Transcriptional Coactivator BOB1 (OBF1, OCA-B) Modulates the Specificity of DNA Recognition by the POU-Domain Factors OCT1 and OCT2 in a Monomeric Configuration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid Constructs, Expression, and Purification of Proteins
2.2. EMSA–SELEX
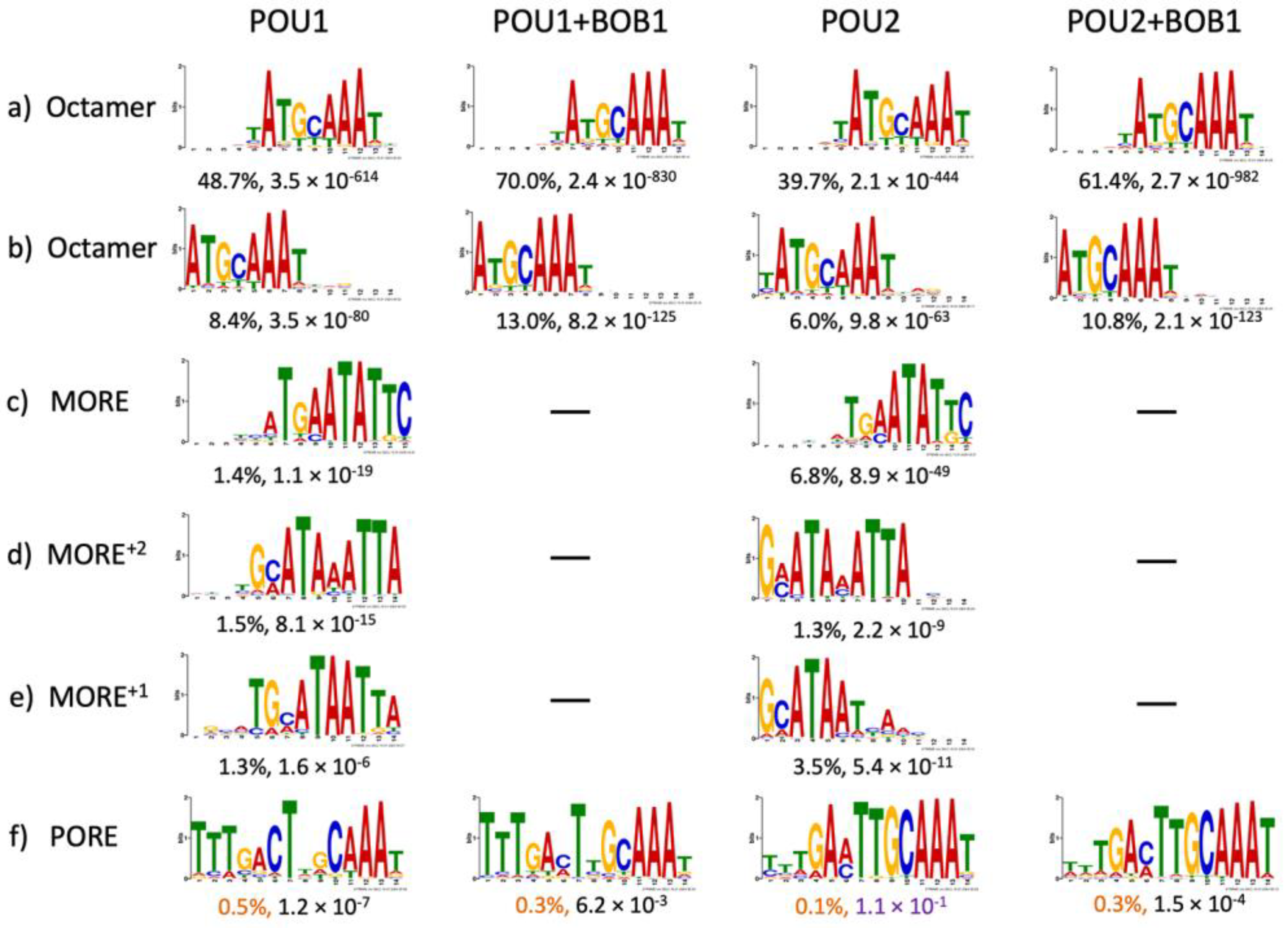
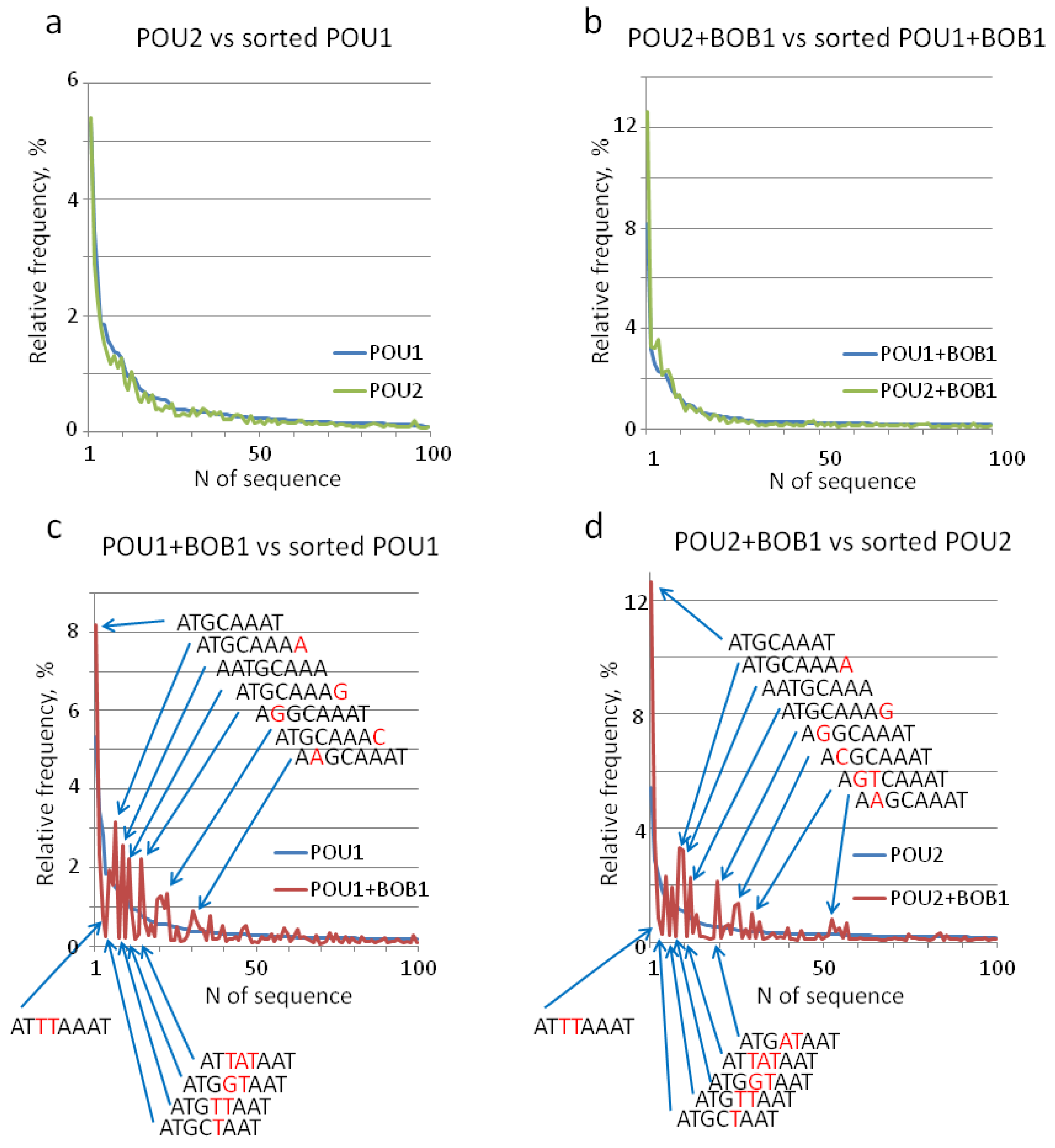
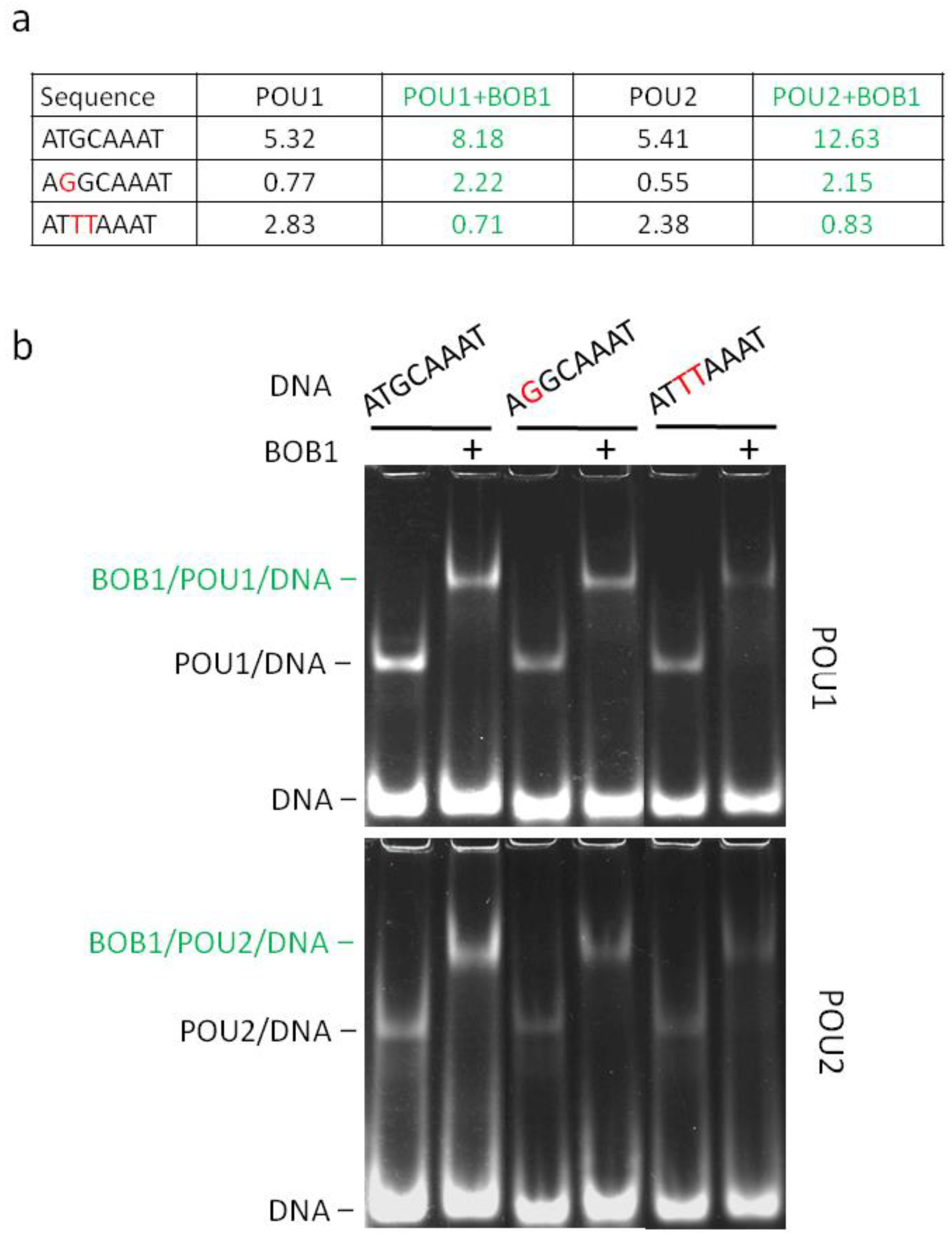
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yeremenko, N.; Danger, R.; Baeten, D.; Tomilin, A.; Brouard, S. Transcriptional regulator BOB.1: Molecular mechanisms and emerging role in chronic inflammation and autoimmunity. Autoimmun. Rev. 2021, 20, 102833. [Google Scholar] [CrossRef] [PubMed]
- Betzler, A.C.; Ushmorov, A.; Brunner, C. The transcriptional program during germinal center reaction—A close view at GC B cells, Tfh cells and Tfr cells. Front. Immunol. 2023, 14, 1125503. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Cao, C.; Choukrallah, M.A.; Tang, F.; Christofori, G.; Kohler, H.; Wu, F.; Fodor, B.D.; Frederiksen, M.; Willis, S.N.; et al. OBF1 and Oct factors control the germinal center transcriptional program. Blood 2021, 137, 2920–2934. [Google Scholar] [CrossRef] [PubMed]
- Doane, A.S.; Chu, C.S.; Di Giammartino, D.C.; Rivas, M.A.; Hellmuth, J.C.; Jiang, Y.; Yusufova, N.; Alonso, A.; Roeder, R.G.; Apostolou, E.; et al. OCT2 pre-positioning facilitates cell fate transition and chromatin architecture changes in humoral immunity. Nat. Immunol. 2021, 22, 1327–1340. [Google Scholar] [CrossRef]
