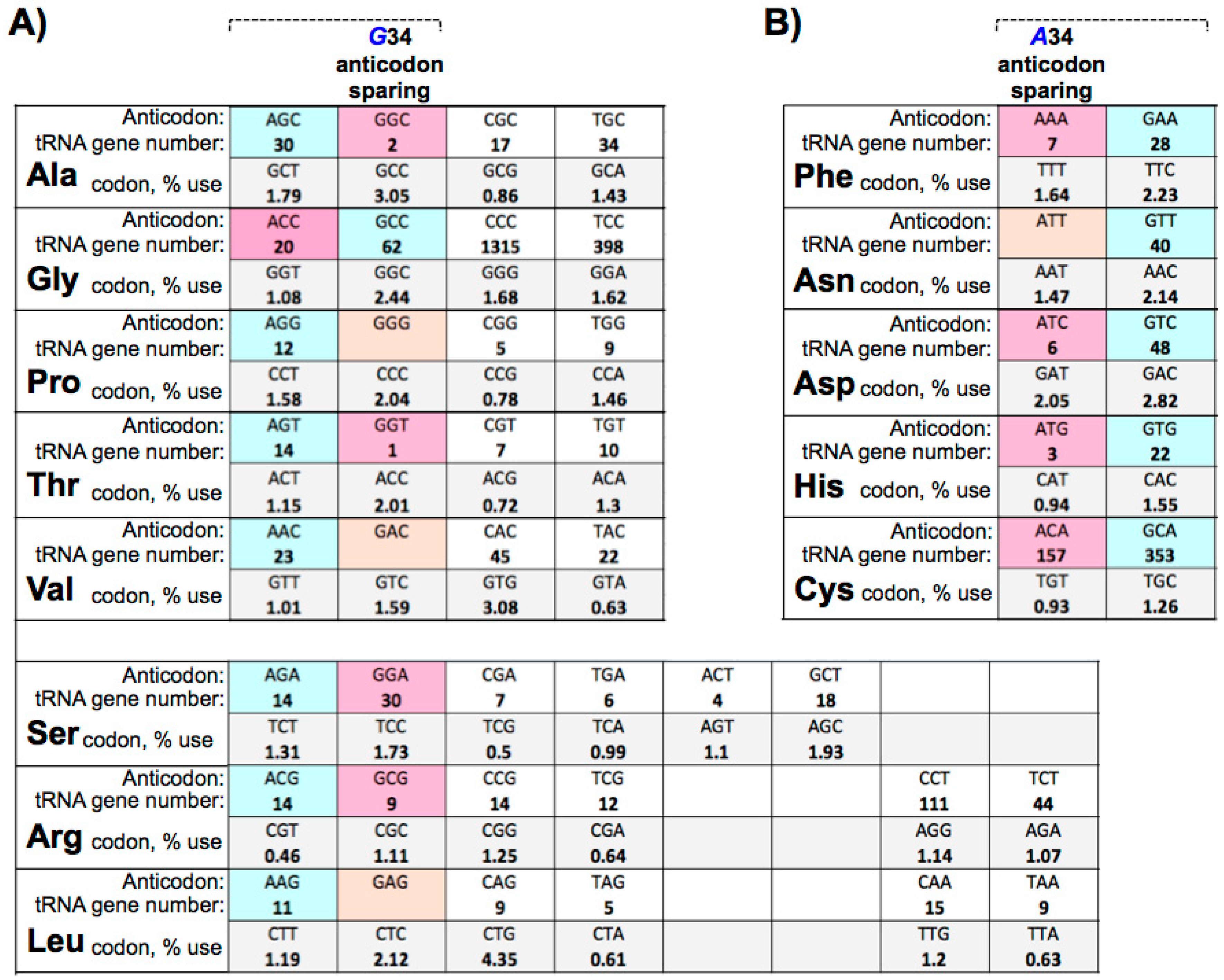
9. Eukaryotic tRNA Guanine 34 Anticodon-Sparing
10. Large tRNAomes Disregard Guanine 34 Anticodon-Sparing and Adenosine 34 tRNA Genes
11. tRNA Adenosine 34 to Inosine May Be Keyed to Differential Synonymous Codon Splitting
12. Targeting Sensitive Synonymous Codons for Cognate Specific Response
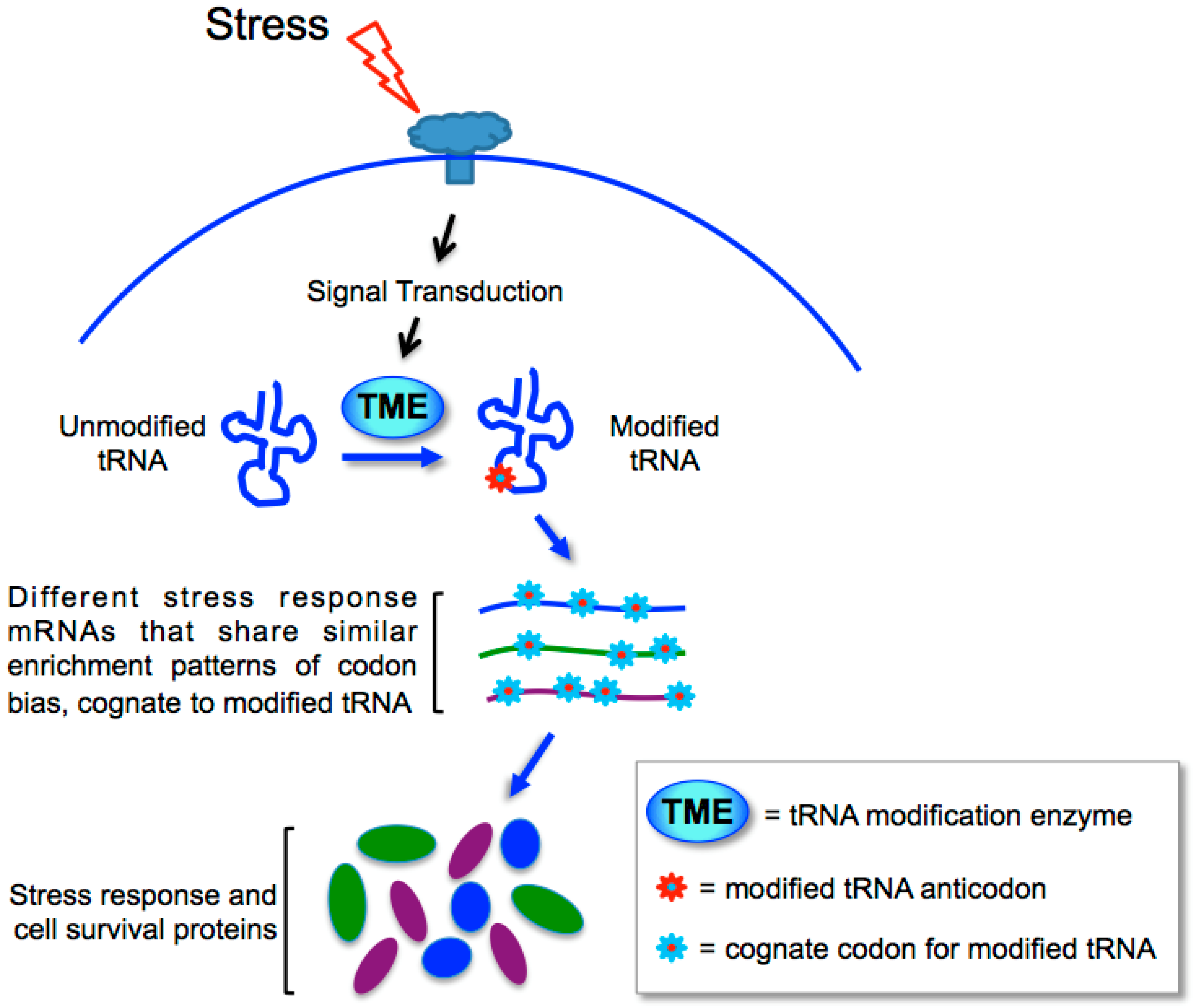
13. Biased Codons Keyed to tRNA Wobble Modification for Programmed Stress Response
14. Deriving Pliable ‘Secondary’ Information from the Redundancy of the Genetic Code
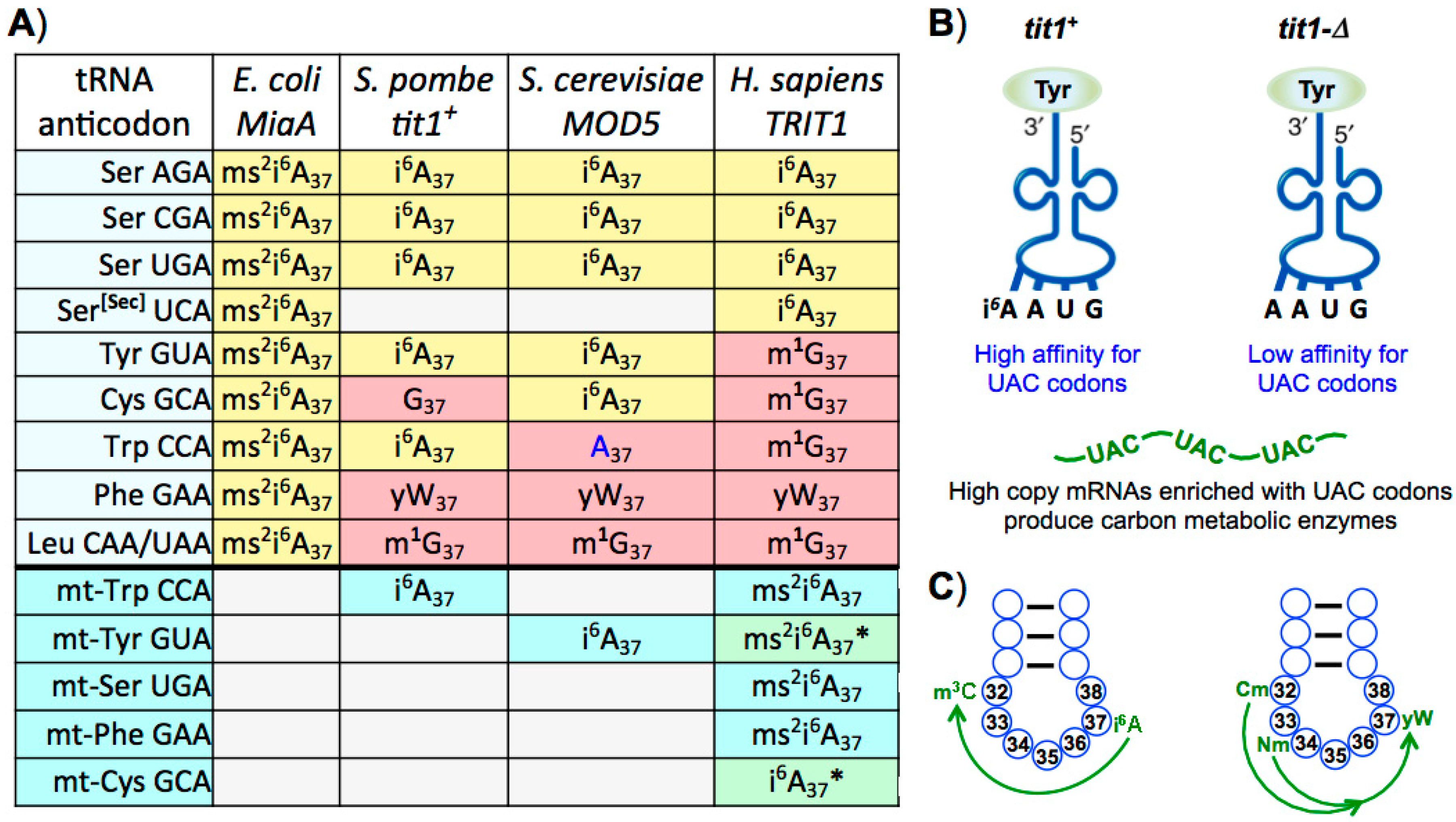
15. Differential Presence and Secondary Code Use of t6A37 and i6A37 Modifications
16. Interdependence of Position 37 Modifications in Eukaryal tRNAs
17. Amplification and Diversification of Eukaryal tRNA Methyltransferases
18. The tRNAs in Health and Disease
19. Conclusions
Conflicts of Interest
References
- Li, S.; Mason, C.E. The pivotal regulatory landscape of RNA modifications. Annu. Rev. Genomics Hum. Genet. 2014, 15, 127–150. [Google Scholar] [CrossRef] [PubMed]
- Helm, M.; Alfonzo, J.D. Posttranscriptional RNA Modifications: Playing Metabolic Games in a Cell’s Chemical Legoland. Chem. Biol. 2014, 21, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.; de Crécy-Lagard, V.; Marck, C. Deciphering synonymous codons in the three domains of life: Co-evolution with specific tRNA modification enzymes. FEBS Lett. 2010, 584, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Novoa, E.M.; Pavon-Eternod, M.; Pan, T.; Ribas de Pouplana, L. A role for tRNA modifications in genome structure and codon usage. Cell 2012, 149, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Marck, C.; Grosjean, H. tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 2002, 8, 1189–1232. [Google Scholar] [CrossRef]
- Saint-Léger, A.; Bello, C.; Dans, P.D.; Torres, A.G.; Novoa, E.M.; Camacho, N.; Orozco, M.; Kondrashov, F.A.; Ribas de Pouplana, L. Saturation of recognition elements blocks evolution of new tRNA identities. Sci Adv. 2016, 2, e1501860. [Google Scholar] [CrossRef] [PubMed]
- Olejniczak, M.; Dale, T.; Fahlman, R.P.; Uhlenbeck, O.C. Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat. Struct. Mol. Biol. 2005, 12, 788–793. [Google Scholar] [CrossRef] [PubMed]
- Ledoux, S.; Uhlenbeck, O.C. Different aa-tRNAs are selected uniformly on the ribosome. Mol. Cell 2008, 31, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Targanski, I.; Cherkasova, V. Analysis of genomic tRNA sets from Bacteria, Archaea, and Eukarya points to anticodon-codon hydrogen bonds as a major determinant of tRNA compositional variations. RNA 2008, 14, 1095–1109. [Google Scholar] [CrossRef] [PubMed]
- Shepotinovskaya, I.; Uhlenbeck, O.C. tRNA residues evolved to promote translational accuracy. RNA 2013, 19, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Bloom-Ackermann, Z.; Navon, S.; Gingold, H.; Towers, R.; Pilpel, Y.; Dahan, O. A comprehensive tRNA deletion library unravels the genetic architecture of the tRNA pool. PLoS Genet. 2014, 10, e1004084. [Google Scholar] [CrossRef] [PubMed]
- Yona, A.H.; Bloom-Ackermann, Z.; Frumkin, I.; Hanson-Smith, V.; Charpak-Amikam, Y.; Feng, Q.; Boeke, J.D.; Dahan, O.; Pilpel, Y. tRNA genes rapidly change in evolution to meet novel translational demands. eLife 2013, 2, e01339. [Google Scholar] [CrossRef] [PubMed]
- Sagi, D.; Rak, R.; Gingold, H.; Adir, I.; Maayan, G.; Dahan, O.; Broday, L.; Pilpel, Y.; Rechavi, O. Tissue- and Time-Specific Expression of Otherwise Identical tRNA Genes. PLoS Genet. 2016, 12, e1006264. [Google Scholar] [CrossRef] [PubMed]
- Ishimura, R.; Nagy, G.; Dotu, I.; Zhou, H.; Yang, X.L.; Schimmel, P.; Senju, S.; Nishimura, Y.; Chuang, J.H.; Ackerman, S.L. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 2014, 345, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Ding, Y.; Niemczyk, M.; Kudla, G.; Plotkin, J.B. Rate-limiting steps in yeast protein translation. Cell 2013, 153, 1589–1601. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, A.R.; Pan, T.; Cluzel, P. Environmental perturbations lift the degeneracy of the genetic code to regulate protein levels in bacteria. Proc. Natl. Acad. Sci. USA 2013, 110, 2419–2424. [Google Scholar] [CrossRef] [PubMed]
- Maraia, R.J.; Iben, J.R. Different types of secondary information in the genetic code. RNA 2014, 20, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Quax, T.E.; Claassens, N.J.; Soll, D.; van der Oost, J. Codon Bias as a Means to Fine-Tune Gene Expression. Mol. Cell 2011, 59, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016, 44, D184–D189. [Google Scholar] [CrossRef] [PubMed]
- Glanzmann, B.; Moller, M.; le Roex, N.; Tromp, G.; Hoal, E.G.; van Helden, P.D. The complete genome sequence of the African buffalo (Syncerus caffer). BMC Genom. 2016, 17, 1001. [Google Scholar] [CrossRef] [PubMed]
- Iben, J.R.; Epstein, J.A.; Bayfield, M.A.; Bruinsma, M.W.; Hasson, S.; Bacikova, D.; Ahmad, D.; Rockwell, D.; Kittler, E.L.W.; Zapp, M.L.; et al. Comparative whole genome sequencing reveals phenotypic tRNA gene duplication in spontaneous Schizosaccharomyces pombe La mutants. Nucleic Acids Res. 2011, 39, 4728–4742. [Google Scholar] [CrossRef] [PubMed]
- Iben, J.R.; Maraia, R.J. Yeast tRNAomics: tRNA gene copy number variation and codon use provide bioinformatics evidence of a new wobble pair in a eukaryote. RNA 2012, 18, 1358–1372. [Google Scholar] [CrossRef]
