Mitochondrial Dysfunction and Protein Homeostasis in Aging: Insights from a Premature-Aging Mouse Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Animals
2.3. Tissue Preparation
2.4. Proteasome Activity Assay
2.5. Western Blot
2.6. Adult Mouse Fibroblast Culture and Chloroquine Treatment
2.7. GFP-LC3B Puncta
2.8. Mitochondrial Analysis
2.9. Lipofuscin Detection
2.10. Statistical Analysis
3. Results
3.1. Effect of Mitochondrial Dysfunction and Aging on the Ubiquitin–Proteasome System
3.2. Effect of Mitochondrial Dysfunction and Aging on the Autophagy-Lysosome System
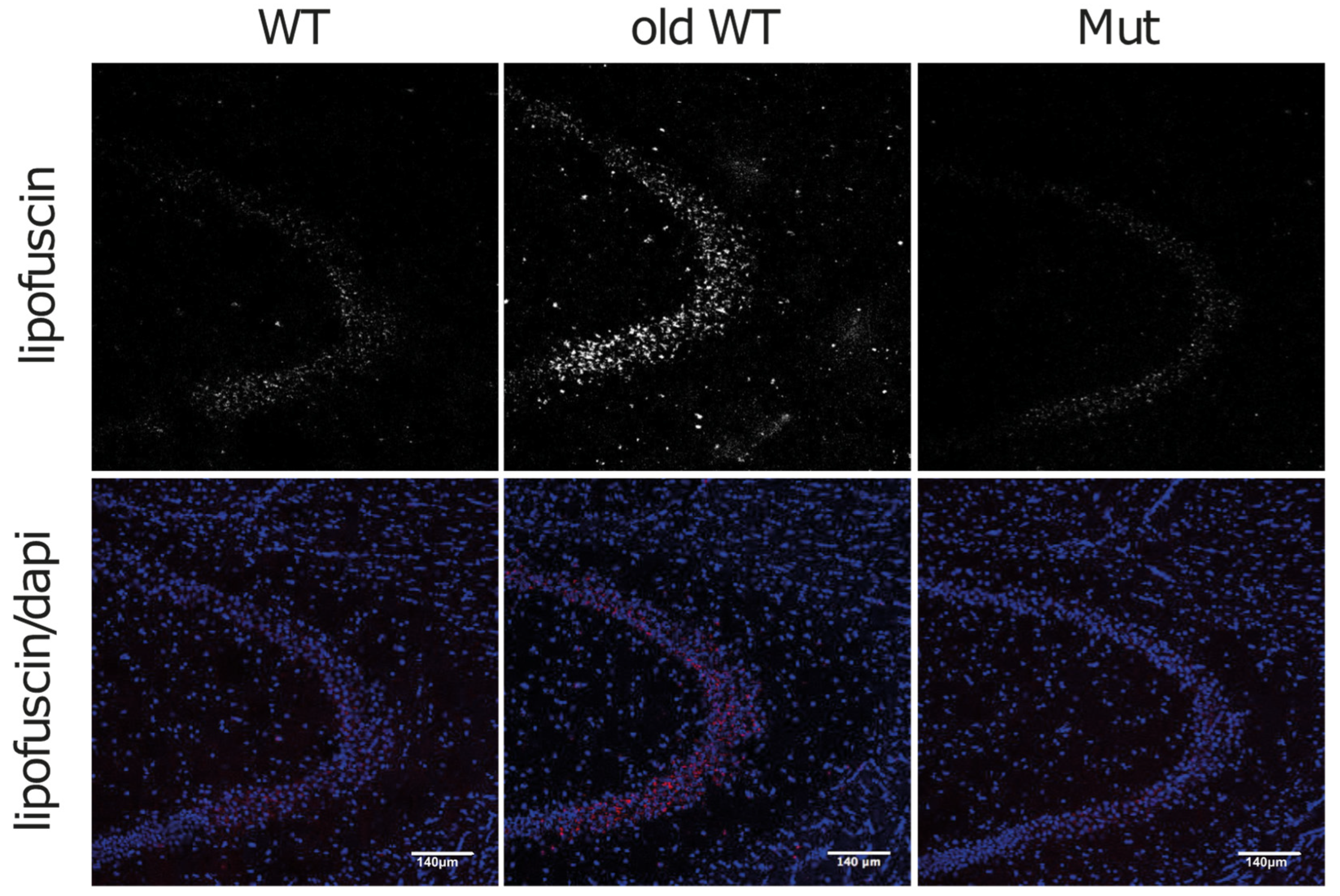
3.3. Lipofuscin and Mitochondrial Dysfunction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- da Silva, P.F.L.; Schumacher, B. Principles of the Molecular and Cellular Mechanisms of Aging. J. Investig. Dermatol. 2021, 141, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Z.; Ren, Y.; Wang, Y.; Fang, J.; Yue, H.; Ma, S.; Guan, F. Aging and Age-related Diseases: From Mechanisms to Therapeutic Strategies. Biogerontology 2021, 22, 165–187. [Google Scholar] [CrossRef]
- Jin, K. Modern Biological Theories of Aging. Aging Dis. 2010, 1, 72–74. [Google Scholar] [PubMed]
- McCay, C.M. Is Longevity Compatible with Optimum Growth? Science 1933, 77, 410–411. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, C.; Chang, J.; Gensch, E.; Rudner, A.; Tabtiang, R.A. C. elegans Mutant That Lives Twice as Long as Wild Type. Nature 1993, 366, 461–464. [Google Scholar] [CrossRef]
- Lamming, D.W.; Ye, L.; Sabatini, D.M.; Baur, J.A. Rapalogs and mTOR Inhibitors as Anti-Aging Therapeutics. J. Clin. Investig. 2013, 123, 980–989. [Google Scholar] [CrossRef]
