Metabolomics Analysis Identifies Differential Metabolites as Biomarkers for Acute Myocardial Infarction
Abstract
:1. Introduction
2. Materials and Methods
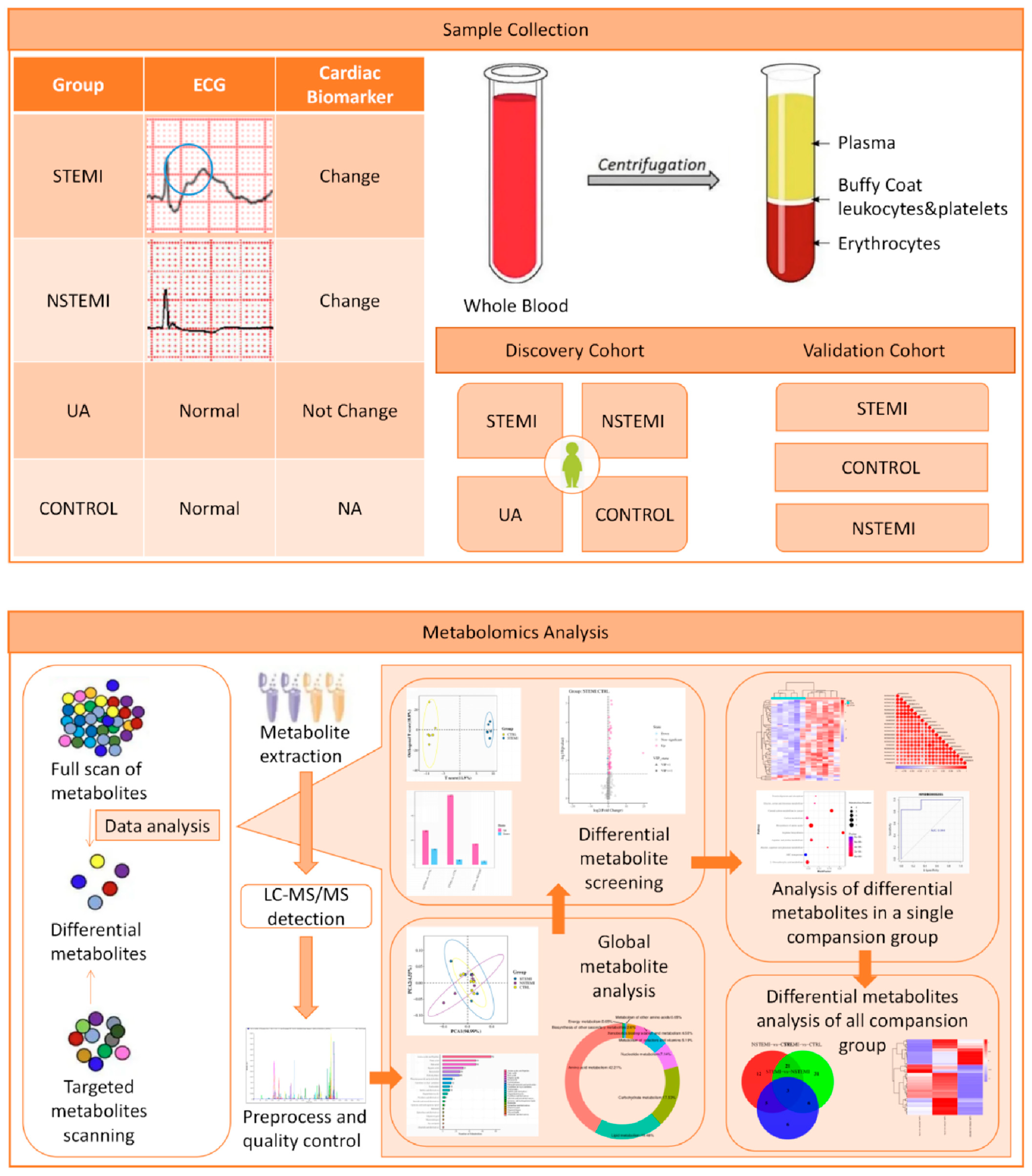
2.1. Study Design and Population
2.2. Samples Collection
2.3. Metabolomics Analysis
2.3.1. Identification of Differential Metabolites (DMs)
2.3.2. Validation of DMs
2.3.3. Bioinformatics Analysis
2.4. Statistical Analysis
3. Results
3.1. Baseline Characteristics of Patients
3.2. Untargeted Metabolomics Profiling of Plasma from Discovery Phase
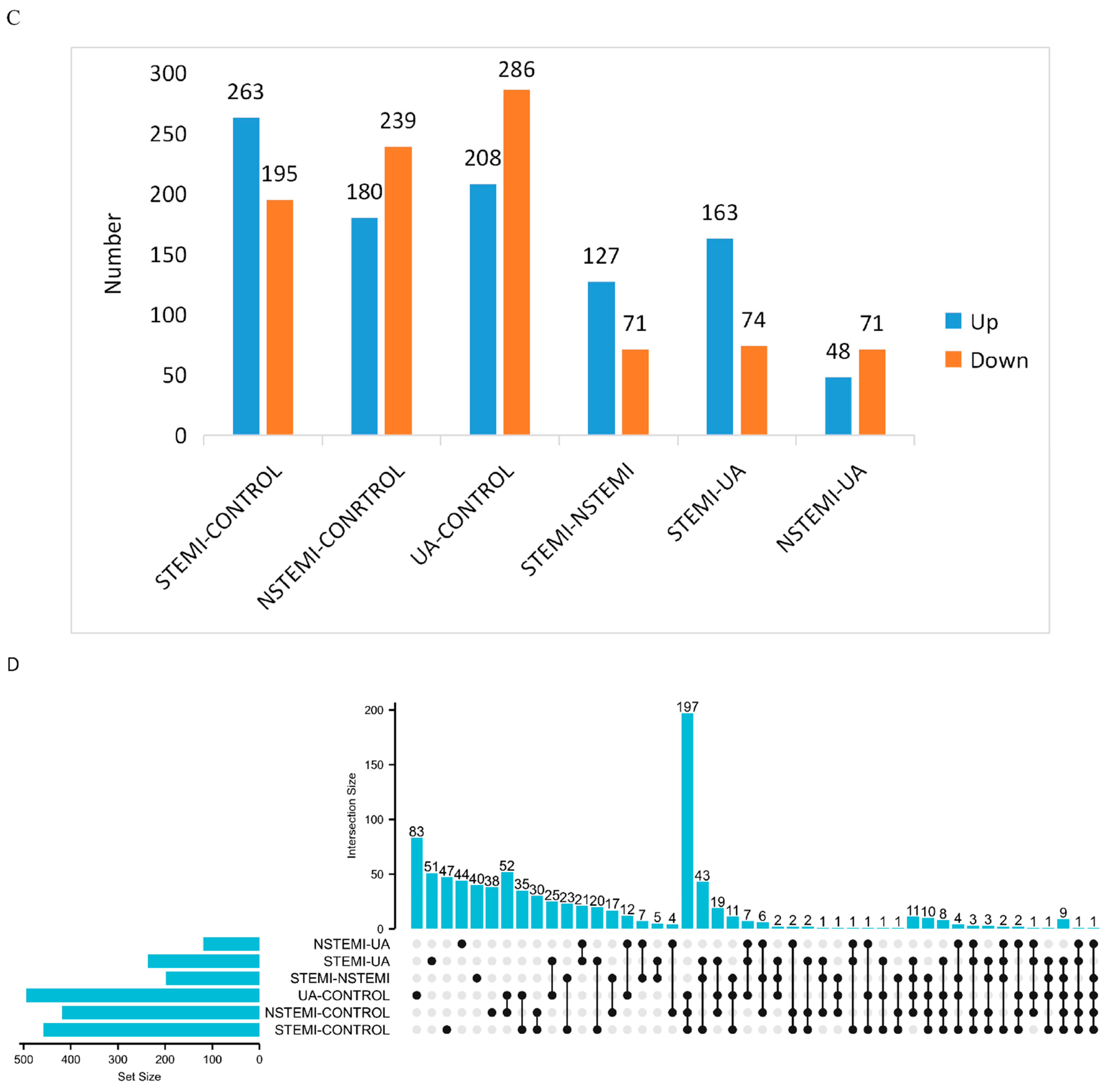
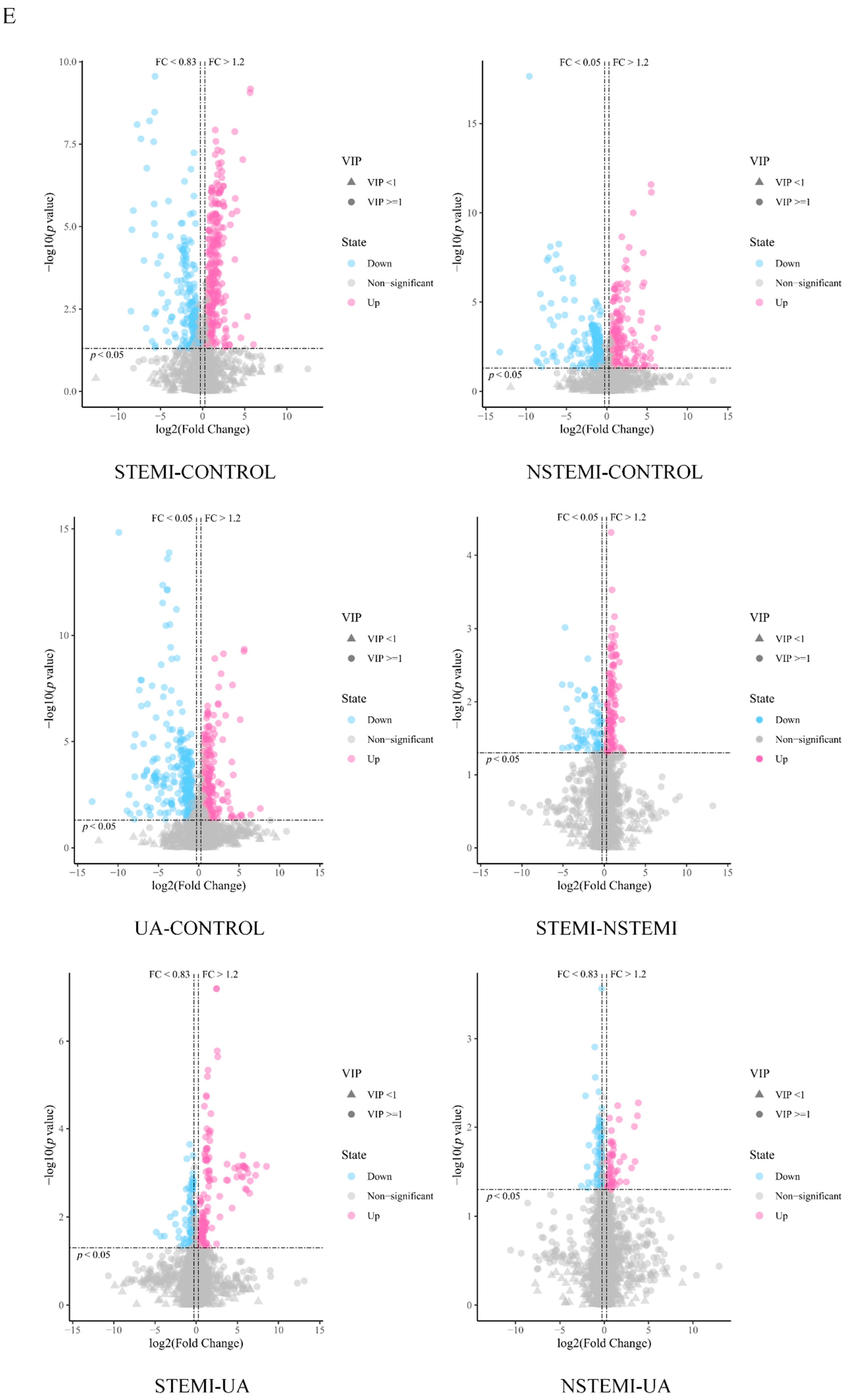
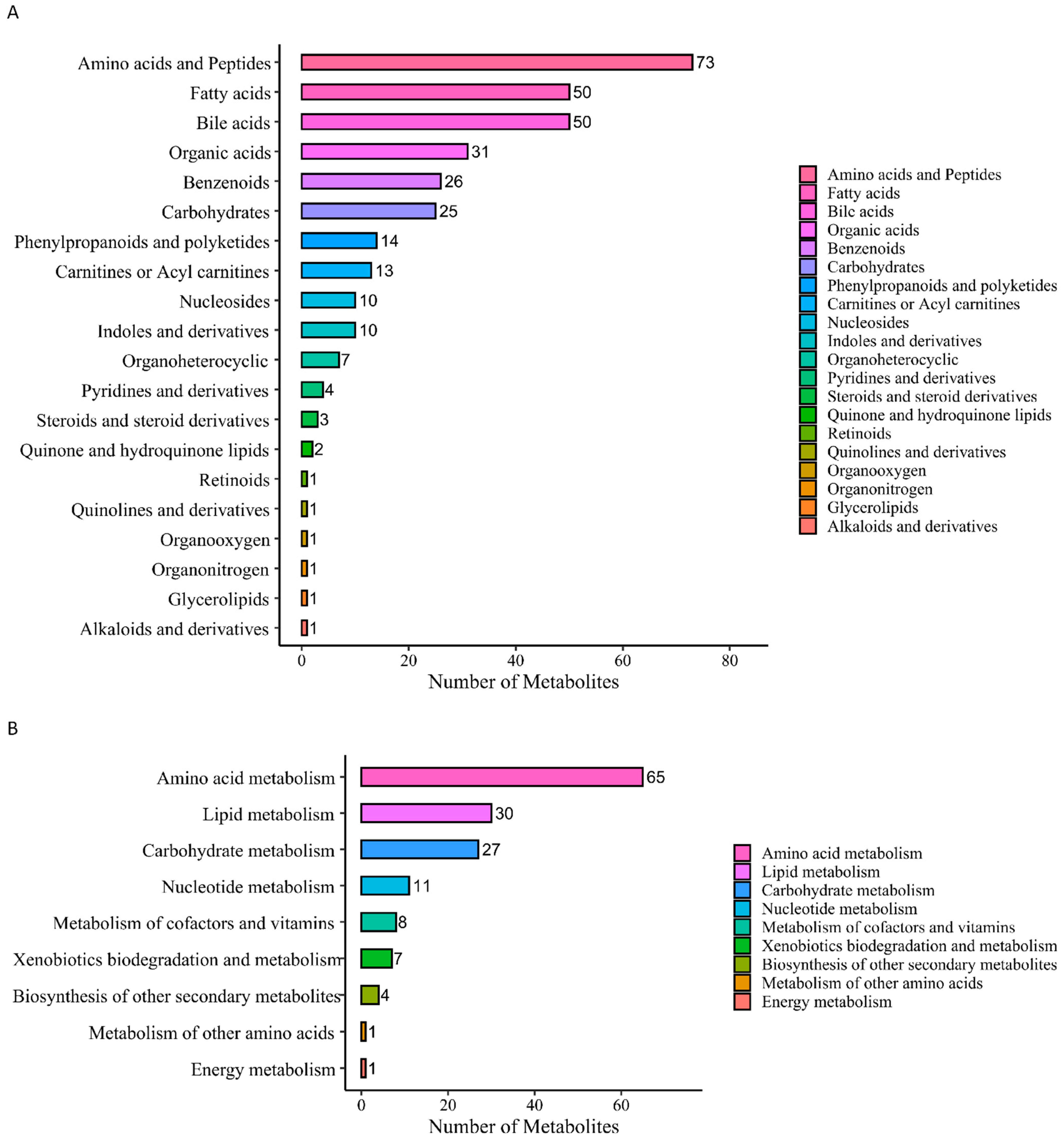
3.2.1. Screening of DMs
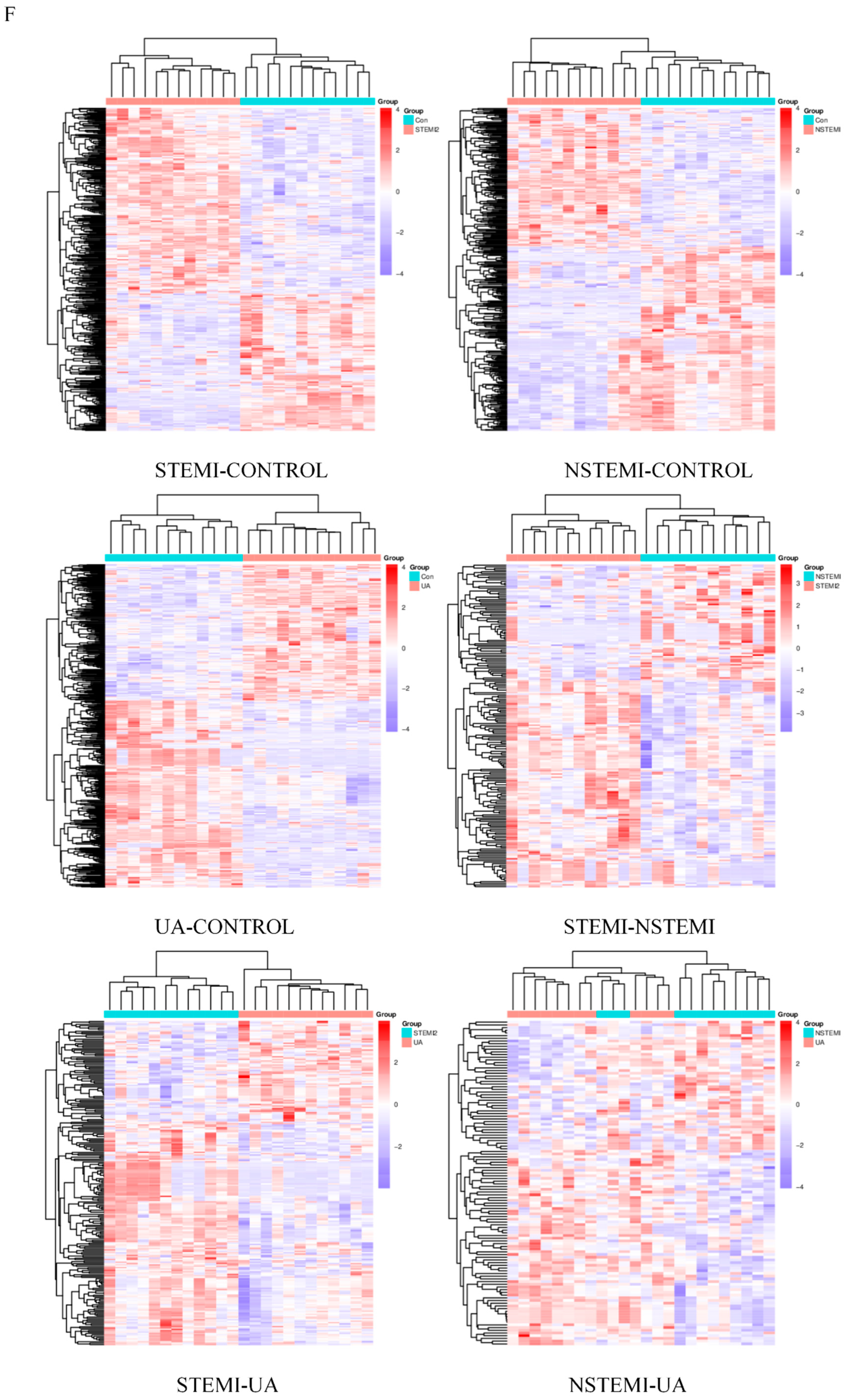
3.2.2. Cluster Analysis and Correlation Analysis of DMs
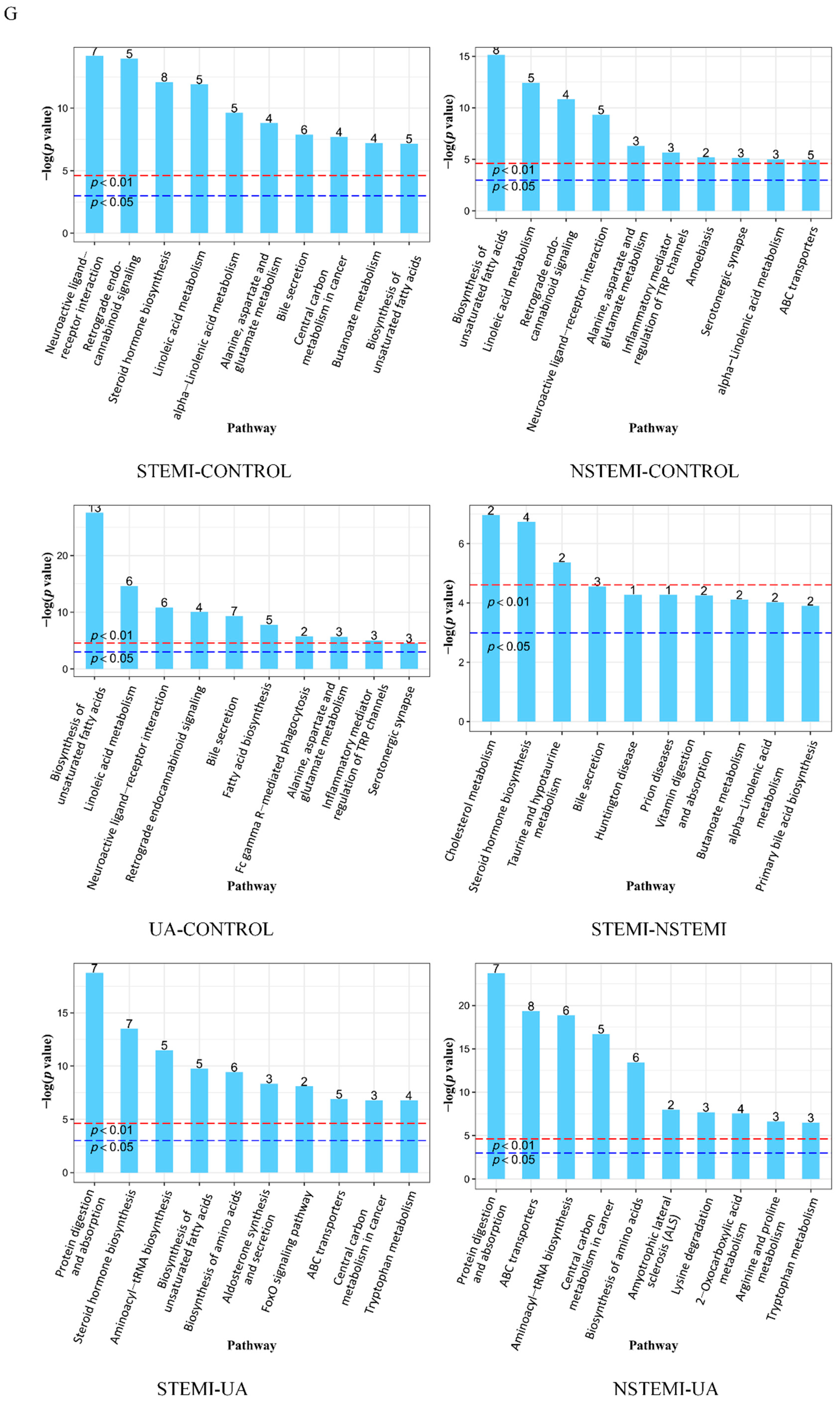
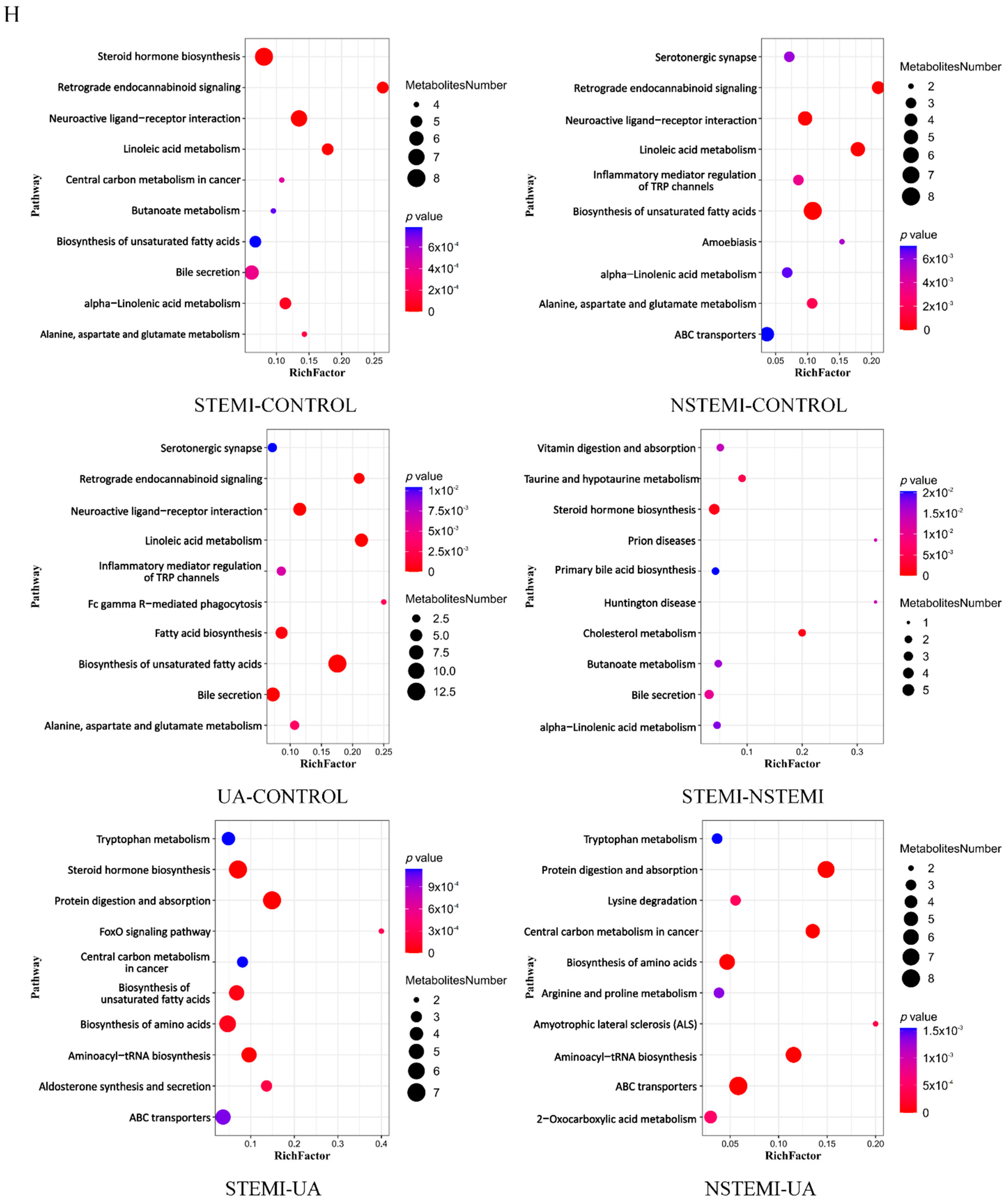
3.2.3. Metabolic Pathway Enrichment Analysis of DMs
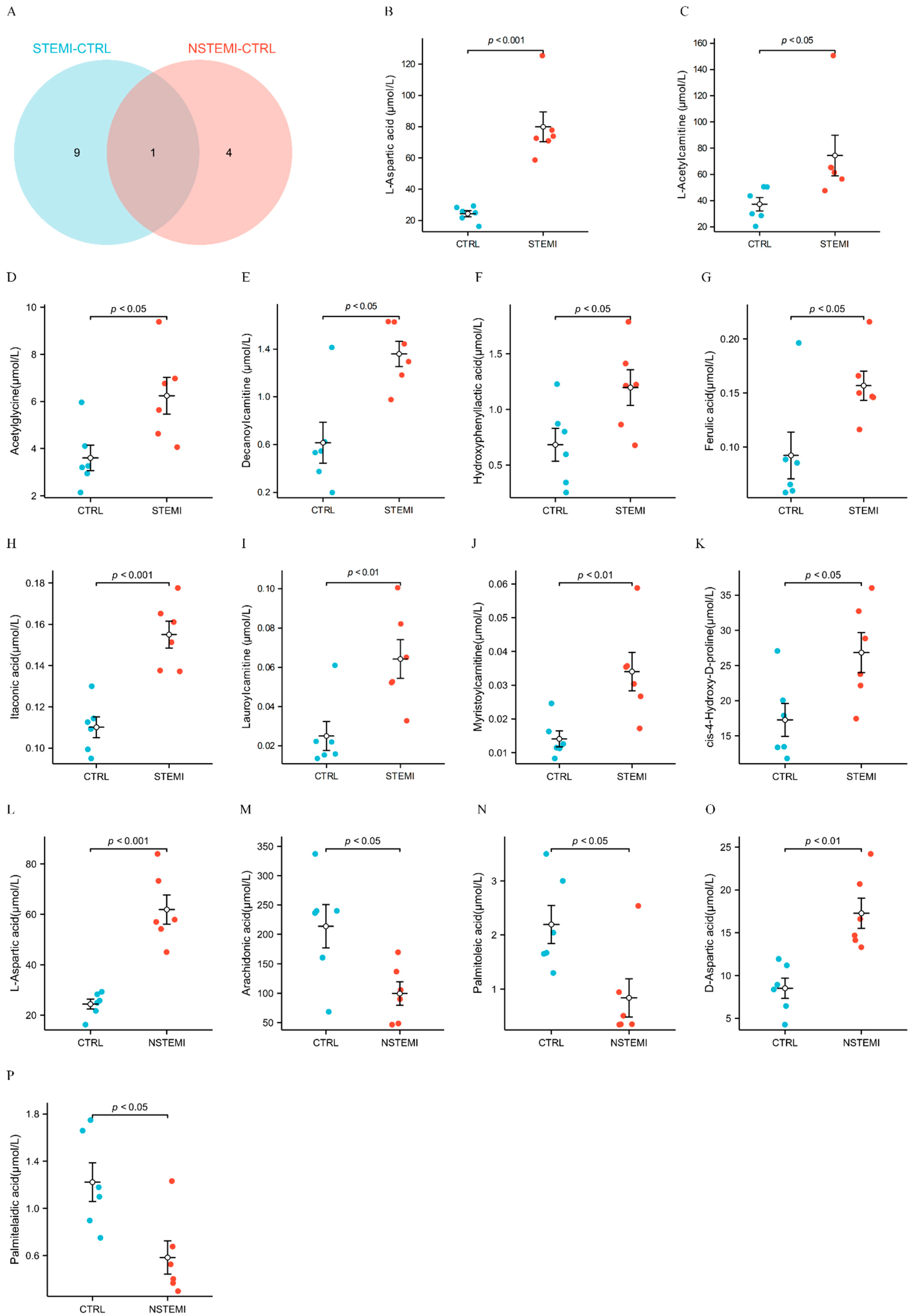
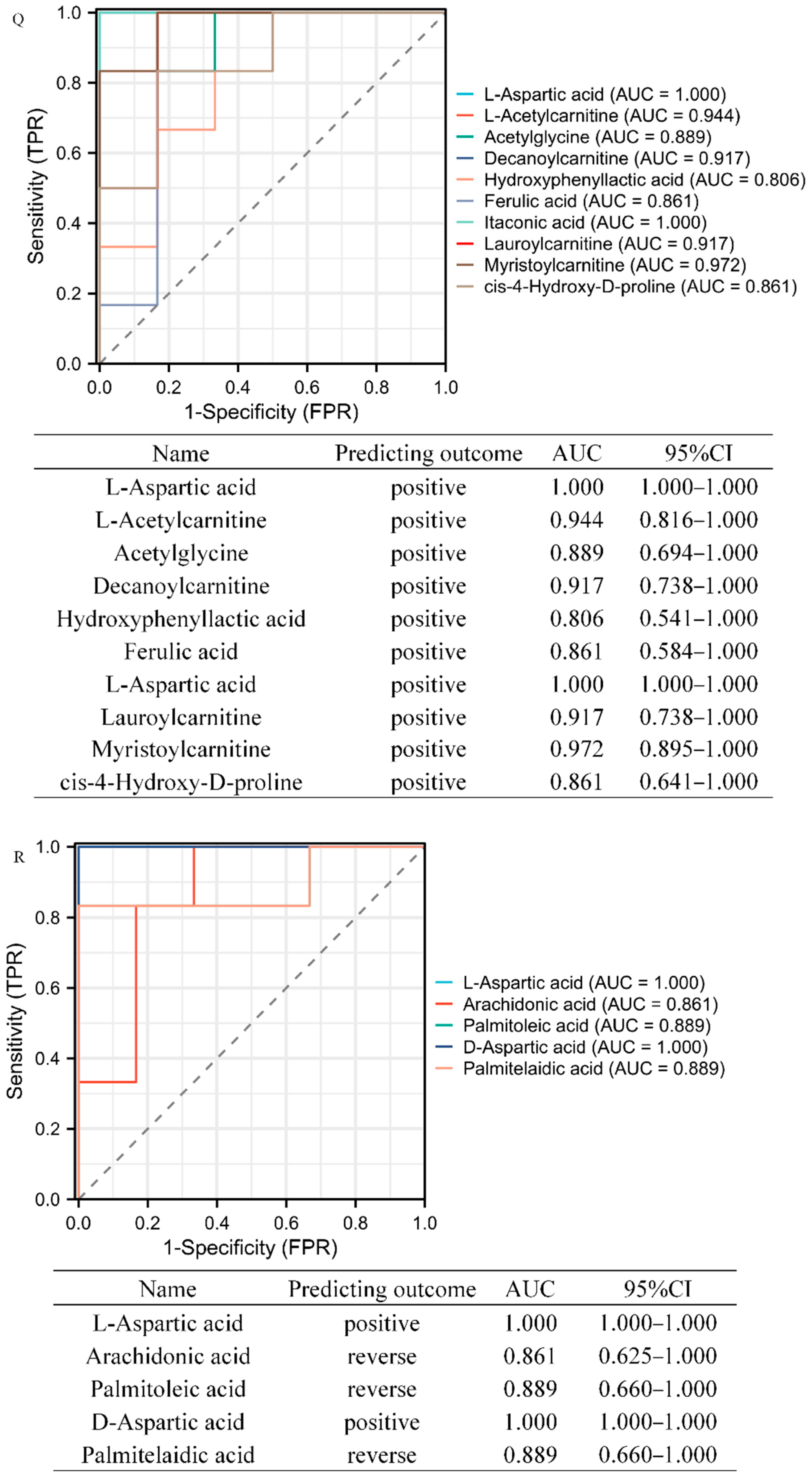
3.3. Target Metabolomics Analysis Revealed the Potential Biomarkers of STEMI and NSTEMI Patients in the Validation Phase
3.4. The Correlation between the DMs and CKMB
3.5. The Correlation between the DMs and the Severity of MI
3.6. The Correlation between the DMs and Risk Factors
4. Discussion
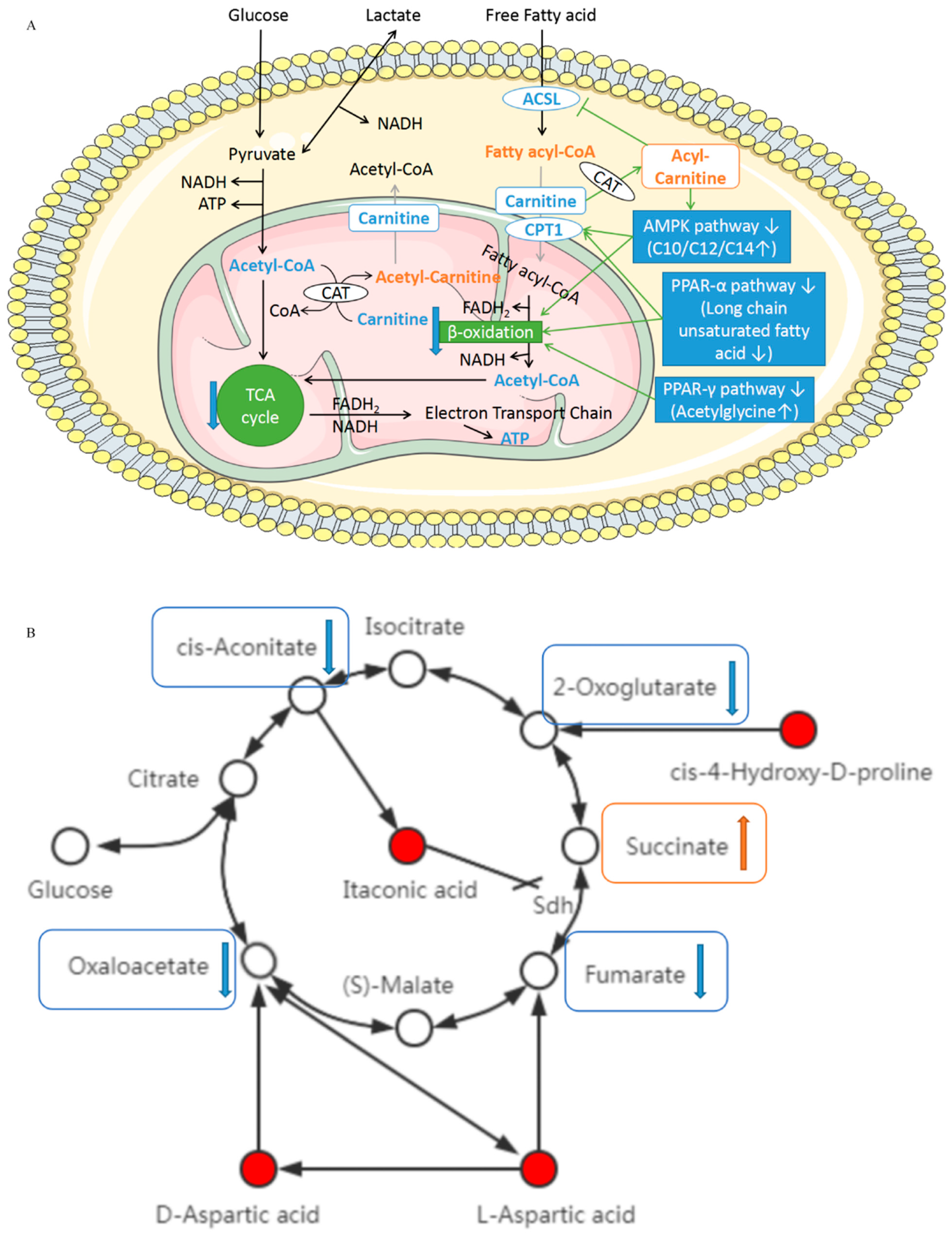
4.1. L-Acetylcarnitine, Decanoylcarnitine (C10), Lauroylcarnitine (C12), and Myristoylcarnitine (C14)
4.2. L-aspartic Acid, D-aspartate, cis-4-hydroxy-D-proline, and Itaconic Acid
4.3. Long Chain Monounsaturated Fatty Acids (Palmitoleic Acid/Palmitelaidic Acid)
4.4. Acetylglycine
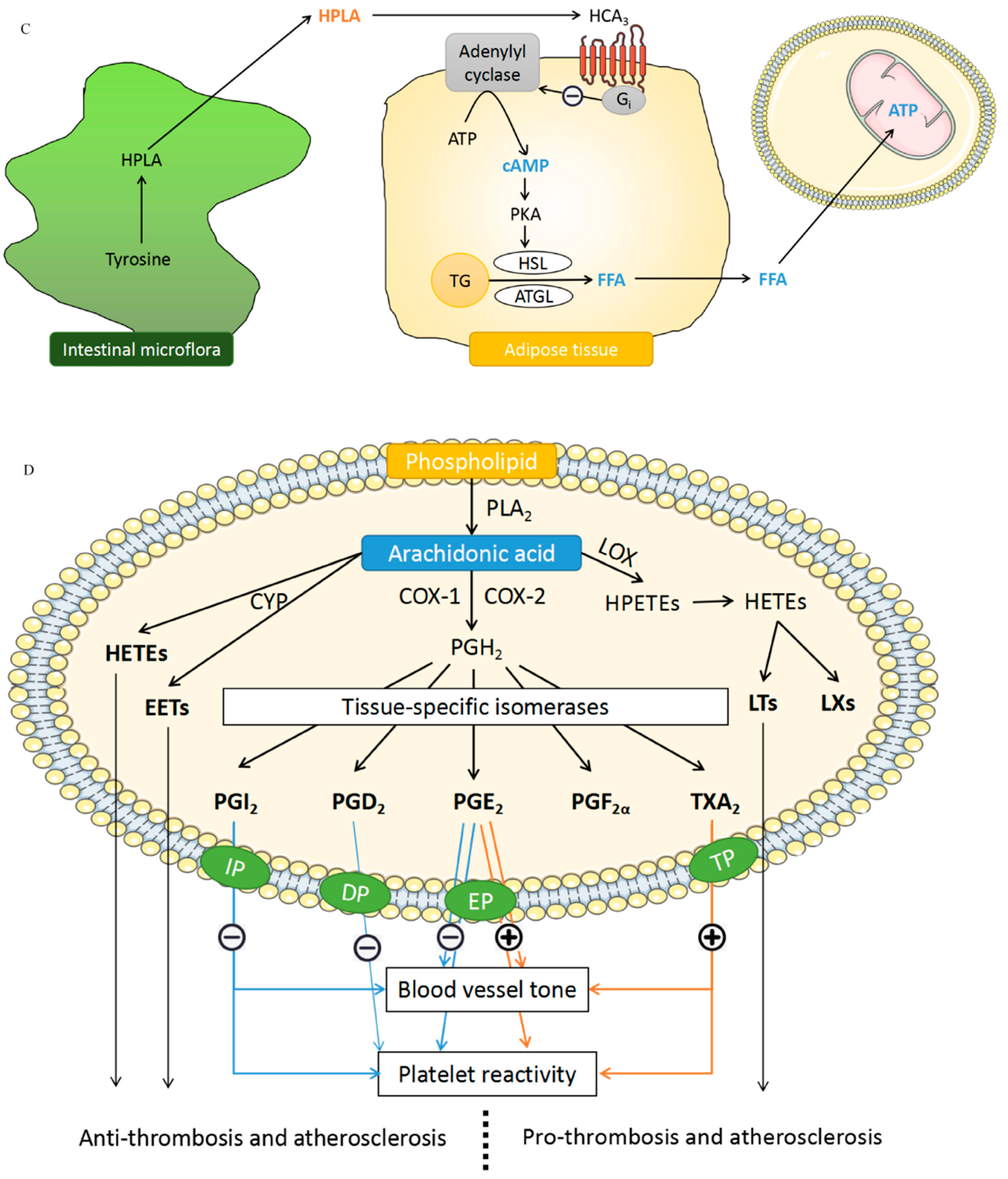
4.5. Hydroxyphenyllactic Acid (HPLA)
4.6. Arachidonic Acid (AA)
4.7. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhatt, D.L.; Lopes, R.D.; Harrington, R.A. Diagnosis and treatment of acute coronary syndromes: A review. JAMA 2022, 327, 662–675. [Google Scholar] [CrossRef] [PubMed]
- American Heart Association. Focus on Quality. Available online: http://www.heart.org/en/professional/quality-improvement (accessed on 28 April 2023).
