An Engineered Laccase from Fomitiporia mediterranea Accelerates Lignocellulose Degradation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cloning and Expression of Laccase Variants
2.2. Purification of Recombinant Fom_lac Enzyme
2.3. Enzyme Reactions with the Fluorous-Tagged Phenolic/Nonphenolic β-O-4 Linked Model Compound
2.4. Biomass Feedstock Preparation
2.5. Enzymatic Saccharification Reactions
2.6. Simulation Methods
3. Results
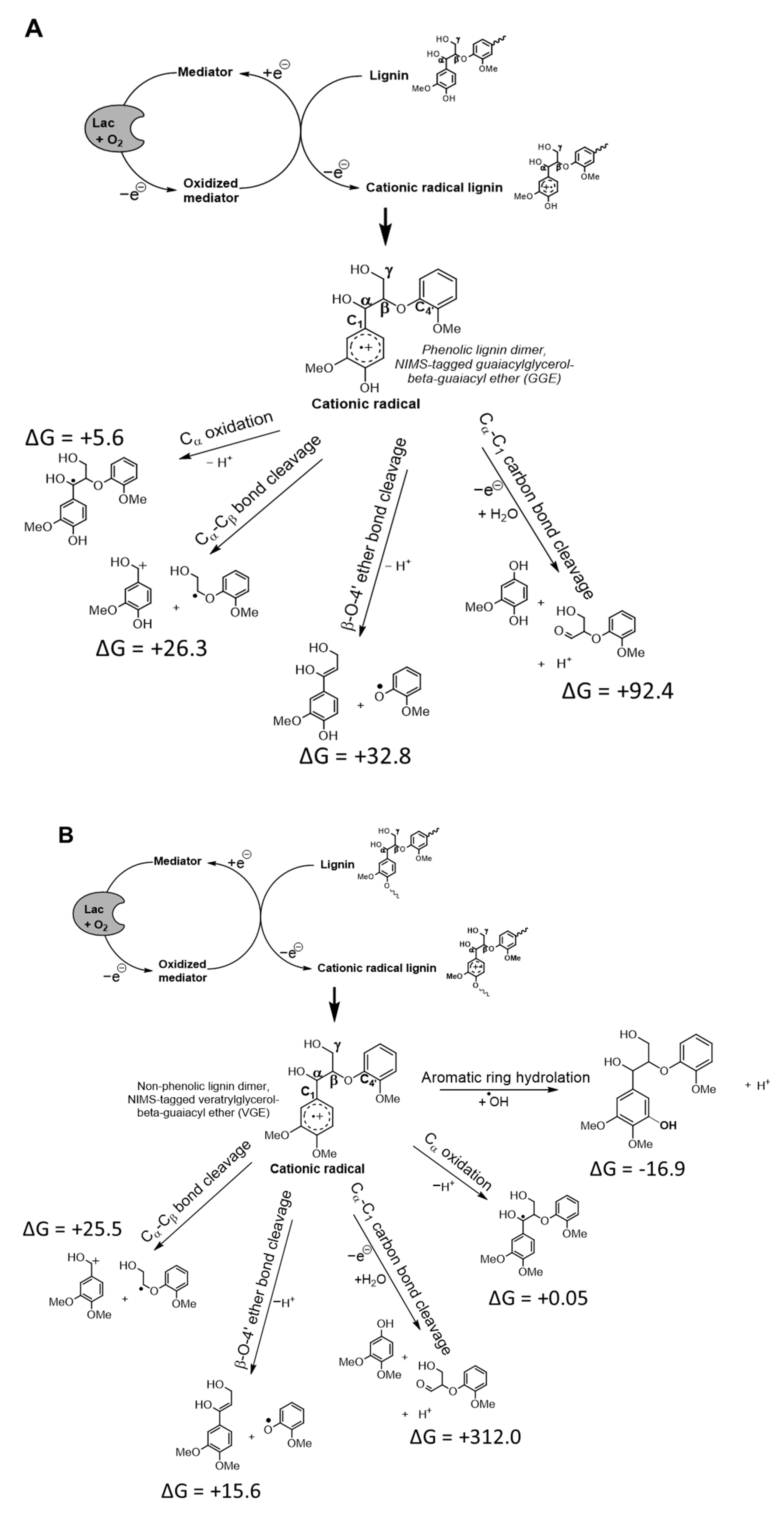
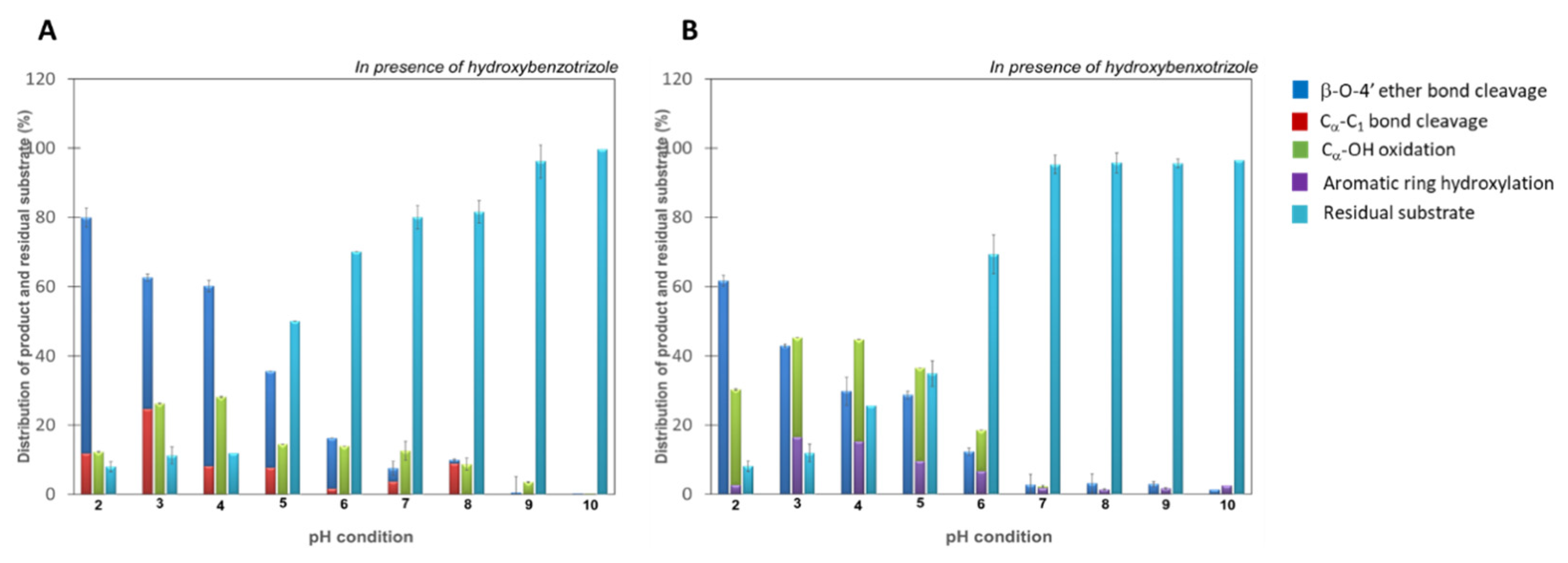
3.1. Low pH Accelerates Catalysis of Bond Cleavage of Lignin Dimers by Wildtype Laccase from F. mediterranea
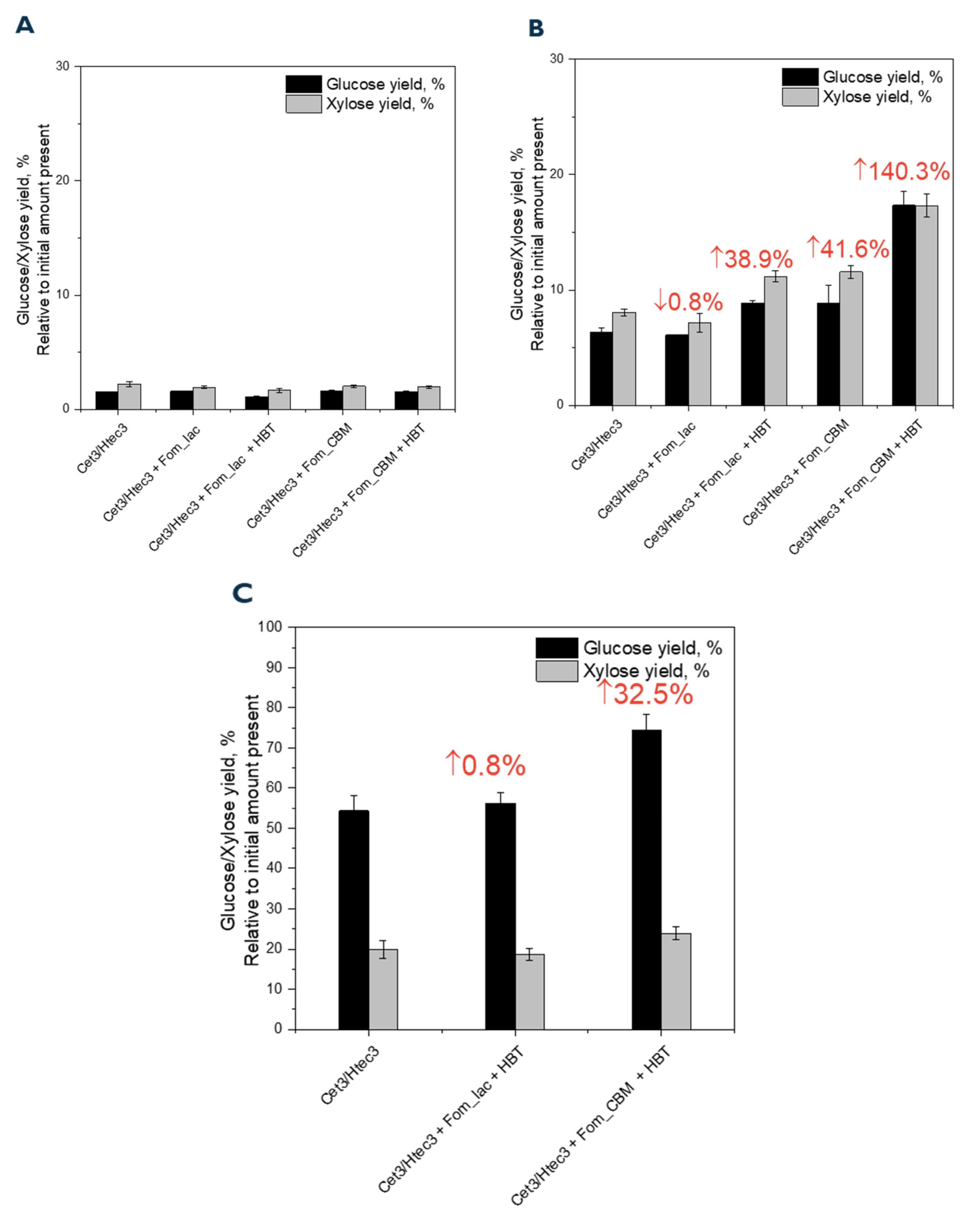
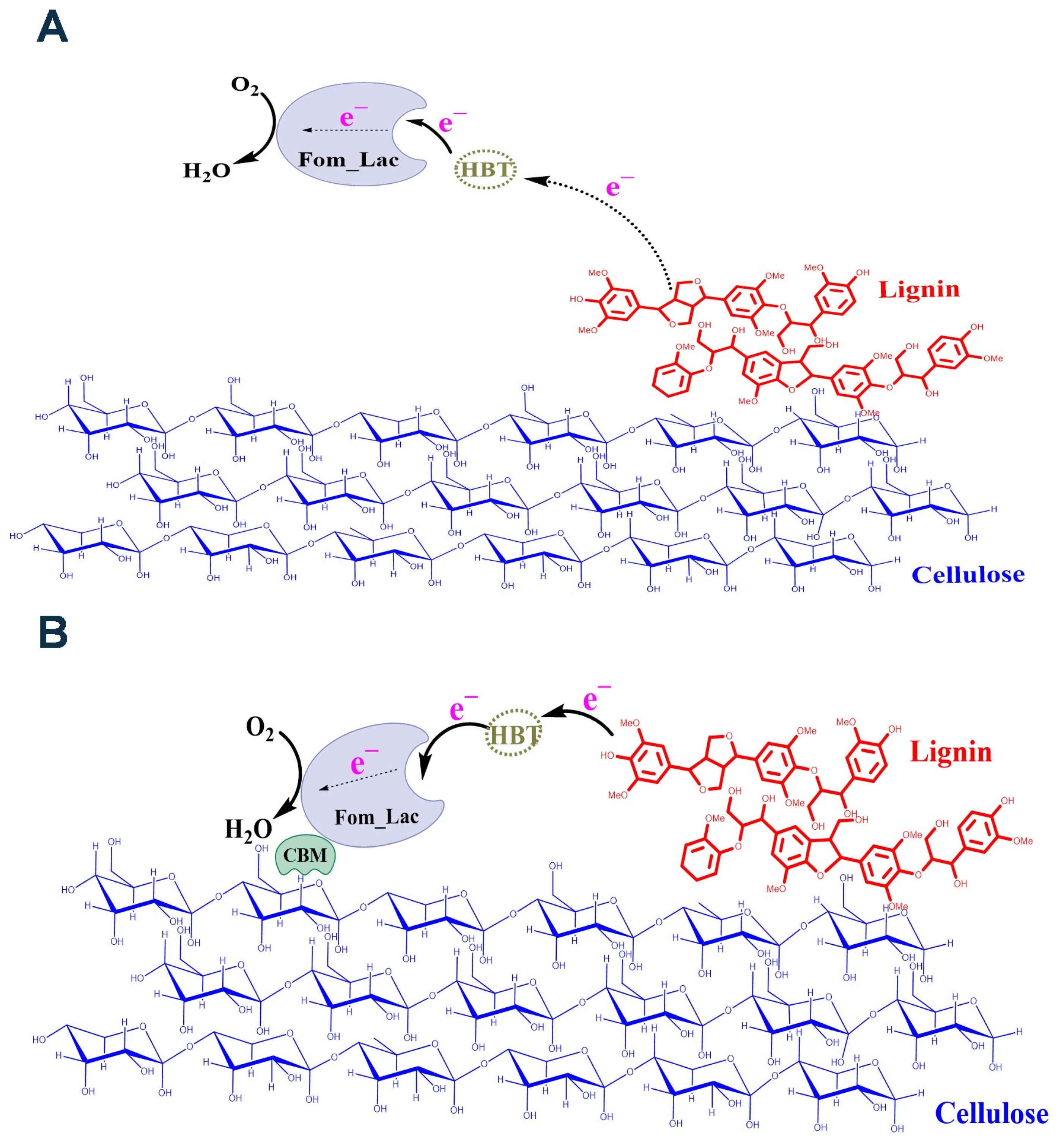
3.2. Enhanced Lignocellulosic Biomass Degradation Was Achieved by Fusing a CBM to the Laccase from F. mediterranea
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xu, Z.; Lei, P.; Zhai, R.; Wen, Z.; Jin, M. Recent advances in lignin valorization with bacterial cultures: Microorganisms, metabolic pathways, and bio-products. Biotechnol. Biofuels 2019, 12, 32. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Cui, H.; Wang, M.; Fu, X.; Wang, X.; Li, X.; Huang, H. Integrated biorefinery approaches for the industrialization of cellulosic ethanol fuel. Bioresour. Technol. 2022, 360, 127516. [Google Scholar] [CrossRef] [PubMed]
