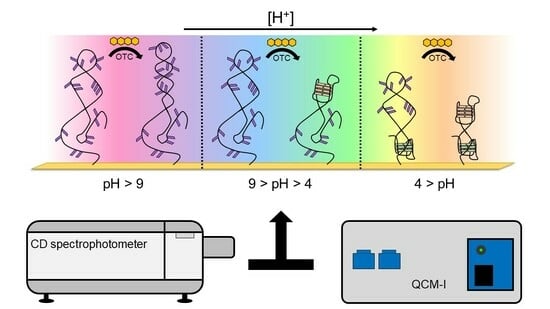
Comparative Analysis of pH and Target-Induced Conformational Changes of an Oxytetracycline Aptamer in Solution Phase and Surface-Immobilized Form
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Buffer Preparation
2.3. Circular Dichroism Spectroscopy Measurement
2.4. Quartz Crystal Microbalance Measurement
3. Results and Discussion
3.1. Induced Conformational Changes in Solution Phase
3.1.1. Thermal Denaturation Study
3.1.2. Effect of pH
3.2. Induced Structural Changes in Surface-Immobilized Aptamer
3.2.1. pH-Induced Responses of the Aptamer
3.2.2. Target-Induced Responses of the Aptamer
Non-Specific OTC Adsorption on Gold and l-Cysteine-Based Antifouling Layer
OTC-Aptamer Binding, the Effect of Surface Density and pH
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Wang, Z.; Wang, S.; Wu, Y.; Ma, Y.; Liu, J. Introduction of SELEX and Important SELEX Variants, in: Aptamers Anal. In Aptamers for Analytical Applications: Affinity Acquisition and Method Design; Dong, Y., Ed.; Wiley-VCH: Weinheim, Germany, 2018; pp. 1–26. [Google Scholar] [CrossRef]
- Birader, K.; Kumar, P.; Tammineni, Y.; Barla, J.A.; Reddy, S.; Suman, P. Colorimetric aptasensor for on-site detection of oxytetracycline antibiotic in milk. Food Chem. 2021, 356, 129659. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ong, S.; Chen, Y.; Jimmy Huang, P.J.; Liu, J. Label-free and Dye-free Fluorescent Sensing of Tetracyclines Using a Capture-Selected DNA Aptamer. Anal. Chem. 2022, 94, 10175–10182. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, A.; Panneerselvam, P. A novel fluorescent sensing platform based on metal-polydopamine frameworks for the dual detection of kanamycin and oxytetracycline. Analyst 2019, 144, 2337–2344. [Google Scholar] [CrossRef]
- He, B.; Wang, L.; Dong, X.; Yan, X.; Li, M.; Yan, S.; Yan, D. Aptamer-based thin film gold electrode modified with gold nanoparticles and carboxylated multi-walled carbon nanotubes for detecting oxytetracycline in chicken samples. Food Chem. 2019, 300, 125179. [Google Scholar] [CrossRef]
- Blidar, A.; Hosu, O.; Feier, B.; Ştefan, G.; Bogdan, D.; Cristea, C. Gold-based nanostructured platforms for oxytetracycline detection from milk by a “signal-on” aptasensing approach. Food Chem. 2022, 371, 131127. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Bai, X.; Xing, C.; Gu, N.; Zhang, B.; Tang, J. Aptamer-based cantilever array sensors for oxytetracycline detection. Anal. Chem. 2013, 85, 2010–2014. [Google Scholar] [CrossRef]
- Spagnolo, S.; De La Franier, B.; Davoudian, K.; Hianik, T.; Thompson, M. Detection of E. coli Bacteria in Milk by an Acoustic Wave Aptasensor with an Anti-Fouling Coating. Sensors 2022, 22, 1853. [Google Scholar] [CrossRef]
- Cai, R.; Chen, X.; Zhang, Y.; Wang, X.; Zhou, N. Systematic bio-fabrication of aptamers and their applications in engineering biology. Syst. Microbiol. Biomanuf. 2023, 3, 223–245. [Google Scholar] [CrossRef]
- Röthlisberger, P.; Hollenstein, M. Aptamer chemistry. Adv. Drug Deliv. Rev. 2018, 134, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Catanante, G.; Mishra, R.K.; Hayat, A.; Marty, J.L. Sensitive analytical performance of folding based biosensor using methylene blue tagged aptamers. Talanta 2016, 153, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Uzawa, T.; White, R.J.; DeMartini, D.; Plaxco, K.W. On the signaling of electrochemical aptamer-based sensors: Collision- and folding-based mechanisms. Electroanalysis 2009, 21, 1267–1271. [Google Scholar] [CrossRef] [PubMed]