- Boateng, S.; Sanborn, T. Acute myocardial infarction. Disease-a-Month 2013, 59, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Tsao, C.W.; Aday, A.W.; Almarzooq, Z.I.; Anderson, C.A.; Arora, P.; Avery, C.L.; Baker-Smith, C.M.; Beaton, A.Z.; Boehme, A.K.; Buxton, A.E.; et al. Heart disease and stroke statistics-2023 update: A report from the American Heart Association. Circulation 2023, 147, e93–e621. [Google Scholar] [CrossRef] [PubMed]
- Surendran, A.; Atefi, N.; Zhang, H.; Aliani, M.; Ravandi, A. Defining acute coronary syndrome through metabolomics. Metabolites 2021, 11, 685. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, T.; West, N.E.; El-Omar, M. ST elevation myocardial infarction. Clin. Med. 2016, 16, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Deda, O.; Panteris, E.; Meikopoulos, T.; Begou, O.; Mouskeftara, T.; Karagiannidis, E.; Papazoglou, A.S.; Sianos, G.; Theodoridis, G.; Gika, H. Correlation of serum acylcarnitines with clinical presentation and severity of coronary artery disease. Biomolecules 2022, 12, 354. [Google Scholar] [CrossRef]
- Liu, J.; Huang, L.; Shi, X.; Gu, C.; Xu, H.; Liu, S. Clinical parameters and metabolomic biomarkers that predict inhospital outcomes in patients with ST-segment elevated myocardial infarctions. Front. Physiol. 2022, 12, 820240. [Google Scholar] [CrossRef]
- Luo, J.; Shaikh, J.A.; Huang, L.; Zhang, L.; Iqbal, S.; Wang, Y.; Liu, B.; Zhou, Q.; Ajmal, A.; Rizvi, M.; et al. Human plasma metabolomics identify 9-cis-retinoic acid and dehydrophytosphingosine levels as novel biomarkers for early ventricular fibrillation after ST-elevated myocardial infarction. Bioengineered 2022, 13, 3334–3350. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, L.; Li, T.; Liu, Y.-W.; Wang, Y.; Liu, B.-J. Human plasma metabolomics implicates modified 9-cis-retinoic acid in the phenotype of left main artery lesions in acute ST-segment elevated myocardial infarction. Sci. Rep. 2018, 8, 12958. [Google Scholar] [CrossRef]
- Chorell, E.; Olsson, T.; Jansson, J.H.; Wennberg, P. Lysophospholipids as predictive markers of ST-elevation myocardial infarction (STEMI) and non-ST-elevation myocardial infarction (NSTEMI). Metabolites 2020, 11, 25. [Google Scholar] [CrossRef]