- Solomon, E.I.; Sundaram, U.M.; Machonkin, T.E. Multicopper Oxidases and Oxygenases. Chem. Rev. 1996, 96, 2563–2606. [Google Scholar] [CrossRef]
- Pawlik, A.; Ruminowicz-Stefaniuk, M.; Frąc, M.; Mazur, A.; Wielbo, J.; Janusz, G. The wood decay fungus Cerrena unicolor adjusts its metabolism to grow on various types of wood and light conditions. PLoS ONE 2019, 14, e0211744. [Google Scholar] [CrossRef] [PubMed]
- Schilling, M.; Maia-Grondard, A.; Baltenweck, R.; Robert, E.; Hugueney, P.; Bertsch, C.; Farine, S.; Gelhaye, E. Wood degradation by Fomitiporia mediterranea M. Fischer: Physiologic, metabolomic and proteomic approaches. Front. Plant Sci. 2022, 13, 988709. [Google Scholar] [CrossRef]
- Abou-Mansour, E.; Polier, J.; Pezet, R.; Tabacchi, R. Purification and partial characterisation of a 60 KDa laccase from Fomitiporia mediterranea. Phytopathol. Mediterr. 2009, 48, 447–453. [Google Scholar]
- Deng, K.; Zeng, J.; Cheng, G.; Gao, J.; Sale, K.L.; Simmons, B.A.; Singh, A.K.; Adams, P.D.; Northen, T.R. Rapid characterization of the activities of lignin-modifying enzymes based on nanostructure-initiator mass spectrometry (NIMS). Biotechnol. Biofuels 2018, 11, 266. [Google Scholar] [CrossRef]
- Mai Pham, L.T.; Deng, K.; Northen, T.; Singer, S.; Adams, P.; Simmons, B.; Sale, K. Heterologous Expression, Characterization, and Comparison of Laccases from the White Rot Causing Basidiomycete Cerrena Unicolor. Catal. Res. 2022, 2, 028. [Google Scholar] [CrossRef]