- Iben, J.R.; Maraia, R.J. tRNA gene copy number variation in humans. Gene 2014, 536, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Parisien, M.; Wang, X.; Pan, T. Diversity of human tRNA genes from the 1000-genomes project. RNA Biol. 2013, 10, 1853–1867. [Google Scholar] [CrossRef] [PubMed]
- Eichinger, L.; Pachebat, J.A.; Glockner, G.; Rajandream, M.A.; Sucgang, R.; Berriman, M.; Song, J.; Olsen, R.; Szafranski, K.; Xu, Q. The genome of the social amoeba Dictyostelium discoideum. Nature 2005, 435, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Goodenbour, J.M.; Pan, T. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006, 34, 6137–6146. [Google Scholar] [CrossRef] [PubMed]
- Inokuchi, H.; Yamao, F. Structure and Expression of Prokaryotic tRNA Genes. In tRNA: Structure, Biosynthesis, and Function; Söll, D., RajBhandary, U.L., Eds.; ASM Press: Washington, DC, USA, 1995. [Google Scholar]
- Thomm, M.; Hausner, W. Genes for Stable RNAs and Their Expression in Archaea. In Genetics and Molecular Biology of Anaerobic Bacteria; Sebald, M., Ed.; Brock Springer Series in Contemporary Bioscience; Springer: New York, NY, USA, 2012. [Google Scholar]
- Werner, F.; Grohmann, D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat. Rev. Microbiol. 2011, 9, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Haag, J.R.; Pikaard, C.S. Multisubunit RNA polymerases IV and V: Purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 2011, 12, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Carter, R.; Drouin, G. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol. Biol. Evol. 2009, 27, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Drouin, G.; Carter, R. Evolution of Eukaryotic RNA Polymerases. eLS 2010. [Google Scholar] [CrossRef]
- French, S.L.; Osheim, Y.N.; Schneider, D.A.; Sikes, M.L.; Fernandez, C.F.; Copela, L.A.; Misra, V.A.; Nomura, M.; Wolin, S.L.; Beyer, A.L. Visual analysis of the yeast 5S rRNA gene transcriptome: Regulation and role of La protein. Mol. Cell Biol. 2008, 28, 4576–4587. [Google Scholar] [CrossRef] [PubMed]
- Hopper, A.K.; Phizicky, E.M. tRNA transfers to the limelight. Genes Dev. 2003, 17, 162–180. [Google Scholar] [CrossRef] [PubMed]
- Phizicky, E.M.; Hopper, A.K. tRNA biology charges to the front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef] [PubMed]
- Arimbasseri, A.G.; Rijal, K.; Maraia, R.J. Transcription termination by the eukaryotic RNA polymerase III. Biochim. Biophys. Acta 2013, 1829, 318–330. [Google Scholar] [CrossRef] [PubMed]
- Arimbasseri, A.G.; Rijal, K.; Maraia, R.J. Comparative overview of RNA polymerase II and III transcription cycles, with focus on RNA polymerase III termination and reinitiation. Transcription 2014, 5, e27639. [Google Scholar] [CrossRef] [PubMed]
- Waldron, C.; Lacroute, F. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J. Bacteriol. 1975, 122, 855–865. [Google Scholar] [PubMed]
- Chedin, S.; Riva, M.; Schultz, P.; Sentenac, A.; Carles, C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998, 12, 3857–3871. [Google Scholar] [CrossRef] [PubMed]
- Arimbasseri, A.G.; Maraia, R.J. Distinguishing core and holoenzyme mechanisms of transcription termination by RNA polymerase III. Mol. Cell. Biol 2013, 33, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Shatalin, K.; Epshtein, V.; Gottesman, M.E.; Nudler, E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell 2011, 146, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Clelland, B.W.; Schultz, M.C. Genome stability control by checkpoint regulation of tRNA gene transcription. Transcription 2010, 1, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.C.; Clelland, B.W.; Hockman, D.J.; Kujat-Choy, S.L.; Mewhort, H.E.; Schultz, M.C. Replication stress checkpoint signaling controls tRNA gene transcription. Nat. Struct. Mol. Biol. 2010, 17, 976–981. [Google Scholar] [CrossRef] [PubMed]
- Bousquet-Antonelli, C.; Deragon, J.M. A comprehensive analysis of the La-motif protein superfamily. RNA 2009, 15, 750–764. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, N.A.; Jakobi, A.J.; Moreno-Morcillo, M.; Glatt, S.; Kosinski, J.; Hagen, W.J.; Sachse, C.; Muller, C.W. Molecular structures of unbound and transcribing RNA polymerase III. Nature 2015, 528, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Maraia, R.J.; Rijal, K. Structural biology: A transcriptional specialist resolved. Nature 2015, 528, 204–205. [Google Scholar] [CrossRef] [PubMed]
- Cozzarelli, N.R.; Gerrard, S.P.; Schlissel, M.; Brown, D.D.; Bogenhagen, D.F. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell 1983, 34, 829–835. [Google Scholar] [CrossRef]
- Arimbasseri, A.G.; Maraia, R.J. Mechanism of Transcription Termination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element. Mol. Cell 2015, 58, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
- Arimbasseri, A.G.; Maraia, R.J. A high density of cis-information terminates RNA Polymerase III on a 2-rail track. RNA Biol. 2016, 13, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Bogenhagen, D.F.; Brown, D.D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell 1981, 24, 261–270. [Google Scholar] [CrossRef]