- Weirauch, M.T.; Cote, A.; Nore, L.R.; Annala, M.; Zhao, Y.; Riley, T.R.; Saez-Rodriguez, J.; Cokelaer, T.; Vedenko, A.; Talukder, S.; et al. Evaluation of methods for modeling transcription factor sequence specificity. Nat. Biotechnol. 2013, 31, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Meers, M.P.; Janssens, D.H.; Henikoff, S. Pioneer Factor-Nucleosome Binding Events during Differentiation Are Motif Encoded. Mol. Cell 2019, 75, 562–575.e5. [Google Scholar] [CrossRef] [PubMed]
- Kibet, C.K.; Machanick, P. Transcription factor motif quality assessment requires systematic comparative analysis. F1000Research 2015, 4, ISCB Comm J-1429. [Google Scholar] [CrossRef]
- Malik, V.; Zimmer, D.; Jauch, R. Diversity among POU transcription factors in chromatin recognition and cell fate reprogramming. Cell Mol. Life Sci. 2018, 75, 1587–1612. [Google Scholar] [CrossRef]
- Jolma, A.; Yin, Y.; Nitta, K.R.; Dave, K.; Popov, A.; Taipale, M.; Enge, M.; Kivioja, T.; Morgunova, E.; Taipale, J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 2015, 527, 384–388. [Google Scholar] [CrossRef]
- Gstaiger, M.; Knoepfel, L.; Georgiev, O.; Schaffner, W.; Hovens, C.M. A B-cell coactivator of octamer-binding transcription factors. Nature 1995, 373, 360–362. [Google Scholar] [CrossRef]
- Gstaiger, M.; Georgiev, O.; van Leeuwen, H.; van der Vliet, P.; Schaffner, W. The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J. 1996, 15, 2781–2790. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.S.; Cheung, S.L.; Gao, Y.; Weinbuch, M.; Hu, H.; Shi, L.; Ti, S.C.; Hutchins, A.P.; Cojocaru, V.; Jauch, R. The homeodomain of Oct4 is a dimeric binder of methylated CpG elements. Nucleic Acids Res. 2023, 51, 1120–1138. [Google Scholar] [CrossRef] [PubMed]