- Zhang, C.; Yan, Z.; Li, X.; Wang, J.; Ren, X.; Liu, X. Comprehensive Analysis of Catalytic Characteristics and Molecular Mechanisms in Mutant Trametes versicolor Strains with Enhanced Laccase Activities. Fermentation 2023, 9, 995. [Google Scholar] [CrossRef]
- Choudhary, H.; Simmons, B.A.; Gladden, J.M. Comparative Study on the Pretreatment of Aspen and Maple with 1-Ethyl-3-methylimidazolium Acetate and Cholinium Lysinate. Front. Energy Res. 2022, 10, 868181. [Google Scholar] [CrossRef]
- Floudas, D.; Binder, M.; Riley, R.; Barry, K.; Blanchette, R.A.; Henrissat, B.; Martínez, A.T.; Otillar, R.; Spatafora, J.W.; Yadav, J.S.; et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 2012, 336, 1715–1719. [Google Scholar] [CrossRef]
- Yao, A.; Choudhary, H.; Mohan, M.; Rodriguez, A.; Magurudeniya, H.; Pelton, J.G.; George, A.; Simmons, B.A.; Gladden, J.M. Can Multiple Ions in an Ionic Liquid Improve the Biomass Pretreatment Efficacy? ACS Sustain. Chem. Eng. 2021, 9, 4371–4376. [Google Scholar] [CrossRef]
- Pham, L.T.M.; Deng, K.; Northen, T.R.; Singer, S.W.; Adams, P.D.; Simmons, B.A.; Sale, K.L. Experimental and theoretical insights into the effects of pH on catalysis of bond-cleavage by the lignin peroxidase isozyme H8 from Phanerochaete chrysosporium. Biotechnol. Biofuels 2021, 14, 108. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 09, Revision A.02; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B Condens. Matter 1988, 37, 785–789. [Google Scholar] [CrossRef]