- Guarente, L.; Kenyon, C. Genetic Pathways That Regulate Ageing in Model Organisms. Nature 2000, 408, 255–262. [Google Scholar] [CrossRef]
- Budovsky, A.; Abramovich, A.; Cohen, R.; Chalifa-Caspi, V.; Fraifeld, V. Longevity Network: Construction and Implications. Mech. Ageing Dev. 2007, 128, 117–124. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of Aging: An Expanding Universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef]
- Taylor, R.C.; Dillin, A. Aging as an Event of Proteostasis Collapse. Cold Spring Harb. Perspect. Biol. 2011, 3, a004440. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The Ubiquitin System. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Codogno, P. The Mechanism and Physiological Function of Macroautophagy. J. Innate Immun. 2013, 5, 427–433. [Google Scholar] [CrossRef]
- Jung, T.; Catalgol, B.; Grune, T. The Proteasomal System. Mol. Aspects Med. 2009, 30, 191–296. [Google Scholar] [CrossRef]
- Morimoto, R.I. Proteotoxic Stress and Inducible Chaperone Networks in Neurodegenerative Disease and Aging. Genes Dev. 2008, 22, 1427–1438. [Google Scholar] [CrossRef]
- Baraibar, M.A.; Friguet, B. Changes of the Proteasomal System during the Aging Process. Prog. Mol. Biol. Transl. Sci. 2012, 109, 249–275. [Google Scholar] [CrossRef]
- Cuanalo-Contreras, K.; Mukherjee, A.; Soto, C. Role of Protein Misfolding and Proteostasis Deficiency in Protein Misfolding Diseases and Aging. Int. J. Cell Biol. 2013, 2013, 638083. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.; Li, L.; Perry, G.; Lee, H.-G.; Zhu, X. Oxidative Stress and Mitochondrial Dysfunction in Alzheimer’s Disease. Biochim. Biophys. Acta 2013, 1842, 1240–1247. [Google Scholar] [CrossRef]