- Thygesen, K.; Alpert, J.S.; Jaffe, A.S.; Chaitman, B.R.; Bax, J.J.; Morrow, D.A.; White, H.D.; Executive Group on behalf of the Joint European Society of Cardiology (ESC)/American College of Cardiology (ACC)/American Heart Association (AHA)/World Heart Federation (WHF) Task Force for the Universal Definition of Myocardial Infarction. Fourth universal definition of myocardial infarction (2018). J. Am. Coll. Cardiol. 2018, 72, 2231–2264. [Google Scholar] [CrossRef] [PubMed]
- Barstow, C.; Rice, M.; McDivitt, J.D. Acute coronary syndrome: Diagnostic evaluation. Am. Fam. Physician 2017, 95, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Ibanez, B.; James, S.; Agewall, S.; Antunes, M.J.; Bucciarelli-Ducci, C.; Bueno, H.; Caforio, A.L.P.; Crea, F.; Goudevenos, J.A.; Halvorsen, S.; et al. 2017 ESC Guidelines for the management of acute myocardial infarction in patients presenting with ST-segment elevation: The Task Force for the management of acute myocardial infarction in patients presenting with ST-segment elevation of the european society of cardiology (ESC). Eur. Heart J. 2018, 39, 119–177. [Google Scholar] [CrossRef] [PubMed]
- Park, J.R.; Ahn, J.H.; Jung, M.H.; Kim, J.H.; Kang, M.G.; Kim, K.H.; Jang, J.Y.; Park, H.W.; Koh, J.-S.; Hwang, S.-J.; et al. Serum microRNA-185 levels and myocardial injury in patients with acute ST-segment elevation myocardial infarction. Intern. Med. 2022, 61, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Y.; Hou, H.-T.; Chen, H.-X.; Liu, X.-C.; Wang, J.; Yang, Q.; He, G.-W. Preoperative plasma biomarkers associated with atrial fibrillation after coronary artery bypass surgery. J. Thorac. Cardiovasc. Surg. 2021, 162, 851–863.e3. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Chen, H.-X.; Hou, H.-T.; Wang, J.; Yang, Q.; He, G.-W. Protein biomarkers and risk scores in pulmonary arterial hypertension associated with ventricular septal defect: Integration of multi-omics and validation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 319, L810–L822. [Google Scholar] [CrossRef] [PubMed]