- Davidson, E.R.; Feller, D. Basis set selection for molecular calculations. Chem. Rev. 1986, 86, 681–696. [Google Scholar] [CrossRef]
- Wiberg, K.B. Ab Initio Molecular Orbital Theory by W. J. Hehre, L. Radom, P. v. R. Schleyer, and J. A. Pople, John Wiley, New York, 548pp. Price: $79.95 (1986). J. Comput. Chem. 1986, 7, 379. [Google Scholar] [CrossRef]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef]
- Ufimtsev, I.S.; Martinez, T.J. Quantum Chemistry on Graphical Processing Units. 3. Analytical Energy Gradients, Geometry Optimization, and First Principles Molecular Dynamics. J. Chem. Theory Comput. 2009, 5, 2619–2628. [Google Scholar] [CrossRef]
- Ufimtsev, I.S.; Martinez, T.J. Quantum Chemistry on Graphical Processing Units. 2. Direct Self-Consistent-Field Implementation. J. Chem. Theory Comput. 2009, 5, 1004–1015. [Google Scholar] [CrossRef]
- Ufimtsev, I.S.; Martínez, T.J. Quantum Chemistry on Graphical Processing Units. 1. Strategies for Two-Electron Integral Evaluation. J. Chem. Theory Comput. 2008, 4, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Luehr, N.; Jin, A.G.; Martinez, T.J. Ab initio interactive molecular dynamics on graphical processing units (GPUs). J. Chem. Theory Comput. 2015, 11, 4536–4544. [Google Scholar] [CrossRef] [PubMed]