- Büeler, H. Impaired Mitochondrial Dynamics and Function in the Pathogenesis of Parkinson’s Disease. Exp. Neurol. 2009, 218, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Trifunovic, A.; Wredenberg, A.; Falkenberg, M.; Spelbrink, J.N.; Rovio, A.T.; Bruder, C.E.; Bohlooly-Y, M.; Gidlöf, S.; Oldfors, A.; Wibom, R.; et al. Premature Ageing in Mice Expressing Defective Mitochondrial DNA Polymerase. Nature 2004, 429, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.M.; Stewart, J.B.; Hagström, E.; Brené, S.; Mourier, A.; Coppotelli, G.; Freyer, C.; Lagouge, M.; Hoffer, B.J.; Olson, L.; et al. Germline Mitochondrial DNA Mutations Aggravate Ageing and Can Impair Brain Development. Nature 2013, 501, 412–415. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.M.; Coppotelli, G.; Hoffer, B.J.; Olson, L. Maternally Transmitted Mitochondrial DNA Mutations Can Reduce Lifespan. Sci. Rep. 2014, 4, 6569. [Google Scholar] [CrossRef] [PubMed]
- Coppotelli, G.; Mughal, N.; Marescotti, D.; Masucci, M.G. High Avidity Binding to DNA Protects Ubiquitylated Substrates from Proteasomal Degradation. J. Biol. Chem. 2011, 286, 19565–19575. [Google Scholar] [CrossRef]
- Martorelli, D.; Coppotelli, G.; Muraro, E.; Dolcetti, R.; Masucci, M.G. Remodeling of the Epitope Repertoire of a Candidate Idiotype Vaccine by Targeting to Lysosomal Degradation in Dendritic Cells. Cancer Immunol. Immunother. 2012, 61, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Owens, G.C.; Walcott, E.C. Extensive Fusion of Mitochondria in Spinal Cord Motor Neurons. PLoS ONE 2012, 7, e38435. [Google Scholar] [CrossRef]