- Li, M.-Y.; Chen, H.-X.; Hou, H.-T.; Wang, J.; Liu, X.-C.; Yang, Q.; He, G.-W. Biomarkers and key pathways in atrial fibrillation associated with mitral valve disease identified by multi-omics study. Ann. Transl. Med. 2021, 9, 393. [Google Scholar] [CrossRef]
- Jia, H.; Liu, C.; Li, D.; Huang, Q.; Liu, D.; Zhang, Y.; Ye, C.; Zhou, D.; Wang, Y.; Tan, Y.; et al. Metabolomic analyses reveal new stage-specific features of COVID-19. Eur. Respir. J. 2022, 59, 2100284. [Google Scholar] [CrossRef]
- Calvani, M.; Reda, E.; Arrigoni-Martelli, E. Regulation by carnitine of myocardial fatty acid and carbohydrate metabolism under normal and pathological conditions. Basic Res. Cardiol. 2000, 95, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-Y.; Liu, Y.-Y.; Liu, G.-H.; Lu, H.-B.; Mao, C.-Y. L-carnitine and heart disease. Life Sci. 2018, 194, 88–97. [Google Scholar] [CrossRef]
- Marcovina, S.M.; Sirtori, C.; Peracino, A.; Gheorghiade, M.; Borum, P.; Remuzzi, G.; Ardehali, H. Translating the basic knowledge of mitochondrial functions to metabolic therapy: Role of L-carnitine. Transl. Res. 2013, 161, 73–84. [Google Scholar] [CrossRef]
- Zheng, D.-M.; An, Z.-N.; Ge, M.-H.; Wei, D.-Z.; Jiang, D.-W.; Xing, X.-J.; Shen, X.-L.; Liu, C. Medium & long-chain acylcarnitine’s relation to lipid metabolism as potential predictors for diabetic cardiomyopathy: A metabolomic study. Lipids Health Dis. 2021, 20, 151. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; An, R.; Li, Z.-G.; Lai, Z.-X.; Li, D.-L.; Cao, J.-X.; Chen, R.-H.; Zhang, W.-J.; Li, Q.-H.; Lai, X.-F.; et al. Citrus maxima and tea regulate AMPK signaling pathway to retard the progress of nonalcoholic fatty liver disease. Food Nutr. Res. 2021, 65, 7652. [Google Scholar] [CrossRef]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef]
- Husted, A.S.; Trauelsen, M.; Rudenko, O.; Hjorth, S.A.; Schwartz, T.W. GPCR-mediated signaling of metabolites. Cell Metab. 2017, 25, 777–796. [Google Scholar] [CrossRef] [PubMed]
- Fatehi-Hassanabad, Z.; Chan, C.B. Transcriptional regulation of lipid metabolism by fatty acids: A key determinant of pancreatic beta-cell function. Nutr. Metab. 2005, 2, 1. [Google Scholar] [CrossRef]