- Phillips, K.; Luisi, B. The virtuoso of versatility: POU proteins that flex to fit. J. Mol. Biol. 2000, 302, 1023–1039. [Google Scholar] [CrossRef] [PubMed]
- Lins, K.; Reményi, A.; Tomilin, A.; Massa, S.; Wilmanns, M.; Matthias, P.; Schöler, H.R. OBF1 enhances transcriptional potential of Oct1. EMBO J. 2003, 22, 2188–2198. [Google Scholar] [CrossRef] [PubMed]
- Botquin, V.; Hess, H.; Fuhrmann, G.; Anastassiadis, C.; Gross, M.K.; Vriend, G.; Schöler, H.R. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 1998, 12, 2073–2090. [Google Scholar] [CrossRef] [PubMed]
- Tomilin, A.; Reményi, A.; Lins, K.; Bak, H.; Leidel, S.; Vriend, G.; Wilmanns, M.; Schöler, H.R. Synergism with the coactivator OBF-1 (OCA-B, BOB-1) is mediated by a specific POU dimer configuration. Cell 2000, 103, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L. STREME: Accurate and versatile sequence motif discovery. Bioinformatics 2021, 37, 2834–2840. [Google Scholar] [CrossRef] [PubMed]
- Sauter, P.; Matthias, P. Coactivator OBF-1 makes selective contacts with both the POU-specific domain and the POU homeodomain and acts as a molecular clamp on DNA. Mol. Cell Biol. 1998, 18, 7397–7409. [Google Scholar] [CrossRef]
- Cepek, K.L.; Chasman, D.I.; Sharp, P.A. Sequence-specific DNA binding of the B-cell-specific coactivator OCA-B. Genes Dev. 1996, 10, 2079–2088. [Google Scholar] [CrossRef]
- Chasman, D.; Cepek, K.; Sharp, P.A.; Pabo, C.O. Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: Specific recognition of a protein-DNA interface. Genes Dev. 1999, 13, 2650–2657. [Google Scholar] [CrossRef]