- Daniel, C.; Roupioz, Y.; Gasparutto, D.; Livache, T.; Buhot, A. Solution-Phase vs Surface-Phase Aptamer-Protein Affinity from a Label-Free Kinetic Biosensor. PLoS ONE 2013, 8, e75419. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.H.; Chen, R.H.; Lee, C.H.; Chang, Y.; Chen, C.S.; Chen, W.Y. Studies of the binding mechanism between aptamers and thrombin by circular dichroism, surface plasmon resonance and isothermal titration calorimetry. Colloids Surf. B Biointerfaces 2011, 88, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Tello, A.; Cao, R.; Marchant, M.J.; Gomez, H. Conformational Changes of Enzymes and Aptamers Immobilized on Electrodes. Bioconjug. Chem. 2016, 27, 2581–2591. [Google Scholar] [CrossRef]
- Kypr, J.; Kejnovská, I.; Renčiuk, D.; Vorlíčková, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Kypr, J.; Kejnovská, I.; Bednářová, K.; Vorlíčková, M. Circular dichroism spectroscopy of nucleic acids. In Comprehensive Chiroptical Spectroscopy: Applications in Stereochemical Analysis of Synthetic Compounds, Natural Products, and Biomolecules; Berova, N., Polavarapu, P.L., Nakanishi, K., Woody, R.W., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012; Volume 2, pp. 575–586. [Google Scholar] [CrossRef]
- Vorlíčková, M.; Kejnovská, I.; Sagi, J.; Renčiuk, D.; Bednářová, K.; Motlová, J.; Kypr, J. Circular dichroism and guanine quadruplexes. Methods 2012, 57, 64–75. [Google Scholar] [CrossRef]
- Kerler, Y.; Sass, S.; Hille, C.; Menger, M.M. Determination of Aptamer Structure Using Circular Dichroism Spectroscopy. In Nucleic Acid Aptamers: Selection, Characterization, and Application, 2nd ed.; Mayer, G., Menger, M.M., Eds.; Humana Press: Totowa, NJ, USA, 2022; pp. 119–128. [Google Scholar] [CrossRef]
- Johannsmann, D. Energy Trapping and Its Consequences. In The Quartz Crystal Microbalance in Soft Matter Research: Fundamentals and Modeling; Springer: Berlin/Heidelberg, Germany, 2015; pp. 169–189. [Google Scholar] [CrossRef]
- Lim, H.J.; Saha, T.; Beng, T.T.; Tan, W.S.; Ooi, C.W. Quartz crystal microbalance-based biosensors as rapid diagnostic devices for infectious diseases. Biosens. Bioelectron. J. 2020, 168, 112513. [Google Scholar] [CrossRef]
- Osypova, A.; Thakar, D.; Dejeu, J.; Bonnet, H.; Van Der Heyden, A.; Dubacheva, G.V.; Richter, R.P.; Defrancq, E.; Spinelli, N.; Coche-Guérente, L.; et al. Sensor Based on Aptamer Folding to Detect Low-Molecular Weight Analytes. Anal. Chem. 2015, 87, 7566–7574. [Google Scholar] [CrossRef] [PubMed]
- Pons, M.; Perenon, M.; Bonnet, H.; Gillon, E.; Vallée, C.; Coche-Guérente, L.; Defrancq, E.; Spinelli, N.; Van der Heyden, A.; Dejeu, J. Conformational transition in SPR experiments: Impact of spacer length, immobilization mode and aptamer density on signal sign and amplitude. Analyst 2022, 147, 4197–4205. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, H.; Bonnet, H.; Van der Heyden, A.; Defrancq, E.; Spinelli, N.; Coche-Guérente, L.; Dejeu, J. Influence of Aptamer Surface Coverage on Small Target Recognition: A SPR and QCM-D Comparative Study. J. Phys. Chem. C 2019, 123, 13561–13568. [Google Scholar] [CrossRef]
- Grossman, T.H. Tetracycline antibiotics and resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef]
- Liu, X.; Huang, D.; Lai, C.; Zeng, G.; Qin, L.; Zhang, C.; Yi, H.; Li, B.; Deng, R.; Liu, S.; et al. Recent advances in sensors for tetracycline antibiotics and their applications. TrAC Trends Anal. Chem. 2018, 109, 260–274. [Google Scholar] [CrossRef]