- Naiman, S.; Huynh, F.K.; Gil, R.; Glick, Y.; Shahar, Y.; Touitou, N.; Nahum, L.; Avivi, M.Y.; Roichman, A.; Kanfi, Y.; et al. SIRT6 promotes hepatic beta-oxidation via activation of PPARα. Cell Rep. 2019, 29, 4127–4143.e8. [Google Scholar] [CrossRef]
- Tahri-Joutey, M.; Andreoletti, P.; Surapureddi, S.; Nasser, B.; Cherkaoui-Malki, M.; Latruffe, N. Mechanisms mediating the regulation of peroxisomal fatty acid beta-oxidation by PPARα. Int. J. Mol. Sci. 2021, 22, 8969. [Google Scholar] [CrossRef] [PubMed]
- Adeva-Andany, M.; Souto-Adeva, G.; Ameneiros-Rodríguez, E.; Fernández-Fernández, C.; Donapetry-García, C.; Domínguez-Montero, A. Insulin resistance and glycine metabolism in humans. Amino Acids 2018, 50, 11–27. [Google Scholar] [CrossRef]
- Almanza-Perez, J.; Alarcon-Aguilar, F.; Blancas-Flores, G.; Campos-Sepulveda, A.; Roman-Ramos, R.; Garcia-Macedo, R.; Cruz, M. Glycine regulates inflammatory markers modifying the energetic balance through PPAR and UCP-2. Biomed. Pharmacother. 2010, 64, 534–540. [Google Scholar] [CrossRef]
- Vallée, A.; Lecarpentier, Y. Crosstalk between peroxisome proliferator-activated receptor gamma and the canonical WNT/β-catenin pathway in chronic inflammation and oxidative stress during carcinogenesis. Front. Immunol. 2018, 9, 745. [Google Scholar] [CrossRef] [PubMed]
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017, 551, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, T.; Horigome, A.; Odamaki, T.; Shimizu, T.; Xiao, J.-Z. Production of hydroxycarboxylic acid receptor 3 (HCA3) ligands by bifidobacterium. Microorganisms 2021, 9, 2397. [Google Scholar] [CrossRef] [PubMed]
- Peters, A.; Krumbholz, P.; Jäger, E.; Heintz-Buschart, A.; Çakir, M.V.; Rothemund, S.; Gaudl, A.; Ceglarek, U.; Schöneberg, T.; Stäubert, C. Metabolites of lactic acid bacteria present in fermented foods are highly potent agonists of human hydroxycarboxylic acid receptor 3. PLoS Genet. 2019, 15, e1008145. [Google Scholar] [CrossRef]