- Klemm, J.D.; Rould, M.A.; Aurora, R.; Herr, W.; Pabo, C.O. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell 1994, 77, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Jolma, A.; Yan, J.; Whitington, T.; Toivonen, J.; Nitta, K.R.; Rastas, P.; Morgunova, E.; Enge, M.; Taipale, M.; Wei, G.; et al. DNA-binding specificities of human transcription factors. Cell 2013, 152, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Reményi, A.; Tomilin, A.; Pohl, E.; Lins, K.; Philippsen, A.; Reinbold, R.; Schöler, H.R.; Wilmanns, M. Differential dimer activities of the transcription factor Oct-1 by DNA-induced interface swapping. Mol. Cell 2001, 8, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Mistri, T.K.; Devasia, A.G.; Chu, L.T.; Ng, W.P.; Halbritter, F.; Colby, D.; Martynoga, B.; Tomlinson, S.R.; Chambers, I.; Robson, P.; et al. Selective influence of Sox2 on POU transcription factor binding in embryonic and neural stem cells. EMBO Rep. 2015, 16, 1177–1191. [Google Scholar] [CrossRef] [PubMed]
- Malik, V.; Glaser, L.V.; Zimmer, D.; Velychko, S.; Weng, M.; Holzner, M.; Arend, M.; Chen, Y.; Srivastava, Y.; Veerapandian, V.; et al. Pluripotency reprogramming by competent and incompetent POU factors uncovers temporal dependency for Oct4 and Sox2. Nat. Commun. 2019, 10, 3477. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.S.; Chen, Y.; Gao, Y.; Bednarz, A.; Wei, Y.; Malik, V.; Ho, D.H.; Weng, M.; Ho, S.Y.; Srivastava, Y.; et al. Directed Evolution of an Enhanced POU Reprogramming Factor for Cell Fate Engineering. Mol. Biol. Evol. 2021, 38, 2854–2868. [Google Scholar] [CrossRef] [PubMed]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef]