- Rijal, K.; Maraia, R.J.; Arimbasseri, A.G. A methods review on use of nonsense suppression to study 3′ end formation and other aspects of tRNA biogenesis. Gene 2015, 556, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Fairley, J.A.; Kantidakis, T.; Kenneth, N.S.; Intine, R.V.; Maraia, R.J.; White, R.J. Human La is Found at RNA Polymerase III-Transcribed Genes In Vivo. Proc. Natl. Acad. Sci. USA 2005, 102, 18350–18355. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Luo, T.; Roeder, R.G. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 1997, 11, 2371–2382. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Intine, R.V.; Mozlin, A.; Hasson, S.; Maraia, R.J. Mutations in the RNA Polymerase III Subunit Rpc11p That Decrease RNA 3′ Cleavage Activity Increase 3′-Terminal Oligo(U) Length and La-Dependent tRNA Processing. Mol. Cell Biol. 2005, 25, 621–636. [Google Scholar] [CrossRef] [PubMed]
- Stefano, J.E. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 1984, 36, 145–154. [Google Scholar] [CrossRef]
- Bayfield, M.A.; Maraia, R.J. Precursor-product discrimination by La protein during tRNA metabolism. Nat. Struct. Mol. Biol. 2009, 16, 430–437. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.; Phan, L.; Cuesta, R.; Carlson, B.A.; Pak, M.; Asano, K.; Björk, G.R.; Tamame, M.; Hinnebusch, A.G. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 1998, 12, 3650–3662. [Google Scholar] [CrossRef] [PubMed]
- Copela, L.A.; Chakshusmathi, G.; Sherrer, R.L.; Wolin, S.L. The La protein functions redundantly with tRNA modification enzymes to ensure tRNA structural stability. RNA 2006, 12, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Maraia, R.J.; Lamichhane, T.N. 3′ processing of eukaryotic precursor tRNAs. WIRES RNA 2011, 2, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Helm, M. tRNA stabilization by modified nucleotides. Biochemistry 2010, 49, 4934–4944. [Google Scholar] [CrossRef] [PubMed]
- Bayfield, M.A.; Yang, R.; Maraia, R.J. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim. Biophys. Acta 2010, 1799, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Bayfield, M.A.; Intine, R.V.; Maraia, R.J. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat. Struct. Mol. Biol. 2006, 13, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Chakshusmathi, G.; Kim, S.D.; Rubinson, D.A.; Wolin, S.L. A La protein requirement for efficient pre-tRNA folding. EMBO J. 2003, 22, 6562–6572. [Google Scholar] [CrossRef] [PubMed]
- Kucera, N.J.; Hodsdon, M.E.; Wolin, S.L. An intrinsically disordered C terminus allows the La protein to assist the biogenesis of diverse noncoding RNA precursors. Proc. Natl. Acad. Sci. USA 2011, 108, 1308–1313. [Google Scholar] [CrossRef] [PubMed]
- Naeeni, A.R.; Conte, M.R.; Bayfield, M.A. RNA chaperone activity of the human La protein is mediated by a variant RNA recognition motif. J. Biol. Chem. 2012, 287, 5472–5482. [Google Scholar] [CrossRef] [PubMed]
- Wolin, S.L.; Cedervall, T. The La protein. Annu. Rev. Biochem. 2002, 71, 375–403. [Google Scholar] [CrossRef] [PubMed]
- Wolin, S.L.; Wurtmann, E.J. Molecular chaperones and quality control in noncoding RNA biogenesis. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Yoo, C.J.; Wolin, S.L. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 1997, 89, 393–402. [Google Scholar] [CrossRef]
- Pannone, B.; Xue, D.; Wolin, S.L. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998, 17, 7442–7453. [Google Scholar] [CrossRef] [PubMed]
- Xue, D.; Rubinson, D.A.; Pannone, B.K.; Yoo, C.J.; Wolin, S.L. U snRNP assembly in yeast involves the La protein. EMBO J. 2000, 19, 1650–1660. [Google Scholar] [CrossRef] [PubMed]
- Copela, L.A.; Fernandez, C.F.; Sherrer, R.L.; Wolin, S.L. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA 2008, 14, 1214–1227. [Google Scholar] [CrossRef] [PubMed]
- Leung, E.; Schneider, C.; Yan, F.; Mohi-El-Din, H.; Kudla, G.; Tuck, A.; Wlotzka, W.; Doronina, V.; Bartley, R.; Watkins, N.J.; et al. Integrity of SRP RNA is ensured by La and the nuclear RNA quality control machinery. Nucleic Acids Res. 2014, 42, 10698–10710. [Google Scholar] [CrossRef] [PubMed]
- Hasler, D.; Lehmann, G.; Murakawa, Y.; Klironomos, F.; Jakob, L.; Grässer, F.A.; Rajewsky, N.; Landthaler, M.; Meister, G. The Lupus Autoantigen La Prevents Mis-channeling of tRNA Fragments into the Human MicroRNA Pathway. Mol. Cell 2016, 63, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Steitz, J.A.; Berg, C.; Hendrick, J.P.; La Branche-Chabot, H.; Metspalu, A.; Rinke, J.; Yario, T. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J. Cell Biol. 1998, 106, 545–556. [Google Scholar] [CrossRef]
- Belisova, A.; Semrad, K.; Mayer, O.; Kocian, G.; Waigmann, E.; Schroeder, R.; Steiner, G. RNA chaperone activity of protein components of human Ro RNPs. RNA 2005, 11, 1084–1094. [Google Scholar] [CrossRef] [PubMed]
- Hopper, A.K.; Huang, H.Y. Quality Control Pathways for Nucleus-Encoded Eukaryotic tRNA Biosynthesis and Subcellular Trafficking. Mol. Cell. Biol. 2015, 35, 2052–2058. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.J.; Bystrom, A.S. Dual function of the tRNA(m5U54)methyltransferase in tRNA maturation. RNA 2002, 8, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Kadaba, S.; Krueger, A.; Trice, T.; Krecic, A.M.; Hinnebusch, A.G.; Anderson, J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004, 18, 1227–1240. [Google Scholar] [CrossRef] [PubMed]