- Brunk, U.T.; Terman, A. The Mitochondrial-Lysosomal Axis Theory of Aging: Accumulation of Damaged Mitochondria as a Result of Imperfect Autophagocytosis. Eur. J. Biochem. 2002, 269, 1996–2002. [Google Scholar] [CrossRef]
- Nakanishi, H.; Tominaga, K.; Amano, T.; Hirotsu, I.; Inoue, T.; Yamamoto, K. Age-Related Changes in Activities and Localizations of Cathepsins D, E, B, and L in the Rat Brain Tissues. Exp. Neurol. 1994, 126, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Ferland, G.; Perea, A.; Audet, M.; Tuchweber, B. Characterization of Liver Lysosomal Enzyme Activity in Hepatocytes, Kupffer and Endothelial Cells during Aging: Effect of Dietary Restriction. Mech. Ageing Dev. 1990, 56, 143–154. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the Use and Interpretation of Assays for Monitoring Autophagy (4th Edition) 1. Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Ross, J.M.; Öberg, J.; Brené, S.; Coppotelli, G.; Terzioglu, M.; Pernold, K.; Goiny, M.; Sitnikov, R.; Kehr, J.; Trifunovic, A.; et al. High Brain Lactate Is a Hallmark of Aging and Caused by a Shift in the Lactate Dehydrogenase A/B Ratio. Proc. Natl. Acad. Sci. USA 2010, 107, 20087–20092. [Google Scholar] [CrossRef]
- Höhn, A.; Grune, T. Lipofuscin: Formation, Effects and Role of Macroautophagy. Redox Biol. 2013, 1, 140–144. [Google Scholar] [CrossRef]
- Terman, A.; Brunk, U.T. Lipofuscin. Int. J. Biochem. Cell Biol. 2004, 36, 1400–1404. [Google Scholar] [CrossRef]
- Veldhuis, J.D. Changes in Pituitary Function with Ageing and Implications for Patient Care. Nat. Rev. Endocrinol. 2013, 9, 205–215. [Google Scholar] [CrossRef]