- Holt, R.J.; Zhang, Y.; Binia, A.; Dixon, A.L.; Vandiedonck, C.; Cookson, W.O.; Knight, J.C.; Moffatt, M.F. Allele-specific transcription of the asthma-associated PHD finger protein 11 gene (PHF11) modulated by octamer-binding transcription factor 1 (Oct-1). J. Allergy Clin. Immunol. 2011, 127, 1054–1062.e1-2. [Google Scholar] [CrossRef]
- Knight, J.C.; Udalova, I.; Hill, A.V.; Greenwood, B.M.; Peshu, N.; Marsh, K.; Kwiatkowski, D. A polymorphism that affects OCT-1 binding to the TNF promoter region is associated with severe malaria. Nat. Genet. 1999, 22, 145–150. [Google Scholar] [CrossRef]
- Hohjoh, H.; Tokunaga, K. Allele-specific binding of the ubiquitous transcription factor OCT-1 to the functional single nucleotide polymorphism (SNP) sites in the tumor necrosis factor-alpha gene (TNFA) promoter. Genes Immun. 2001, 2, 105–109. [Google Scholar] [CrossRef]
- Kiesler, P.; Shakya, A.; Tantin, D.; Vercelli, D. An allergy-associated polymorphism in a novel regulatory element enhances IL13 expression. Hum. Mol. Genet. 2009, 18, 4513–4520. [Google Scholar] [CrossRef]
- Vince, N.; Li, H.; Ramsuran, V.; Naranbhai, V.; Duh, F.M.; Fairfax, B.P.; Saleh, B.; Knight, J.C.; Anderson, S.K.; Carrington, M. HLA-C Level Is Regulated by a Polymorphic Oct1 Binding Site in the HLA-C Promoter Region. Am. J. Hum. Genet. 2016, 99, 1353–1358. [Google Scholar] [CrossRef]
- Apps, R.; Qi, Y.; Carlson, J.M.; Chen, H.; Gao, X.; Thomas, R.; Yuki, Y.; Del Prete, G.Q.; Goulder, P.; Brumme, Z.L.; et al. Influence of HLA-C expression level on HIV control. Science 2013, 340, 87–91. [Google Scholar] [CrossRef]