- Kadaba, S.; Wang, X.; Anderson, J.T. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 2006, 12, 508–521. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Hu, C.; Anderson, J.; Björk, G.; Sarkar, S.; Hopper, A.; Hinnebusch, A.G. Defects in tRNA Processing and Nuclear Export Induce GCN4 Translation Independently of Phosphorylation of the α Subunit of Eukaryotic Translation Initiation Factor 2. Mol. Cell. Biol. 2000, 20, 2505–2516. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Dahlberg, J.E. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 1998, 282, 2082–2085. [Google Scholar] [CrossRef] [PubMed]
- Jarosz, D.F.; Taipale, M.; Lindquist, S. Protein homeostasis and the phenotypic manifestation of genetic diversity: Principles and mechanisms. Annu. Rev. Genet. 2010, 44, 189–216. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.L.; Lindquist, S. Hsp90 as a capacitor for morphological evolution. Nature 1998, 396, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Arimbasseri, A.G.; Maraia, R.J. RNA Polymerase III Advances: Structural and tRNA Functional Views. Trends Biochem. Sci. 2016, 41, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Marck, C.; Kachouri-Lafond, R.; Lafontaine, I.; Westhof, E.; Dujon, B.; Grosjean, H. The RNA polymerase III-dependent family of genes in hemiascomycetes: Comparative RNomics, decoding strategies, transcription and evolutionary implications. Nucleic Acids Res. 2006, 34, 1816–1835. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.G.; Piñeyro, D.; Filonava, L.; Stracker, T.H.; Batlle, E.; Ribas de Pouplana, L. A-to-I editing on tRNAs: biochemical, biological and evolutionary implications. FEBS Lett. 2014, 588, 4279–4286. [Google Scholar] [CrossRef] [PubMed]
- Haumont, E.; Fournier, M.; de Henau, S.; Grosjean, H. Enzymatic conversion of adenosine to inosine in the wobble position of yeast tRNAAsp: the dependence on the anticodon sequence. Nucleic Acids Res. 1984, 12, 2705–2715. [Google Scholar] [CrossRef] [PubMed]
- Auxilien, S.; Crain, P.F.; Trewyn, R.W.; Grosjean, H. Mechanism, specificity and general properties of the yeast enzyme catalysing the formation of inosine 34 in the anticodon of transfer RNA. J. Mol. Biol. 1996, 262, 437–458. [Google Scholar] [CrossRef] [PubMed]
- Ohira, T.; Suzuki, T. Retrograde nuclear import of tRNA precursors is required for modified base biogenesis in yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 10502–10507. [Google Scholar] [CrossRef] [PubMed]
- Droogmans, L.; Grosjean, H. Enzymatic conversion of guanosine 3′ adjacent to the anticodon of yeast tRNAPhe to N1-methylguanosine and the wye nucleoside: Dependence on the anticodon sequence. EMBO J. 1987, 6, 477–483. [Google Scholar] [PubMed]
- Agris, P.F.; Vendeix, F.A.; Graham, W.D. tRNA’s wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007, 366, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Begley, U.; Dyavaiah, M.; Patil, A.; Rooney, J.P.; Direnzo, D.; Young, C.M.; Conklin, D.S.; Zitomer, R.S.; Begley, T.J. Trm9-Catalyzed tRNA Modifications Link Translation to the DNA Damage Response. Mol. Cell 2007, 28, 860–870. [Google Scholar] [CrossRef] [PubMed]
- Arimbasseri, A.G.; Blewett, N.H.; Iben, J.R.; Lamichhane, T.N.; Cherkasova, V.; Hafner, M.; Maraia, R.J. RNA Polymerase III Output Is Functionally Linked to tRNA Dimethyl-G26 Modification. PLoS Genet. 2015, 11, e1005671. [Google Scholar] [CrossRef] [PubMed]
- Martignetti, J.A.; Brosius, J. Neural BC1 RNA as an evolutionary marker: Guinea pig remains a rodent. Proc. Natl. Acad. Sci. USA 1993, 90, 9698–9702. [Google Scholar] [CrossRef]
- Rozhdestvensky, T.S.; Kopylov, A.M.; Brosius, J.; Huttenhofer, A. Neuronal BC1 RNA structure: evolutionary conversion of a tRNAAla domain into an extended stem-loop structure. RNA 2001, 7, 722–730. [Google Scholar] [CrossRef] [PubMed]
- Robeck, T.; Skryabin, B.V.; Rozhdestvensky, T.S.; Skryabin, A.B.; Brosius, J. BC1 RNA motifs required for dendritic transport in vivo. Sci Rep. 2016, 6, 28300. [Google Scholar] [CrossRef] [PubMed]
- Saint-Léger, A.; Ribas de Pouplana, L. The importance of codon-anticodon interactions in translation elongation. Biochimie 2015, 114, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.G.; Piñeyro, D.; Rodriguez-Escribà, M.; Camacho, N.; Reina, O.; Saint-Léger, A.; Filonava, L.; Batlle, E.; Ribas de Pouplana, L. Inosine modifications in human tRNAs are incorporated at the precursor tRNA level. Nucleic Acids Res. 2015, 43, 5145–5157. [Google Scholar] [CrossRef] [PubMed]
- Gerber, A.P.; Keller, W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 1999, 286, 1146–1149. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, M.A.; Olchowik, A.; Grosjean, H.; Bujnicki, J.M. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol. 2014, 11, 1619–1629. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, T.N.; Arimbasseri, A.G.; Rijal, K.; Iben, J.R.; Wei, F.Y.; Tomizawa, K.; Maraia, R.J. Lack of tRNA-i6A modification causes mitochondrial-like metabolic deficiency in S. pombe by limiting activity of cytosolic tRNATyr, not mito-tRNA. RNA 2016, 22, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Pechmann, S.; Frydman, J. Evolutionary conservation of codon optimality reveals hidden signatures of cotranslational folding. Nat. Struct. Mol. Biol. 2013, 20, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Marguerat, S.; Schmidt, A.; Codlin, S.; Chen, W.; Aebersold, R.; Bahler, J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 2012, 151, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Elf, J.; Nilsson, D.; Tenson, T.; Ehrenberg, M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science 2003, 300, 1718–1722. [Google Scholar] [CrossRef] [PubMed]