- Kikis, E.A.; Gidalevitz, T.; Morimoto, R.I. Protein Homeostasis in Models of Aging and Age-Related Conformational Disease. Adv. Exp. Med. Biol. 2010, 694, 138–159. [Google Scholar]
- Zeng, B.-Y.; Medhurst, A.D.; Jackson, M.; Rose, S.; Jenner, P. Proteasomal Activity in Brain Differs between Species and Brain Regions and Changes with Age. Mech. Ageing Dev. 2005, 126, 760–766. [Google Scholar] [CrossRef] [PubMed]
- Keller, J.N.; Hanni, K.B.; Markesbery, W.R. Possible Involvement of Proteasome Inhibition in Aging: Implications for Oxidative Stress. Mech. Ageing Dev. 2000, 113, 61–70. [Google Scholar] [CrossRef]
- Shibatani, T.; Ward, W.F. Effect of Age and Food Restriction on Alkaline Protease Activity in Rat Liver. J. Gerontol. A Biol. Sci. Med. Sci. 1996, 51, B175–B178. [Google Scholar] [CrossRef]
- Petersen, A.; Honarvar, A.; Zetterberg, M. Changes in Activity and Kinetic Properties of the Proteasome in Different Rat Organs during Development and Maturation. Curr. Gerontol. Geriatr. Res. 2010, 2010, 230697. [Google Scholar] [CrossRef] [PubMed]
- Dasuri, K.; Zhang, L.; Ebenezer, P.; Liu, Y.; Fernandez-Kim, S.O.; Keller, J.N. Aging and Dietary Restriction Alter Proteasome Biogenesis and Composition in the Brain and Liver. Mech. Ageing Dev. 2009, 130, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Goto, S. Age-Related Changes in the 20S and 26S Proteasome Activities in the Liver of Male F344 Rats. Mech. Ageing Dev. 1998, 102, 55–66. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, H.; Perry, S.W.; Figueiredo-Pereira, M.E. Negative Regulation of 26S Proteasome Stability via Calpain-Mediated Cleavage of Rpn10 upon Mitochondrial Dysfunction in Neurons. J. Biol. Chem. 2013, 288, 12161–12174. [Google Scholar] [CrossRef]