- Zhou, Y.; Khan, H.; Xiao, J.; Cheang, W.S. Effects of arachidonic acid metabolites on cardiovascular health and disease. Int. J. Mol. Sci. 2021, 22, 12029. [Google Scholar] [CrossRef]



| Control | STEMI | NSTEMI | UA | p | |
|---|---|---|---|---|---|
| (n = 18) | (n = 18) | (n = 18) | (n = 12) | ||
| Age, median (IQR) | 64.00 (55.75–69.25) | 67.00 (58.00–70.50) | 66.50 (60.00–72.00) | 63.50 (54.75–69.25) | 0.728 |
| Male, n (%) | 9 (50%) | 9 (50%) | 9 (50%) | 6 (50%) | 1.000 |
| BMI, kg/m2, median (IQR) | 24.49 (23.43–25.55) | 25.24 (21.17–28.04) | 25.71 (22.63–27.89) | 26.30 (23.89–29.84) | 0.312 |
| Risk factors, n (%) | |||||
| Hypertension, n (%) | 11 (61.1%) | 12 (66.7%) | 11 (61.1%) | 6 (50.0%) | 0.839 |
| Diabetes, n (%) | 1 (5.6%) | 7 (38.9%) | 9 (50.0%) | 5 (41.7%) | 0.020 |
| Dyslipidemia, n (%) | 2 (11.1%) | 8 (44.4%) | 5 (27.8%) | 5 (41.7%) | 0.112 |
| Current smoker, n (%) | 4 (22.2%) | 6 (33.3%) | 4 (22.2%) | 3 (25.0%) | 0.878 |
| Cardiac Biomarkers, median (IQR) | |||||
| Myo (ng/mL) | NA | 333.00 (190.00–500.00) | 213.50 (123.25–408.50) | 74.20 (62.60–86.55) | <0.001 |
| CKMB (ng/mL) | NA | 4.50 (2.85–26.22) | 6.25 (2.50–8.55) | 1.25 (1.00–1.68) | 0.001 |
| hs-cTnI (ng/mL) | NA | 0.145 (0.055–1.308) | 0.425 (0.180–0.958) | 0.050 (0.050–0.050) | 0.002 |
| BNP (pg/mL) | NA | 30.15 (21.00–48.58) | 57.75 (44.62–221.75) | 32.40 (8.50–95.00) | 0.062 |
| D-Dimer (ng/mL) | NA | 124.5 (100.0–409.2) | 164.5 (100.0–545.5) | 134.5 (100.0–224.0) | 0.608 |
| Vessel lesion, n (%) | |||||
| Single-vessel disease | 0 (0%) | 1 (5.6%) | 8 (44.4%) | 2 (16.7%) | 0.020 |
| Multi-vessel disease | 0 (0%) | 17 (94.4%) | 10 (55.6%) | 10 (83.3%) | 0.020 |
| Laboratory data | |||||
| Blood glucose (mmol/L) | 5.25 (5.10–5.68) | 8.05 (6.70–10.18) | 7.35 (6.32–9.48) | 7.25 (5.88–7.92) | <0.001 |
| ALT (U/L) | 16.50 (13.25–30.25) | 34.50 (29.00–80.25) | 17.50 (13.00–24.50) | 16.50 (10.00–20.00) | <0.001 |
| AST (U/L) | 17.00 (16.00–23.25) | 114.00 (72.50–349.00) | 31.00 (24.50–58.00) | 18.00 (14.50–19.50) | <0.001 |
| AST/ALT | 1.10 (0.80–1.20) | 3.45 (3.05–4.60) | 2.15 (1.30–3.60) | 1.05 (0.90–1.35) | <0.001 |
| GGT (U/L) | 22.50 (13.25–51.50) | 20.50 (16.00–30.50) | 21.00 (17.00–31.00) | 25.50 (20.7–32.50) | 0.824 |