- Mehlhorn, A.; Rahimi, P.; Joseph, Y. Aptamer-based biosensors for antibiotic detection: A review. Biosensors 2018, 8, 54. [Google Scholar] [CrossRef] [PubMed]
- Evtugyn, G.; Porfireva, A.; Tsekenis, G.; Oravczova, V.; Hianik, T. Electrochemical Aptasensors for Antibiotics Detection: Recent Achievements and Applications for Monitoring Food Safety. Sensors 2022, 22, 3684. [Google Scholar] [CrossRef] [PubMed]
- Niazi, J.H.; Lee, S.J.; Kim, Y.S.; Gu, M.B. ssDNA aptamers that selectively bind oxytetracycline. Bioorganic Med. Chem. 2008, 16, 1254–1261. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Liu, Y.; Yang, Y.; Zhang, J. A Cathodic “signal-off” Photoelectrochemical Aptasensor for Ultrasensitive and Selective Detection of Oxytetracycline. Anal. Chem. 2015, 87, 12215–12220. [Google Scholar] [CrossRef]
- Yildirim-Tirgil, N.; Lee, J.; Cho, H.; Lee, H.; Somu, S.; Busnaina, A.; Gu, A.Z. A SWCNT based aptasensor system for antibiotic oxytetracycline detection in water samples. Anal. Methods 2019, 11, 2692–2699. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; Yan, P.; Ouyang, Q.; Dong, J.; Qian, J.; Chen, J.; Xu, L.; Li, H. Co3O4 nanoparticles/graphitic carbon nitride heterojunction for photoelectrochemical aptasensor of oxytetracycline. Anal. Chim. Acta 2020, 1125, 299–307. [Google Scholar] [CrossRef]
- Fischer, L.M.; Tenje, M.; Heiskanen, A.R.; Masuda, N.; Castillo, J.; Bentien, A.; Émneus, J.; Jakobsen, M.H.; Boisen, A. Gold cleaning methods for electrochemical detection applications. Microelectron. Eng. 2009, 86, 1282–1285. [Google Scholar] [CrossRef]
- Kejnovská, I.; Renčiuk, D.; Palacký, J.; Vorlíčková, M. CD Study of the G-Quadruplex Conformation. Methods Mol. Biol. 2019, 2035, 25–44. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, A.; Spada, G.P.; da Silva, M.W. Circular Dichroism of Quadruplex Structures. In Quadruplex Nucleic Acids; Springer: Berlin/Heidelberg, Germany, 2012; Volume 330, pp. 67–86. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Arachchilage, G.M.; Basu, S. Metal cations in G-quadruplex folding and stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Del Villar-Guerra, R.; Gray, R.D.; Chaires, J.B. Characterization of quadruplex DNA structure by circular dichroism. Curr. Protoc. Nucleic Acid Chem. 2017, 68, 17. [Google Scholar] [CrossRef] [PubMed]
- Novoseltseva, A.A.; Ivanov, N.M.; Novikov, R.A.; Tkachev, Y.V.; Bunin, D.A.; Gambaryan, A.S.; Tashlitsky, V.N.; Arutyunyan, A.M.; Kopylov, A.M.; Zavyalova, E.G. Structural and functional aspects of G-quadruplex aptamers which bind a broad range of influenza a viruses. Biomolecules 2020, 10, 119. [Google Scholar] [CrossRef]
- Bishop, G.R.; Ren, J.; Polander, B.C.; Jeanfreau, B.D.; Trent, J.O.; Chaires, J.B. Energetic basis of molecular recognition in a DNA aptamer. Biophys. Chem. 2007, 126, 165–175. [Google Scholar] [CrossRef]
- Liu, D.; Bruckbauer, A.; Abell, C.; Balasubramanian, S.; Kang, D.J.; Klenerman, D.; Zhou, D. A reversible pH-driven DNA nanoswitch array. J. Am. Chem. Soc. 2006, 128, 2067–2071. [Google Scholar] [CrossRef]
- Choi, J.; Majima, T. Reversible conformational switching of i-motif DNA studied by fluorescence spectroscopy. Photochem. Photobiol. 2013, 89, 513–522. [Google Scholar] [CrossRef]
- Li, L.; Jiang, Y.; Cui, C.; Yang, Y.; Zhang, P.; Stewart, K.; Pan, X.; Li, X.; Yang, L.; Qiu, L.; et al. Modulating Aptamer Specificity with pH-Responsive DNA Bonds. J. Am. Chem. Soc. 2018, 140, 13335–13339. [Google Scholar] [CrossRef]