- Kujoth, G.C.; Hiona, A.; Pugh, T.D.; Someya, S.; Panzer, K.; Wohlgemuth, S.E.; Hofer, T.; Seo, A.Y.; Sullivan, R.; Jobling, W.A.; et al. Mitochondrial DNA Mutations, Oxidative Stress, and Apoptosis in Mammalian Aging. Science 2005, 309, 481–484. [Google Scholar] [CrossRef]
- Kolesar, J.E.; Safdar, A.; Abadi, A.; MacNeil, L.G.; Crane, J.D.; Tarnopolsky, M.A.; Kaufman, B.A. Defects in Mitochondrial DNA Replication and Oxidative Damage in Muscle of mtDNA Mutator Mice. Free Radic. Biol. Med. 2014, 75, 241–251. [Google Scholar] [CrossRef]
- Logan, A.; Shabalina, I.G.; Prime, T.A.; Rogatti, S.; Kalinovich, A.V.; Hartley, R.C.; Budd, R.C.; Cannon, B.; Murphy, M.P. In Vivo Levels of Mitochondrial Hydrogen Peroxide Increase with Age in mtDNA Mutator Mice. Aging Cell 2014, 13, 765–768. [Google Scholar] [CrossRef]
- Livnat-Levanon, N.; Kevei, É.; Kleifeld, O.; Krutauz, D.; Segref, A.; Rinaldi, T.; Erpapazoglou, Z.; Cohen, M.; Reis, N.; Hoppe, T.; et al. Reversible 26S Proteasome Disassembly upon Mitochondrial Stress. Cell Rep. 2014, 7, 1371–1380. [Google Scholar] [CrossRef]
- Schmidt, M.; Finley, D. Regulation of Proteasome Activity in Health and Disease. Biochim. Biophys. Acta 2014, 1843, 13–25. [Google Scholar] [CrossRef]
- Ross, J.M.; Olson, L.; Coppotelli, G. Mitochondrial and Ubiquitin Proteasome System Dysfunction in Ageing and Disease: Two Sides of the Same Coin? Int. J. Mol. Sci. 2015, 16, 19458–19476. [Google Scholar] [CrossRef] [PubMed]
