X-ray Crystallographic Study of Preferred Spacing by the NF-κB p50 Homodimer on κB DNA
Abstract
:1. Introduction
2. Materials and Methods
2.1. DNA Oligonucleotides
2.2. Protein Expression and Purification
2.3. NF-κB p50:DNA Complex Formation and Co-Crystallization
2.4. X-ray Diffraction Data Collection
2.5. Structure Solution and Refinement
| p50:IL-6 | p50:(NGAL)2 | p50:Test | |
|---|---|---|---|
| Data Collection | |||
| X-ray source | NSLS X25 | ALS 8.2.1 | APS ID-24-C |
| Wavelength (Å) | 1.0000 | 0.9999 | 0.9792 |
| Space group | P65 | P212121 | P21 |
| Unit cell (Å) | |||
| a | 162.24 | 68.07 | 47.15 |
| b | 162.24 | 103.34 | 146.45 |
| c | 60.43 | 128.50 | 67.69 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 90 | 90, 98.62, 90 |
| Complex/asymm. unit | 1 | 1 | 1 |
| Resolution range (Å) 1 | 50–2.80 (2.85–2.80) | 50–2.80 (2.85–2.80) | 146.45–3.00 (3.16–3.00) |
| Rsym (%) | 7.9 (59.4) | 8.3 (76.4) | 9.4 (63.1) |
| Observations | 112,366 | 138,228 | 61,285 |
| Unique reflections | 22,645 | 23,123 | 17,962 |