- YYan, Y.; Tan, J.H.; Lu, Y.J.; Yan, S.C.; Wong, K.Y.; Li, D.; Gu, L.Q.; Huang, Z.S. G-Quadruplex conformational change driven by pH variation with potential application as a nanoswitch. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 4935–4942. [Google Scholar] [CrossRef]
- Nishio, M.; Tsukakoshi, K.; Ikebukuro, K. G-quadruplex: Flexible conformational changes by cations, pH, crowding and its applications to biosensing. Biosens. Bioelectron. 2021, 178, 113030. [Google Scholar] [CrossRef] [PubMed]
- Hianik, T.; Ostatná, V.; Sonlajtnerova, M.; Grman, I. Influence of ionic strength, pH and aptamer configuration for binding affinity to thrombin. Bioelectrochemistry 2007, 70, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Miranda, G.; Feng, L.; Shiu, S.C.C.; Dirkzwager, R.M.; Cheung, Y.W.; Tanner, J.A.; Schöning, M.J.; Offenhäusser, A.; Mayer, D. Aptamer-based electrochemical biosensor for highly sensitive and selective malaria detection with adjustable dynamic response range and reusability. Sens. Actuators B Chem. 2018, 255, 235–243. [Google Scholar] [CrossRef]
- Pellitero, M.A.; Kundu, N.; Sczepanski, J.; Arroyo-Currás, N. Os(ii/iii) complex supports pH-insensitive electrochemical DNA-based sensing with superior operational stability than the benchmark methylene blue reporter. Analyst 2023, 148, 806–813. [Google Scholar] [CrossRef]
- Jang, Y.H.; Goddard, W.A.; Noyes, K.T.; Sowers, L.C.; Hwang, S.; Chung, D.S. pKa Values of Guanine in Water: Density Functional Theory Calculations Combined with Poisson-Boltzmann Continuum-Solvation Model. J. Phys. Chem. B 2003, 107, 344–357. [Google Scholar] [CrossRef]
- Bucek, P.; Jaumot, J.; Aviñó, A.; Eritja, R.; Gargallo, R. PH-modulated Watson-Crick duplex-quadruplex equilibria of guanine-rich and cytosine-rich DNA sequences 140 base pairs upstream of the c-kit transcription initiation site. Chem. Eur. J. 2009, 15, 12663–12671. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Hou, Y.; Ngai, T.; Zhang, G. pH induced DNA folding at interface. J. Phys. Chem. B 2010, 114, 775–779. [Google Scholar] [CrossRef]
- Lin, P.; Ding, L.; Lin, C.W.; Gu, F. Nonfouling property of zwitterionic cysteine surface. Langmuir 2014, 30, 6497–6507. [Google Scholar] [CrossRef]
- Lin, P.; Chuang, T.L.; Chen, P.Z.; Lin, C.W.; Gu, F.X. Low-Fouling Characteristics of Ultrathin Zwitterionic Cysteine SAMs. Langmuir 2019, 35, 1756–1767. [Google Scholar] [CrossRef]
- Simon, L.; Bognár, Z.; Gyurcsányi, R.E. Finding the Optimal Surface Density of Aptamer Monolayers by SPR Imaging Detection-based Aptamer Microarrays. Electroanalysis 2020, 32, 851–858. [Google Scholar] [CrossRef]











Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jakab, K.; Melios, N.; Tsekenis, G.; Shaban, A.; Horváth, V.; Keresztes, Z. Comparative Analysis of pH and Target-Induced Conformational Changes of an Oxytetracycline Aptamer in Solution Phase and Surface-Immobilized Form. Biomolecules 2023, 13, 1363. https://doi.org/10.3390/biom13091363
Jakab K, Melios N, Tsekenis G, Shaban A, Horváth V, Keresztes Z. Comparative Analysis of pH and Target-Induced Conformational Changes of an Oxytetracycline Aptamer in Solution Phase and Surface-Immobilized Form. Biomolecules. 2023; 13(9):1363. https://doi.org/10.3390/biom13091363
Chicago/Turabian StyleJakab, Kristóf, Nikitas Melios, George Tsekenis, Abdul Shaban, Viola Horváth, and Zsófia Keresztes. 2023. "Comparative Analysis of pH and Target-Induced Conformational Changes of an Oxytetracycline Aptamer in Solution Phase and Surface-Immobilized Form" Biomolecules 13, no. 9: 1363. https://doi.org/10.3390/biom13091363