| TP (g/L) | 68.50 (67.00–71.50) | 64.00 (60.00–67.75) | 64.00 (62.25–67.25) | 63.00 (62.00–64.25) | 0.022 |
| TBIL (μmol/L) | 9.15 (7.02–14.48) | 12.40 (10.52–20.72) | 9.55 (8.42–12.15) | 10.10 (6.05–13.42) | 0.094 |
| ALP (U/L) | 78.50 (66.00–87.50) | 72.00 (66.00–91.00) | 70.00 (62.50–95.25) | 66.50 (63.25–81.75) | 0.741 |
| Urea (mmol/L) | 6.00 (4.90–7.05) | 5.55 (4.98–6.32) | 7.00 (5.70–7.70) | 5.80 (4.90–7.30) | 0.138 |
| Uric acid (μmol/L) | 299.50 (255.00–326.25) | 323.00 (249.25–355.75) | 285.00 (246.25–379.75) | 296.00 (252.00–381.50) | 0.982 |
| CREA (μmol/L) | 57.50 (50.75–65.75) | 61.00 (48.00–76.00) | 58.00 (50.25–94.25) | 63.00 (60.25–67.25) | 0.866 |
| Total cholesterol (mmol/L) | 4.70 (4.08–5.00) | 4.90 (4.18–5.65) | 4.30 (3.50–5.10) | 4.15 (3.80–4.80) | 0.080 |
| TG (mmol/L) | 1.16 (0.94–1.40) | 1.28 (1.16–1.87) | 1.61 (1.06–2.44) | 1.91 (1.54–2.18) | 0.032 |
| HDL-C (mmol/L) | 1.18 (0.99–1.39) | 1.11 (1.01–1.17) | 0.92 (0.70–1.08) | 0.88 (0.80–0.94) | 0.001 |
| LDL-C (mmol/L) | 2.98 (2.60–3.25) | 3.10 (2.72–3.80) | 2.86 (2.06–3.52) | 2.47 (2.21–3.10) | 0.111 |
| PLT (×109/L) | 223.00 (209.00–272.50) | 244.00 (206.25–270.00) | 224.50 (198.00–250.25) | 243.00 (203.00–261.50) | 0.733 |
| Length of stay (days) | 1.00 (1.00–2.00) | 8.00 (6.25–9.75) | 7.00 (6.00–8.00) | 3.00 (2.75–6.50) | <0.001 |
| Group | HMDB ID | Name | Discovery Group | Validation Group | ||||
|---|---|---|---|---|---|---|---|---|
| FC | p Value | VIP | FC | p Value | VIP | |||
| CONTROL-STEMI | HMDB0000191 | L-Aspartic acid | 2.11358712 | 8.21 × 10−7 | 1.577611655 | 3.279994663 | 7.22 × 10−6 | 1.896413607 |
| HMDB0000201 | L-Acetylcarnitine | 1.965571929 | 5.54 × 10−5 | 1.340201316 | 1.997169295 | 0.013503462 | 1.253073946 | |
| HMDB0000532 | Acetylglycine | 1.725948017 | 0.002633113 | 1.245356672 | 1.733124121 | 0.013522338 | 1.431596275 | |
| HMDB0000651 | Decanoylcarnitine | 4.27951669 | 0.000127514 | 1.844505879 | 2.213434982 | 0.013596305 | 1.628532799 | |
| HMDB0000755 | Hydroxyphenyllactic acid | 2.714464103 | 1.94 × 10−6 | 1.813677974 | 1.752930832 | 0.049981208 | 1.281225837 | |