- Elf, J.; Ehrenberg, M. What makes ribosome-mediated transcriptional attenuation sensitive to amino acid limitation? PLoS Comput. Biol. 2005, 1, e2. [Google Scholar] [CrossRef] [PubMed]
- Chionh, Y.H.; McBee, M.; Babu, I.R.; Hia, F.; Lin, W.; Zhao, W.; Cao, J.; Dziergowska, A.; Malkiewicz, A.; Begley, T.J.; et al. tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 2016, 7, 13302. [Google Scholar] [CrossRef] [PubMed]
- Dirheimer, G.; Keith, G.; Dumas, P.; Westhof, E. Primary, Secondary and Tertiary Structures of tRNAs. In tRNA: Structure, biosynthesis, and function; Söll, D., RajBhandary, U.L., Eds.; ASM Press: Washington, DC, USA, 1995. [Google Scholar]
- Yokoyama, S.; Nishimura, S. Modified Nucleosides and Codon Recognition. In tRNA: Structure, biosynthesis, and function; Söll, D., RajBhandary, U.L., Eds.; ASM Press: Washington, DC, USA, 1995; pp. 207–223. [Google Scholar]
- Agris, P.F. Decoding the genome: A modified view. Nucleic Acids Res. 2004, 32, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.J.; Esberg, A.; Huang, B.; Bjork, G.R.; Bystrom, A.S. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell Biol. 2008, 28, 3301–3312. [Google Scholar] [CrossRef] [PubMed]
- Kalhor, H.R.; Clarke, S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell Biol. 2003, 23, 9283–9292. [Google Scholar] [CrossRef] [PubMed]
- Weissenbach, J.; Dirheimer, G. Pairing properties of the methylester of 5-carboxymethyl uridine in the wobble position of yeast tRNA3Arg. Biochim. Biophys. Acta 1978, 518, 530–534. [Google Scholar] [CrossRef]
- Maraia, R.J.; Blewett, N.H.; Bayfield, M.A. It’s a mod mod tRNA world. Nat. Chem. Biol. 2008, 4, 162–164. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.Y.; Dyavaiah, M.; DeMott, M.S.; Taghizadeh, K.; Dedon, P.C.; Begley, T.J. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010, 6, e1001247. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.Y.; Pang, Y.L.J.; Deng, W.; Babu, I.R.; Dyavaiah, M.; Begley, T.J.; Dedon, P.C. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012, 3, 937. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Vázquez, J.; Vargas-Pérez, I.; Sansó, M.; Buhne, K.; Carmona, M.; Paulo, E.; Hermand, D.; Rodríguez-Gabriel, M.; Ayté, J.; Leidel, S.; et al. Modification of tRNALys UUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet. 2013, 9, e1003647. [Google Scholar] [CrossRef] [PubMed]
- Endres, L.; Dedon, P.C.; Begley, T.J. Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol. 2015, 12, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Dedon, P.C.; Begley, T.J. A System of RNA Modifications and Biased Codon Use Controls Cellular Stress Response at the Level of Translation. Chem Res Toxicol. 2014, 27, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Begley, T.J.; Dedon, P.C. tRNA modifications regulate translation during cellular stress. FEBS Lett. 2014, 588, 4287–4296. [Google Scholar] [CrossRef] [PubMed]
- Yarus, M. Translational efficiency of transfer RNA’s: Uses of an extended anticodon. Science 1982, 218, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Bjork, G.R.; Hagervall, T.G. Transfer RNA Modification: Presence, Synthesis, and Function. EcoSal Plus 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, K.; Kimura, S.; Suzuki, T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 2013, 9, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Thiaville, P.C.; Iwata-Reuyl, D.; de Crécy-Lagard, V. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t6A), a universal modification of tRNA. RNA Biol. 2014, 11, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Thiaville, P.C.; Legendre, R.; Rojas-Benítez, D.; Baudin-Baillieu, A.; Hatin, I.; Chalancon, G.; Glavic, A.; Namy, O.; de Crécy-Lagard, V. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb. Cell 2016, 3, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Thiaville, P.C.; El Yacoubi, B.; Köhrer, C.; Thiaville, J.J.; Deutsch, C.; Iwata-Reuyl, D.; Bacusmo, J.M.; Armengaud, J.; Bessho, Y.; Wetzel, C.; et al. Essentiality of threonylcarbamoyladenosine (t6A), a universal tRNA modification, in bacteria. Mol. Microbiol. 2015, 98, 1199–1221. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, T.N.; Blewett, N.H.; Maraia, R.J. Plasticity and diversity of tRNA anticodon determinants of substrate recognition by eukaryotic A37 isopentenyltransferases. RNA 2011, 17, 1846–1857. [Google Scholar] [CrossRef] [PubMed]
- Juhling, F.; Morl, M.; Hartmann, R.K.; Sprinzl, M.; Stadler, P.F.; Putz, J. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009, 37, D159–D162. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, M.A.; Milanowska, K.; Osman Oglou, O.; Purta, E.; Kurkowska, M.; Olchowik, A.; Januszewski, W.; Kalinowski, S.; Dunin-Horkawicz, S.; Rother, K.M.; et al. MODOMICS: A database of RNA modification pathways--2013 update. Nucleic Acids Res. 2013, 41, D262–D267. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, T.N.; Mattijssen, S.; Maraia, R.J. Human cells have a limited set of tRNA anticodon loop substrates of the tRNA isopentenyltransferase TRIT1 tumor suppressor. Mol. Cell Biol. 2013, 33, 4900–4908. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Suzuki, T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014, 42, 7346–7357. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, T.N.; Blewett, N.H.; Crawford, A.K.; Cherkasova, V.A.; Iben, J.R.; Begley, T.J.; Farabaugh, P.J.; Maraia, R.J. Lack of tRNA modification isopentenyl-A37 alters mRNA decoding and causes metabolic deficiencies in fission yeast. Mol. Cell Biol. 2013, 33, 2918–2929. [Google Scholar] [CrossRef] [PubMed]