- Van Heel, D.A.; Udalova, I.A.; De Silva, A.P.; McGovern, D.P.; Kinouchi, Y.; Hull, J.; Lench, N.J.; Cardon, L.R.; Carey, A.H.; Jewell, D.P.; et al. Inflammatory bowel disease is associated with a TNF polymorphism that affects an interaction between the OCT1 and NF(−kappa)B transcription factors. Hum. Mol. Genet. 2002, 11, 1281–1289. [Google Scholar] [CrossRef]
- Farh, K.K.; Marson, A.; Zhu, J.; Kleinewietfeld, M.; Housley, W.J.; Beik, S.; Shoresh, N.; Whitton, H.; Ryan, R.J.H.; Shishkin, A.A.; et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015, 518, 337–343. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nazarov, I.B.; Zilov, D.S.; Gordeev, M.N.; Potapenko, E.V.; Yeremenko, N.; Tomilin, A.N. Transcriptional Coactivator BOB1 (OBF1, OCA-B) Modulates the Specificity of DNA Recognition by the POU-Domain Factors OCT1 and OCT2 in a Monomeric Configuration. Biomolecules 2024, 14, 123. https://doi.org/10.3390/biom14010123
Nazarov IB, Zilov DS, Gordeev MN, Potapenko EV, Yeremenko N, Tomilin AN. Transcriptional Coactivator BOB1 (OBF1, OCA-B) Modulates the Specificity of DNA Recognition by the POU-Domain Factors OCT1 and OCT2 in a Monomeric Configuration. Biomolecules. 2024; 14(1):123. https://doi.org/10.3390/biom14010123
Chicago/Turabian StyleNazarov, Igor B., Danil S. Zilov, Mikhail N. Gordeev, Evgenii V. Potapenko, Nataliya Yeremenko, and Alexey N. Tomilin. 2024. "Transcriptional Coactivator BOB1 (OBF1, OCA-B) Modulates the Specificity of DNA Recognition by the POU-Domain Factors OCT1 and OCT2 in a Monomeric Configuration" Biomolecules 14, no. 1: 123. https://doi.org/10.3390/biom14010123