- Ernzerhof, M.; Scuseria, G.E. Assessment of the Perdew–Burke–Ernzerhof exchange-correlation functional. J. Chem. Phys. 1999, 110, 5029–5036. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Søndergaard, C.R.; Olsson, M.H.M.; Rostkowski, M.; Jensen, J.H. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. J. Chem. Theory Comput. 2011, 7, 2284–2295. [Google Scholar] [CrossRef]
- Feller, S.E.; Zhang, Y.; Pastor, R.W.; Brooks, B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995, 103, 4613–4621. [Google Scholar] [CrossRef]
- Leeuw, S.W.d.; Perram, J.W.; Smith, E.R.; Rowlinson, J.S. Simulation of electrostatic systems in periodic boundary conditions. III. Further theory and applications. Proc. R. Soc. Lond. A Math. Phys. Sci. 1983, 388, 177–193. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Gollan, M.; Black, G.; Munoz-Munoz, J. A computational approach to optimising laccase-mediated polyethylene oxidation through carbohydrate-binding module fusion. BMC Biotechnol. 2023, 23, 18. [Google Scholar] [CrossRef] [PubMed]
- Kurniati, A.; Puspaningsih, N.N.T.; Putri, K.D.A.; Damayanti, M.; Purwani, N.N.; Rahmah, S.A.; Purkan; Fujiyama, K.; Sakka, M.; Sakka, K.; et al. Heterologous fusion gene expression and characterization of a novel carbohydrate binding module (Cbm36) to laccase (Lcc2). Biocatal. Agric. Biotechnol. 2022, 42, 102377. [Google Scholar] [CrossRef]