- Arimbasseri, A.G.; Iben, J.; Wei, F.Y.; Rijal, K.; Tomizawa, K.; Hafner, M.; Maraia, R.J. Evolving specificity of tRNA 3-methyl-cytidine-32 (m3C32) modification: A subset of tRNAsSer requires N6-isopentenylation of A37. RNA 2016, 22, 1400–1410. [Google Scholar] [CrossRef] [PubMed]
- Guy, M.P.; Phizicky, E.M. Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. RNA 2015, 21, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Guy, M.P.; Shaw, M.; Weiner, C.L.; Hobson, L.; Stark, Z.; Rose, K.; Kalscheuer, V.M.; Gecz, J.; Phizicky, E.M. Defects in tRNA Anticodon Loop 2′-O-Methylation Are Implicated in Nonsyndromic X-Linked Intellectual Disability due to Mutations in FTSJ1. Hum. Mutat. 2015, 36, 1176–1187. [Google Scholar] [CrossRef] [PubMed]
- Aubee, J.I.; Olu, M.; Thompson, K.M. The i6A37 tRNA modification is essential for proper decoding of UUX-Leucine codons during rpoS and iraP translation. RNA 2016, 22, 729–742. [Google Scholar] [CrossRef] [PubMed]
- Thompson, K.M.; Gottesman, S. The MiaA tRNA Modification Enzyme Is Necessary for Robust RpoS Expression in Escherichia coli. J. Bacteriol. 2014, 196, 754–761. [Google Scholar] [CrossRef] [PubMed]
- Guy, M.P.; Phizicky, E.M. Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification. RNA Biol. 2014, 11, 1608–1618. [Google Scholar] [CrossRef] [PubMed]
- Noma, A.; Kirino, Y.; Ikeuchi, Y.; Suzuki, T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006, 25, 2142–2154. [Google Scholar] [CrossRef] [PubMed]
- Morl, M.; Dorner, M.; Paabo, S. C to U editing and modifications during the maturation of the mitochondrial tRNAAsp in marsupials. Nucleic Acids Res. 1995, 23, 3380–3384. [Google Scholar] [CrossRef] [PubMed]
- Tomikawa, C.; Yokogawa, T.; Kanai, T.; Hori, H. N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability at high temperatures through a tRNA modification network. Nucleic Acids Res. 2010, 38, 942–957. [Google Scholar] [CrossRef] [PubMed]
- Ishida, K.; Kunibayashi, T.; Tomikawa, C.; Ochi, A.; Kanai, T.; Hirata, A.; Iwashita, C.; Hori, H. Pseudouridine at position 55 in tRNA controls the contents of other modified nucleotides for low-temperature adaptation in the extreme-thermophilic eubacterium Thermus thermophilus. Nucleic Acids Res. 2011, 39, 2304–2318. [Google Scholar] [CrossRef] [PubMed]
- Rubio, M.A.; Ragone, F.L.; Gaston, K.W.; Ibba, M.; Alfonzo, J.D. C to U Editing Stimulates A to I Editing in the Anticodon Loop of a Cytoplasmic Threonyl tRNA in Trypanosoma brucei. J. Biol. Chem. 2006, 281, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Hartmann, M.; Schuster, I.; Bender, S.; Thüring, K.L.; Helm, M.; Katze, J.R.; Nellen, W.; Lyko, F.; Ehrenhofer-Murray, A.E. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res. 2015, 43, 10952–10962. [Google Scholar] [CrossRef] [PubMed]
- Kernohan, K.D.; Dyment, D.A.; Pupavac, M.; Cramer, Z.; McBride, A.; Bernard, G.; Straub, I.; Tetreault, M.; Hartley, T.; Huang, L.; et al. Matchmaking facilitates the diagnosis of an autosomal-recessive mitochondrial disease caused by biallelic mutation of the tRNA isopentenyltransferase (TRIT1) gene. Hum Mutat. 2017. [Google Scholar] [CrossRef] [PubMed]
- Yarham, J.W.; Lamichhane, T.N.; Pyle, A.; Mattijssen, S.; Baruffini, E.; Bruni, F.; Donnini, C.; Vassilev, A.; He, L.; Blakely, E.L.; et al. Defective i6A37 Modification of Mitochondrial and Cytosolic tRNAs Results from Pathogenic Mutations in TRIT1 and Its Substrate tRNA. PLoS Genet. 2014, 10, e1004424. [Google Scholar] [CrossRef] [PubMed]
- D’Silva, S.; Haider, S.J.; Phizicky, E.M. A domain of the actin binding protein Abp140 is the yeast methyltransferase responsible for 3-methylcytidine modification in the tRNA anti-codon loop. RNA 2011, 17, 1100–1110. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Marcus, E.; D’Silva, S.; Phizicky, E.M. S. cerevisiae Trm140 has two recognition modes for 3-methylcytidine modification of the anticodon loop of tRNA substrates. RNA 2016, 406–419. [Google Scholar] [CrossRef] [PubMed]
- Swinehart, W.E.; Jackman, J.E. Diversity in mechanism and function of tRNA methyltransferases. RNA Biol. 2015, 12, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Towns, W.L.; Begley, T.J. Transfer RNA methytransferases and their corresponding modifications in budding yeast and humans: Activities, predications, and potential roles in human health. DNA Cell Biol. 2012, 31, 434–454. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Noma, A.; Yi, S.; Katoh, T.; Takai, Y.; Suzuki, T.T.; Suzuki, T.T. Actin-binding protein ABP140 is a methyltransferase for 3-methylcytidine at position 32 of tRNAs in Saccharomyces cerevisiae. RNA 2011, 17, 1111–1119. [Google Scholar] [CrossRef] [PubMed]
- Farabaugh, P.J.; Kramer, E.; Vallabhaneni, H.; Raman, A. Evolution of +1 Programmed Frameshifting Signals and Frameshift-Regulating tRNAs in the Order Saccharomycetales. J. Mol. Evol. 2006, 63, 545–561. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Grosjean, H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: Identification of the gene and substrate specificity of the enzyme. RNA 1999, 5, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Dietmann, S.; Flores, J.V.; Hussain, S.; Kutter, C.; Humphreys, P.; Lukk, M.; Lombard, P.; Treps, L.; Popis, M.; et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014, 33, 2020–2039. [Google Scholar] [CrossRef] [PubMed]