| HMDB0000954 | Ferulic acid | 6.08440531 | 0.024210222 | 1.290022785 | 1.699855439 | 0.019853603 | 1.375369857 | |
| HMDB0002092 | Itaconic acid | 1.598398693 | 2.34 × 10−5 | 1.168632741 | 1.407598002 | 0.000256761 | 1.869606847 | |
| HMDB0002250 | Lauroylcarnitine | 2.749056744 | 0.000898345 | 1.466835554 | 2.565912117 | 0.004524507 | 1.522012879 | |
| HMDB0005066 | Myristoylcarnitine | 2.166262401 | 2.10 × 10−6 | 1.516728633 | 2.413711584 | 0.002971447 | 1.533423213 | |
| HMDB0060460 | cis-4-Hydroxy-D-proline | 1.86876251 | 4.64 × 10−6 | 1.416576047 | 1.553709509 | 0.023058005 | 1.359347074 | |
| CONTROL-NSTEMI | HMDB0000191 | L-Aspartic acid | 1.989941591 | 4.99 × 10−6 | 1.411097512 | 2.540095524 | 2.64 × 10−5 | 2.282586177 |
| HMDB0001043 | Arachidonic acid | 0.409790387 | 0.000367677 | 1.480786535 | 0.465412831 | 0.034645824 | 1.677437241 | |
| HMDB0003229 | Palmitoleic acid | 0.407686414 | 0.002118188 | 1.451292166 | 0.382412824 | 0.01157985 | 1.670575527 | |
| HMDB0006483 | D-Aspartic acid | 1.532946834 | 6.03 × 10−5 | 1.053107988 | 2.024616281 | 0.003515567 | 2.032273973 | |
| HMDB0012328 | Palmitelaidic acid | 0.557037574 | 0.001707426 | 1.109067827 | 0.476871871 | 0.01046257 | 1.749465707 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Hou, H.-T.; Song, Y.; Zhou, X.-L.; Chen, H.-X.; Zhang, L.-L.; Xue, H.-M.; Yang, Q.; He, G.-W. Metabolomics Analysis Identifies Differential Metabolites as Biomarkers for Acute Myocardial Infarction. Biomolecules 2024, 14, 532. https://doi.org/10.3390/biom14050532
Zhou J, Hou H-T, Song Y, Zhou X-L, Chen H-X, Zhang L-L, Xue H-M, Yang Q, He G-W. Metabolomics Analysis Identifies Differential Metabolites as Biomarkers for Acute Myocardial Infarction. Biomolecules. 2024; 14(5):532. https://doi.org/10.3390/biom14050532
Chicago/Turabian StyleZhou, Jie, Hai-Tao Hou, Yu Song, Xiao-Lin Zhou, Huan-Xin Chen, Li-Li Zhang, Hong-Mei Xue, Qin Yang, and Guo-Wei He. 2024. "Metabolomics Analysis Identifies Differential Metabolites as Biomarkers for Acute Myocardial Infarction" Biomolecules 14, no. 5: 532. https://doi.org/10.3390/biom14050532