- Ravalason, H.; Herpoël-Gimbert, I.; Record, E.; Bertaud, F.; Grisel, S.; de Weert, S.; van den Hondel, C.A.; Asther, M.; Petit-Conil, M.; Sigoillot, J.C. Fusion of a family 1 carbohydrate binding module of Aspergillus niger to the Pycnoporus cinnabarinus laccase for efficient softwood kraft pulp biobleaching. J. Biotechnol. 2009, 142, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, P.; Tian, J.; Seidi, F.; Guo, J.; Zhu, W.; Xiao, H.; Song, J. Carbohydrate-Binding Modules of Potential Resources: Occurrence in Nature, Function, and Application in Fiber Recognition and Treatment. Polymers 2022, 14, 1806. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pham, L.T.M.; Deng, K.; Choudhary, H.; Northen, T.R.; Singer, S.W.; Adams, P.D.; Simmons, B.A.; Sale, K.L. An Engineered Laccase from Fomitiporia mediterranea Accelerates Lignocellulose Degradation. Biomolecules 2024, 14, 324. https://doi.org/10.3390/biom14030324
Pham LTM, Deng K, Choudhary H, Northen TR, Singer SW, Adams PD, Simmons BA, Sale KL. An Engineered Laccase from Fomitiporia mediterranea Accelerates Lignocellulose Degradation. Biomolecules. 2024; 14(3):324. https://doi.org/10.3390/biom14030324
Chicago/Turabian StylePham, Le Thanh Mai, Kai Deng, Hemant Choudhary, Trent R. Northen, Steven W. Singer, Paul D. Adams, Blake A. Simmons, and Kenneth L. Sale. 2024. "An Engineered Laccase from Fomitiporia mediterranea Accelerates Lignocellulose Degradation" Biomolecules 14, no. 3: 324. https://doi.org/10.3390/biom14030324