- Haag, S.; Warda, A.S.; Kretschmer, J.; Gunnigmann, M.A.; Hobartner, C.; Bohnsack, M.T. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA 2015, 21, 1532–1543. [Google Scholar] [CrossRef] [PubMed]
- Pintard, L.; Lecointe, F.; Bujnicki, J.M.; Bonnerot, C.; Grosjean, H.; Lapeyre, B. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J. 2002, 21, 1811–1820. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.G.; Batlle, E.; Ribas de Pouplana, L. Role of tRNA modifications in human diseases. Trends Mol. Med. 2014, 20, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.W.; Chen, H.L.; Lin, K.Y.; Liu, F.C.; Chong, K.Y.; Cheng, W.T.; Chen, C.M. FTSJ2, a heat shock-inducible mitochondrial protein, suppresses cell invasion and migration. PLoS One 2014, 9, e90818. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.; Okot-Kotber, C.; LaComb, J.F.; Bogenhagen, D.F. Mitochondrial ribosomal RNA (rRNA) methyltransferase family members are positioned to modify nascent rRNA in foci near the mitochondrial DNA nucleoid. J. Biol. Chem. 2013, 288, 31386–31399. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, Q.; Huang, Y. Partitioning of the nuclear and mitochondrial tRNA 3′-end processing activities between two different proteins in Schizosaccharomyces pombe. J. Biol. Chem. 2013, 288, 27415–27422. [Google Scholar] [CrossRef] [PubMed]
- Dittmar, K.A.; Goodenbour, J.M.; Pan, T. Tissue-Specific Differences in Human Transfer RNA Expression. PLoS Genet. 2006, 2, e221. [Google Scholar] [CrossRef] [PubMed]
- Ishimura, R.; Nagy, G.; Dotu, I.; Chuang, J.H.; Ackerman, S.L. Activation of GCN2 kinase by ribosome stalling links translation elongation with translation initiation. eLife 2016, 5, e14295. [Google Scholar] [CrossRef] [PubMed]
- Jackman, J.E.; Montange, R.K.; Malik, H.S.; Phizicky, E.M. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA 2003, 9, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Vilardo, E.; Nachbagauer, C.; Buzet, A.; Taschner, A.; Holzmann, J.; Rossmanith, W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase—extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012, 40, 11583–11593. [Google Scholar] [CrossRef] [PubMed]
- Metodiev, M.D.; Thompson, K.; Alston, C.L.; Morris, A.A.; He, L.; Assouline, Z.; Rio, M.; Bahi-Buisson, N.; Pyle, A.; Griffin, H.; et al. Recessive Mutations in TRMT10C Cause Defects in Mitochondrial RNA Processing and Multiple Respiratory Chain Deficiencies. Am. J. Human Genet. 2016, 99, 246. [Google Scholar] [CrossRef] [PubMed]
- Igoillo-Esteve, M.; Genin, A.; Lambert, N.; Desir, J.; Pirson, I.; Abdulkarim, B.; Simonis, M.; Drielsma, A.; Marselli, L.; Marchetti, P.; et al. tRNA methyltransferase homolog gene TRMT10A mutation in young onset diabetes and primary microcephaly in humans. PLoS Genet. 2013, 9, e1003888. [Google Scholar] [CrossRef] [PubMed]
- Gillis, D.; Krishnamohan, A.; Yaacov, B.; Shaag, A.; Jackman, J.E.; Elpeleg, O. TRMT10A dysfunction is associated with abnormalities in glucose homeostasis, short stature and microcephaly. J. Med. Genet. 2014, 51, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, M.; Ramsey, K.; Grebe, T.; Schrauwen, I.; Szelinger, S.; Huentelman, M.; Craig, D.; Narayanan, V. Case Report: Compound heterozygous nonsense mutations in TRMT10A are associated with microcephaly, delayed development, and periventricular white matter hyperintensities. F1000Research 2015, 4, 912. [Google Scholar] [CrossRef] [PubMed]
- Yew, T.W.; McCreight, L.; Colclough, K.; Ellard, S.; Pearson, E.R. tRNA methyltransferase homologue gene TRMT10A mutation in young adult-onset diabetes with intellectual disability, microcephaly and epilepsy. Diabet. Med. 2016, 33, e21–e25. [Google Scholar] [CrossRef] [PubMed]
- Zung, A.; Kori, M.; Burundukov, E.; Ben-Yosef, T.; Tatoor, Y.; Granot, E. Homozygous deletion of TRMT10A as part of a contiguous gene deletion in a syndrome of failure to thrive, delayed puberty, intellectual disability and diabetes mellitus. Am. J. Med. Genet. Part A 2015, 167A, 3167–3173. [Google Scholar] [CrossRef] [PubMed]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Goll, M.G.; Bestor, T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005, 74, 481–514. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, S.; Jurkowski, T.P.; Kellner, S.; Schneider, D.; Jeltsch, A.; Helm, M. The RNA methyltransferase Dnmt2 methylates DNA in the structural context of a tRNA. RNA Biol. 2016, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Durdevic, Z.; Mobin, M.B.; Hanna, K.; Lyko, F.; Schaefer, M. The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep. 2013, 4, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Elhardt, W.; Shanmugam, R.; Jurkowski, T.P.; Jeltsch, A. Somatic cancer mutations in the DNMT2 tRNA methyltransferase alter its catalytic properties. Biochimie 2015, 112, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Nagao, A.; Suzuki, T. Human mitochondrial tRNAs: Biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011, 45, 299–329. [Google Scholar] [CrossRef] [PubMed]