- Li-Harms, X.; Milasta, S.; Lynch, J.; Wright, C.; Joshi, A.; Iyengar, R.; Neale, G.; Wang, X.; Wang, Y.-D.; Prolla, T.A.; et al. Mito-Protective Autophagy Is Impaired in Erythroid Cells of Aged mtDNA-Mutator Mice. Blood 2015, 125, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Gomes, L.C.; Scorrano, L. Mitochondrial Morphology in Mitophagy and Macroautophagy. Biochim. Et Biophys. Acta (BBA) Mol. Cell Res. 2013, 1833, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Apostolova, N.; Gomez-Sucerquia, L.J.; Gortat, A.; Blas-Garcia, A.; Esplugues, J.V. Compromising Mitochondrial Function with the Antiretroviral Drug Efavirenz Induces Cell Survival-Promoting Autophagy. Hepatology 2011, 54, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Chen, H.-F.; Gi, S.-J.; Chi, T.-H.; Cheng, C.-K.; Hsu, C.-F.; Ma, Y.-S.; Wei, Y.-H.; Liu, C.-S.; Hsieh, M. Decreased Heat Shock Protein 27 Expression and Altered Autophagy in Human Cells Harboring A8344G Mitochondrial DNA Mutation. Mitochondrion 2011, 11, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.J.; Quijano, C.; Chen, E.; Liu, H.; Cao, L.; Fergusson, M.M.; Rovira, I.I.; Gutkind, S.; Daniels, M.P.; Komatsu, M.; et al. Mitochondrial Dysfunction and Oxidative Stress Mediate the Physiological Impairment Induced by the Disruption of Autophagy. Aging 2009, 1, 425–437. [Google Scholar] [CrossRef]
- Krebiehl, G.; Ruckerbauer, S.; Burbulla, L.F.; Kieper, N.; Maurer, B.; Waak, J.; Wolburg, H.; Gizatullina, Z.; Gellerich, F.N.; Woitalla, D.; et al. Reduced Basal Autophagy and Impaired Mitochondrial Dynamics Due to Loss of Parkinson’s Disease-Associated Protein DJ-1. PLoS ONE 2010, 5, e9367. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; George, S.; Roy, U.; Ramachandran, D.; Kolthur-Seetharam, U. NAD: A Master Regulator of Transcription. Biochim. Biophys. Acta 2010, 1799, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.P.; Price, N.L.; Ling, A.J.Y.; Moslehi, J.J.; Montgomery, M.K.; Rajman, L.; White, J.P.; Teodoro, J.S.; Wrann, C.D.; Hubbard, B.P.; et al. Declining NAD(+) Induces a Pseudohypoxic State Disrupting Nuclear-Mitochondrial Communication during Aging. Cell 2013, 155, 1624–1638. [Google Scholar] [CrossRef] [PubMed]
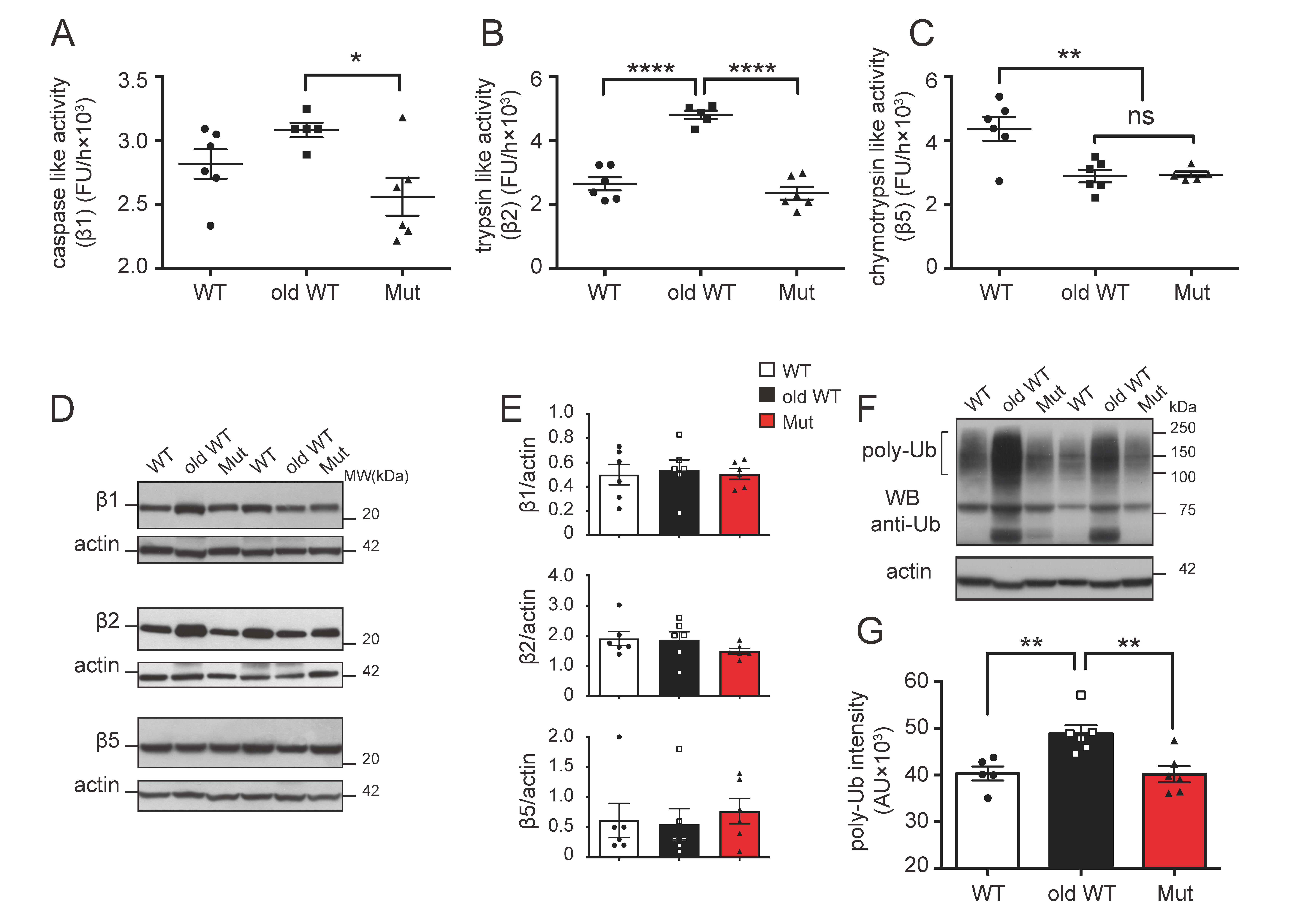

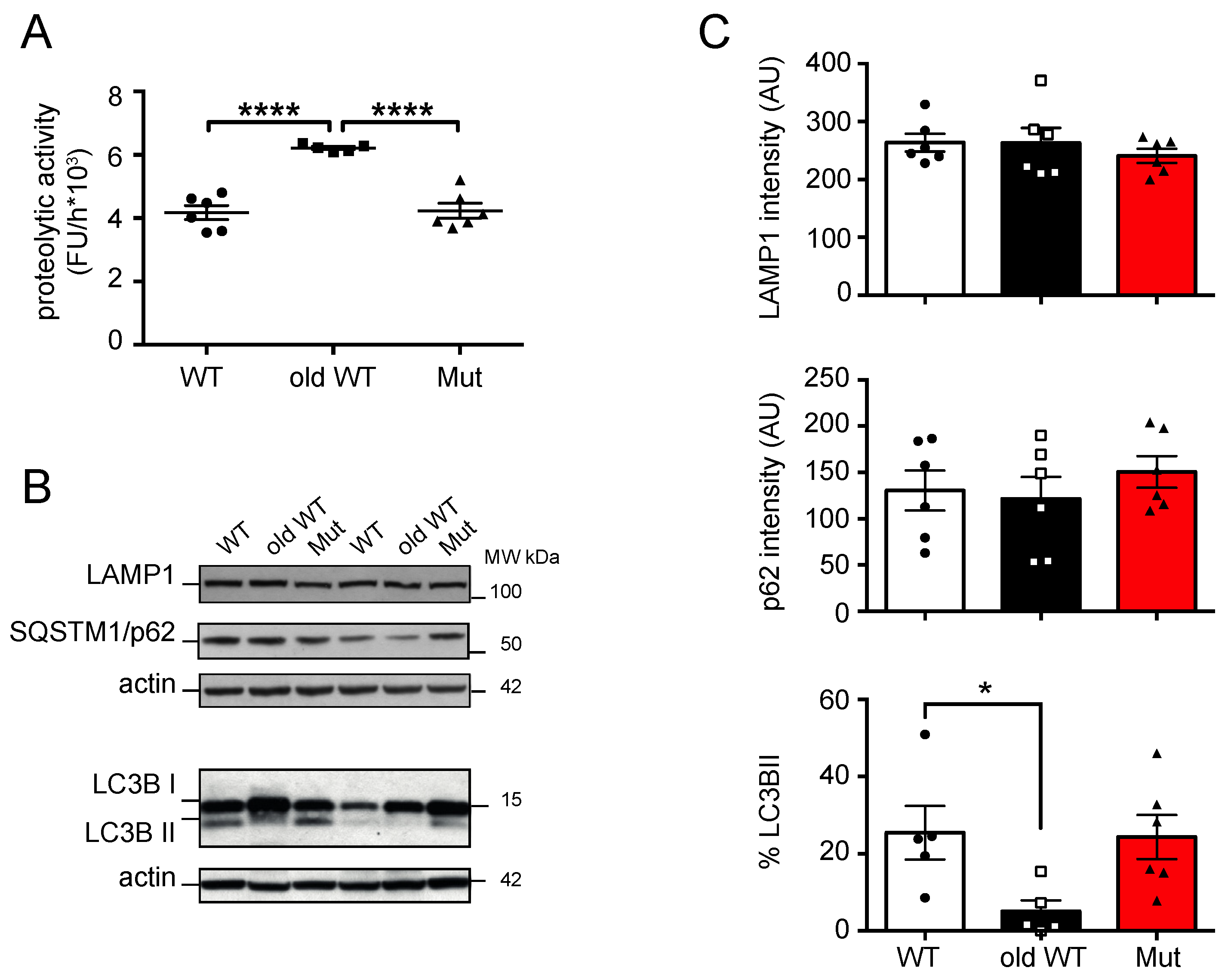

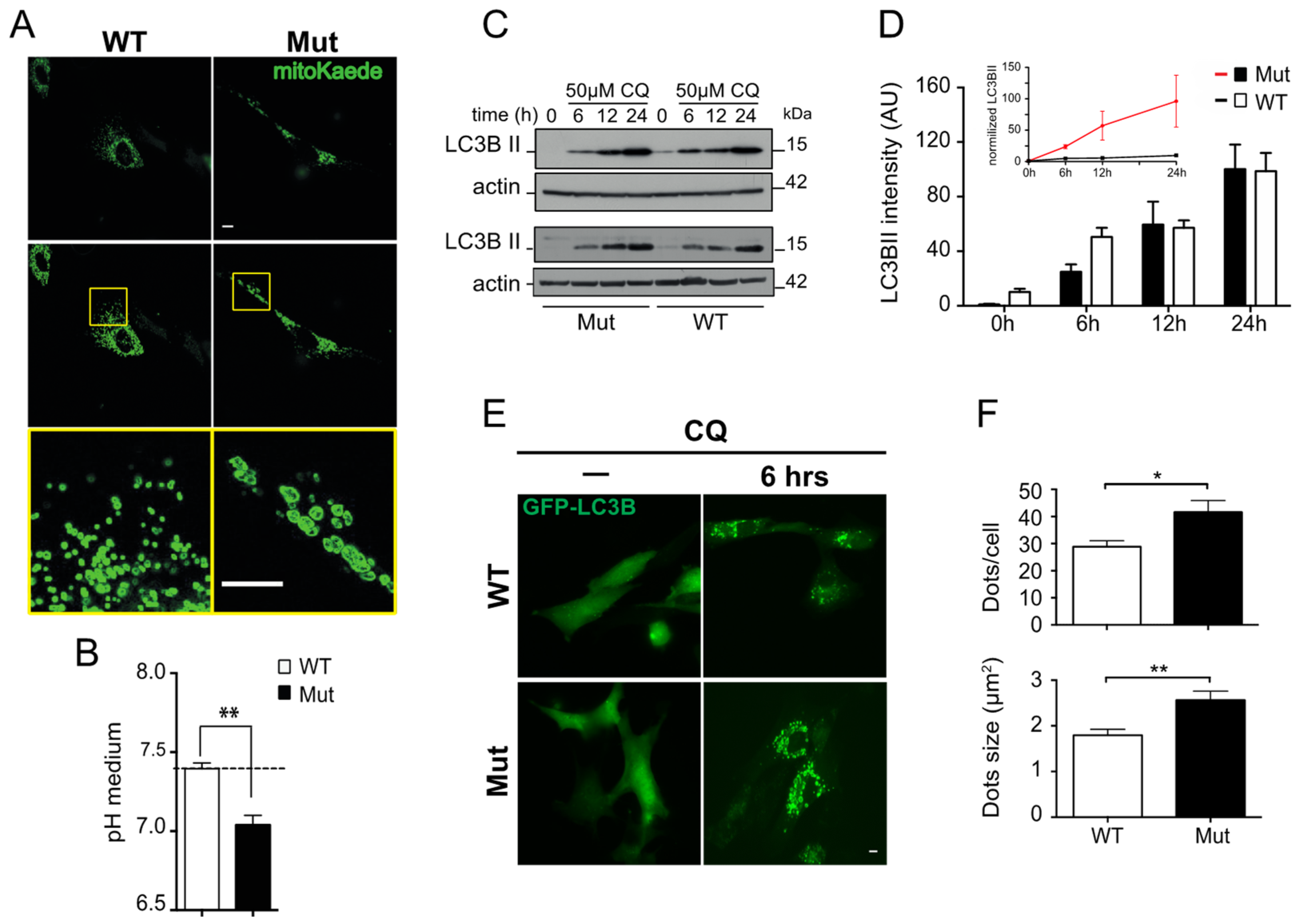
| PROTEASOME | AUTOPHAGY-LYSOSOME | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LIVER | CEREBELLUM | LIVER | CEREBELLUM | |||||||||
| Condition | C | T | CT | p-Ub | C | T | CT | p-Ub | PA | LC3B | PA | LC3B |
| Mitochondrial dysfunction | − | 0 | −− | 0 | (−) | 0 | (−) | (+) | 0 | (−) | (+) | 0 |
| Aging | (+) | +++ | −− | + | (−) | 0 | − | + | +++ | −− | 0 | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ross, J.M.; Olson, L.; Coppotelli, G. Mitochondrial Dysfunction and Protein Homeostasis in Aging: Insights from a Premature-Aging Mouse Model. Biomolecules 2024, 14, 162. https://doi.org/10.3390/biom14020162
Ross JM, Olson L, Coppotelli G. Mitochondrial Dysfunction and Protein Homeostasis in Aging: Insights from a Premature-Aging Mouse Model. Biomolecules. 2024; 14(2):162. https://doi.org/10.3390/biom14020162
Chicago/Turabian StyleRoss, Jaime M., Lars Olson, and Giuseppe Coppotelli. 2024. "Mitochondrial Dysfunction and Protein Homeostasis in Aging: Insights from a Premature-Aging Mouse Model" Biomolecules 14, no. 2: 162. https://doi.org/10.3390/biom14020162