- Yarham, J.W.; Elson, J.L.; Blakely, E.L.; McFarland, R.; Taylor, R.W. Mitochondrial tRNA mutations and disease. Wiley Interdiscip. Rev. RNA 2010, 1, 304–324. [Google Scholar] [CrossRef] [PubMed]
- Abbott, J.A.; Francklyn, C.S.; Robey-Bond, S.M. Transfer RNA and human disease. Front. Genet. 2014, 5, 158. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, S.; Ignatova, Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015, 16, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Yao, P.; Fox, P.L. Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol. Med. 2013, 5, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, N.M.; Lazazzera, B.A.; Ibba, M. Cellular mechanisms that control mistranslation. Nat. Rev. Microbiol. 2010, 8, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Drummond, D.A.; Bloom, J.D.; Adami, C.; Wilke, C.O.; Arnold, F.H. Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. USA 2005, 102, 14338–14343. [Google Scholar] [CrossRef]
- Mukherjee, A.; Morales-Scheihing, D.; Butler, P.C.; Soto, C. Type 2 diabetes as a protein misfolding disease. Trends Mol. Med. 2015, 21, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.Y.; Tomizawa, K. Functional loss of Cdkal1, a novel tRNA modification enzyme, causes the development of type 2 diabetes. Endocr. J. 2010, 58, 819–825. [Google Scholar] [CrossRef]
- Wei, F.; Suzuki, T.; Watanabe, S.; Kimura, S.; Kaitsuka, T.; Fujimura, A.; Matsui, H.; Atta, M.; Michiue, H.; Fontecave, M.; et al. Deficit of tRNALys modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Invest. 2011, 121, 3598–3608. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, J.B.; Robins, H.; Levine, A.J. Tissue-specific codon usage and the expression of human genes. Proc. Natl. Acad. Sci. USA 2004, 101, 12588–12591. [Google Scholar] [CrossRef] [PubMed]

| GO Molecular Function Complete | #A | #B | Expected | Fold Enrichment | p-Value (Only p < 0.05 Shown) | |
|---|---|---|---|---|---|---|
| Top 41 Score 1.0 All UCU, no UCC | rRNA binding | 38 | 5 | 0.33 | 15.37 | 1.33 × 10−2 |
| Structural constitutent of ribosome | 213 | 24 | 1.82 | 13.16 | 3.62 × 10−19 | |
| Structural molecule activity | 246 | 25 | 2.11 | 11.87 | 4.05 × 10−19 | |
| Middle 41 Score 0.5 UCU = UCC | Unclassified | 72 | 3 | 0.59 | 5.08 | 0.00 |
| Bottom 41 Score 0 All UCC, no UCU | Unclassified | 74 | 1 | 0.59 | 1.69 | 0.00 |
| Saccharomyces cerevisiae | Schizosaccharomyces pombe | Homo sapiens | Mus musculus | Modification |
|---|---|---|---|---|
| CCA1 | cca1+ | CCA1 | CCA1 | CCA |
| DUS1 | dus1+ | DUS1L | DUS1L | D (16 and 17) |
| DUS2 | dus2+ | DUS2 | DUS2 | D (20) |
| DUS3 | dus3+ | DUS3L | DUS3L | D (47) |
| DUS4 | dus4+ | DUS4L | DUS4 | D (20a and 20b) |
| MOD5 * | tit1+ | TRIT1 | TRIT1 | i6A (37) |
| PUS1 * | pus1+ | PUS1 | PUS1 | Ψ (26–28, 34–36, 65, 67) ** |
| PUS3/DEG1 * | deg1+ | PUS1 | PUS1 | Ψ (38, 39) |
| PUS4 | pus4+ (predicted) | PUS4 (predicted) | PUS4 (predicted) | Ψ (55) |
| PUS6 | SPCC4G3.16 | RPUSD2 | RPUSD2 | Ψ (31) |
| PUS8 * | SPAC18B11.02c | RPUSD4 | RPUSD4 | Ψ (32) |
| PUS7 * | SPBC1A4.09 | PUS7 | PUS7 | Ψ (13, 35) |
| RIT1 | Ar(P) (64) | |||
| TAD1 * | SPBC16A3.06 | ADARB1/ADAR2 | ADARB1/ADAR2 | I (37) |
| TAD2 * | tad2+ | ADAT2 | ADAT2 | I (34) |
| TAD3 * | tad3+ | ADAT3 | ADAT3 | |
| TAN1 | SPBC25H2.10c | THUMPD1 | THUMPD1 | ac4C (12) |
| Kre33 | SPAC20G8.09c | NAT10 | NAT10 | |
| THG1 | thg1+ | THG1L | THG1L | G (-1) |
| TRM1 | trm1+ | TRMT1 TRMT1L | TRMT1 TRMT1L | m22G26 |
| TRM2 | trm2+ | TRMT2A TRMT2B | TRMT2A TRMT2B | m5U (54) |
| TRM3 | TARBP1 | TARBP1 | Gm (18) | |
| TRM4 * | SPAC17D4.04 SPAC23C4.17 | NSUN2 NOP2/NSUN1 NSUN5 NSUN4 NSUN6 NSUN3 | NSUN2 NOP2/NSUN1 NSUN5 NSUN4 NSUN6 NSUN3 | m5C (C34, C40, C48 and C49) ** |
| TRMT5 * | trm5+ | TRMT5 | TRMT5 | m1G (37) |
| TRM6 | gcd10+ | TRMT6 | TRMT6 | m1A (58) |
| TRM61 | cpd1+ | TRMT61A TRMT61B | TRMT61A TRMT61B | |
| TRM7 * | trm7+ | FTSJ1 FTSJ2 FTSJ3 | FTSJ1 FTSJ2 FTSJ3 | Cm (32) & Nm (34) *** |
| TRM8 | trm8+ | METTL1 | METTL1 | m7G (46) |
| TRM9 * | trm9+ | KIAA1456/Trm9L ALKBH8 | KIAA1456/Trm9L ALKBH8 | mcm5U (34) |
| TRM10 | trm10+ | TRMT10A TRMT10B TRMT10C | TRMT10A TRMT10B TRMT10C | m1G (9) |
| TRM11 | trm11+ | TRMT11 | TRMT11 | m2G (10) |
| TRM12 * | trm12+ | TRMT12 | TRMT12 | Wyosine (37) |
| TRM13 | trm13+ | TRMT13 | TRMT13 | Nm (4) *** |
| TRM44 | SPCC663.10 | TRMT44 | TRMT44 | Um (44) |
| TRM140 * | trm140+ trm141+ | METTL2 METTL6 METTL8 | METTL2 METTL2B METTL6 METTL8 | m3C (32) |
| TYW1 * | tyw1+ | TYW1 | TYW1 | yW |
| TYW3 * | tyw3+ | TYW3 | TYW3 | |
| TYW4 * | tyw4+ | TYW4 | TYW4 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maraia, R.J.; Arimbasseri, A.G. Factors That Shape Eukaryotic tRNAomes: Processing, Modification and Anticodon–Codon Use. Biomolecules 2017, 7, 26. https://doi.org/10.3390/biom7010026
Maraia RJ, Arimbasseri AG. Factors That Shape Eukaryotic tRNAomes: Processing, Modification and Anticodon–Codon Use. Biomolecules. 2017; 7(1):26. https://doi.org/10.3390/biom7010026
Chicago/Turabian StyleMaraia, Richard J., and Aneeshkumar G. Arimbasseri. 2017. "Factors That Shape Eukaryotic tRNAomes: Processing, Modification and Anticodon–Codon Use" Biomolecules 7, no. 1: 26. https://doi.org/10.3390/biom7010026
APA StyleMaraia, R. J., & Arimbasseri, A. G. (2017). Factors That Shape Eukaryotic tRNAomes: Processing, Modification and Anticodon–Codon Use. Biomolecules, 7(1), 26. https://doi.org/10.3390/biom7010026