| Completeness (%) | 100 (100) | 100 (99.9) | 98.6 (99.1) |
| Redundancy | 5.0 (5.0) | 6.0 (6.1) | 3.4 (3.5) |
| <I/σ> | 17.9 (2.0) | 19.0 (2.3) | 10.1 (2.0) |
| Refinement | |||
| Number of reflections | 22,567 (2238) | 21,798 (1749) | 17,018 |
| Rwork (%) | 22.4 (38.8) | 23.8 (30.0) | 23.6 (33.9) |
| Rfree (%) 2 | 27.2 (40.1) | 28.7 (35.8) | 30.8 (42.2) |
| Protein/DNA/H2O atom | 4903/690/55 | 4906/805/0 | 4906/691/0 |
| Geometry (R.m.s.d.) | |||
| Bond lengths (Å) | 0.003 | 0.004 | 0.003 |
| Bond angles (°) | 0.630 | 0.594 | 0.587 |
| Mean B (Å2) | 60.3 | 81.3 | 84.1 |
| Ramachandran plot 3 | |||
| Favored (%) | 91.5 | 93.6 | 93.4 |
| Allowed (%) | 7.8 | 5.7 | 5.9 |
| Outliers (%) | 0.7 | 0.7 | 0.7 |
| MolProbity score 4 | 1.84 | 1.78 | 1.78 |
| PDB accession code | 8TKM | 8TKN | 8TKL |
2.6. Structure Analysis and Figures
3. Results
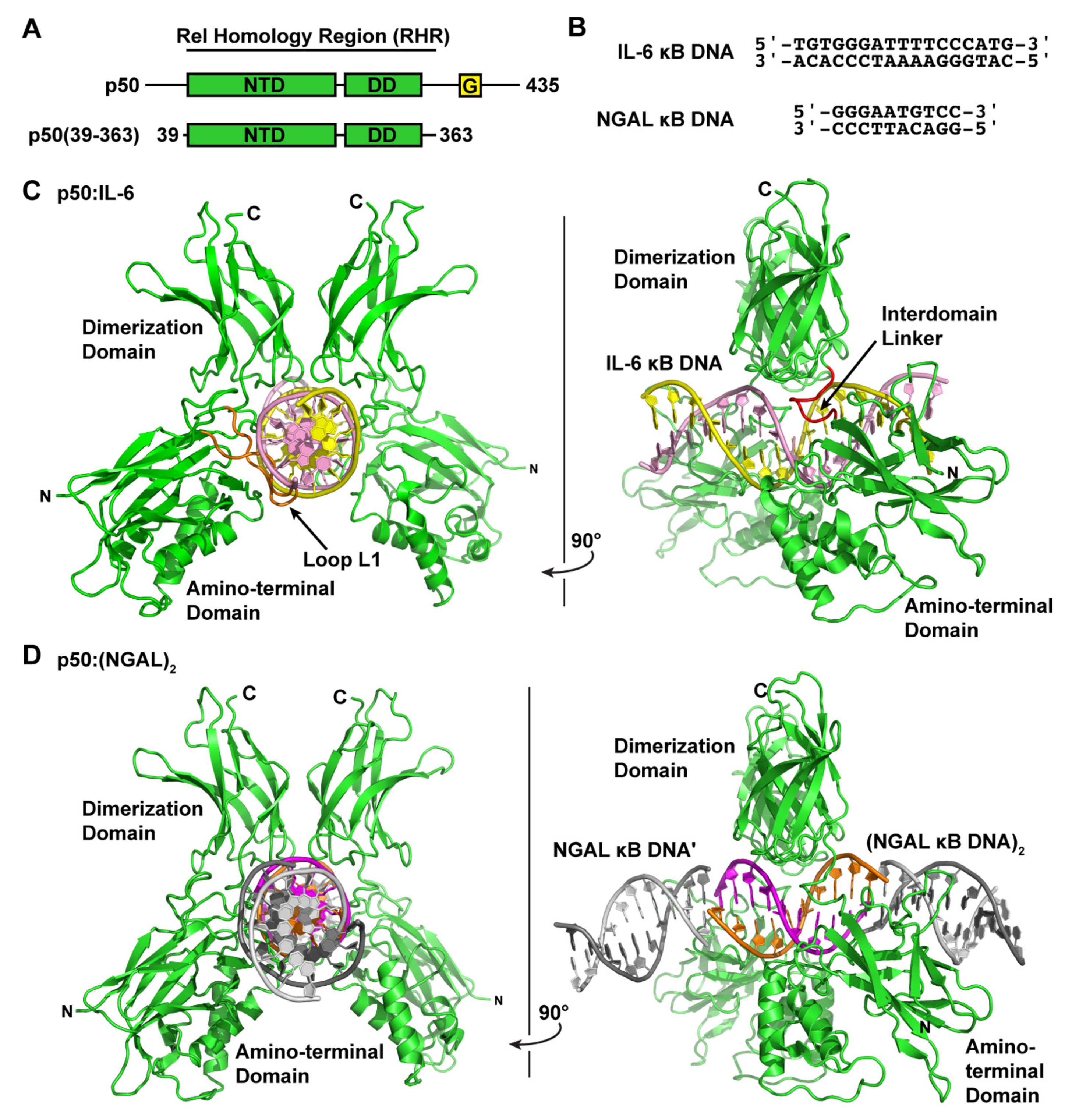
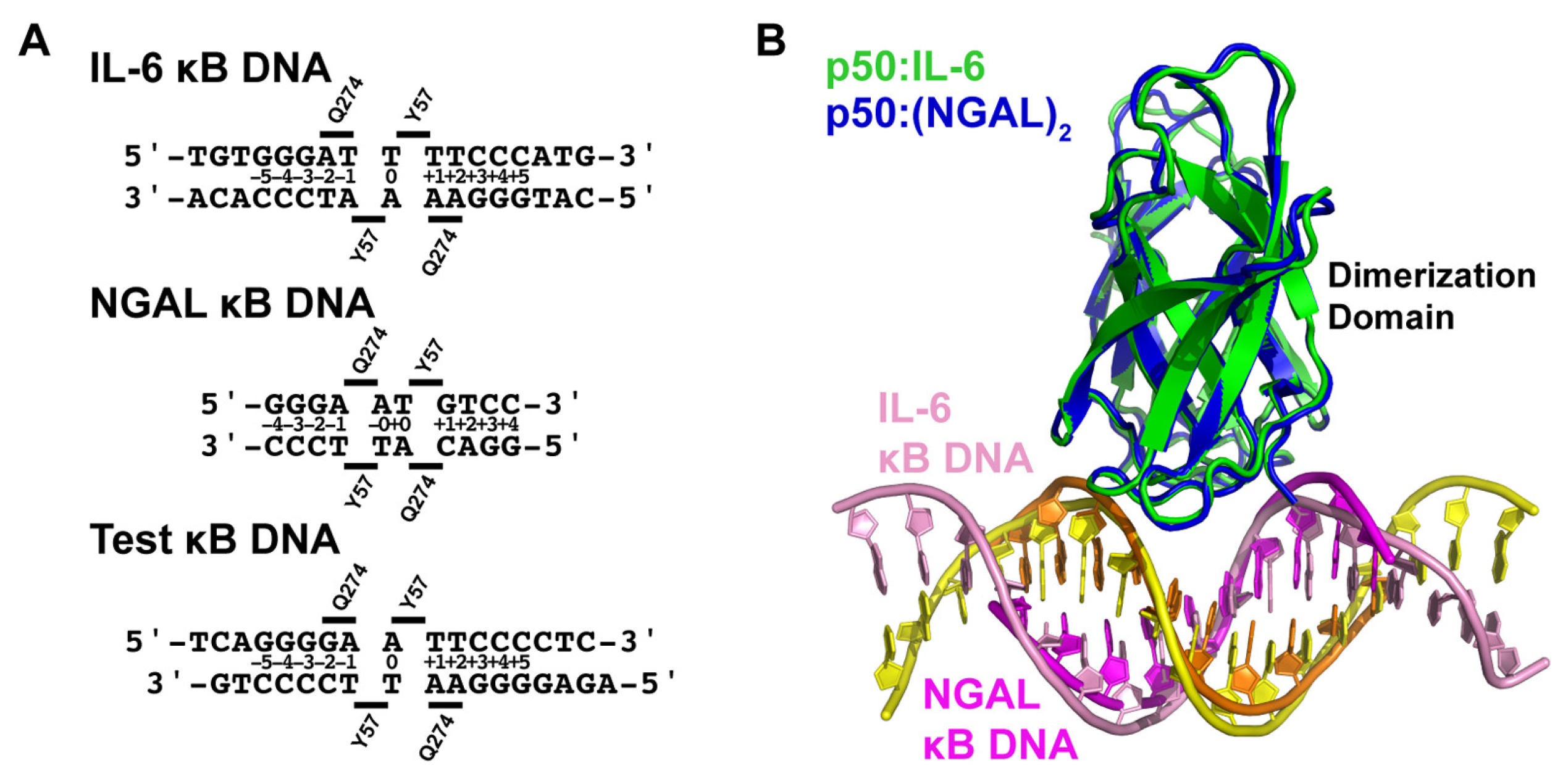
3.1. NF-κB p50 Homodimer:IL-6 κB DNA Complex Co-Crystal Structure
3.2. NF-κB p50 Homodimer:NGAL κB DNA Complex Co-Crystal Structure
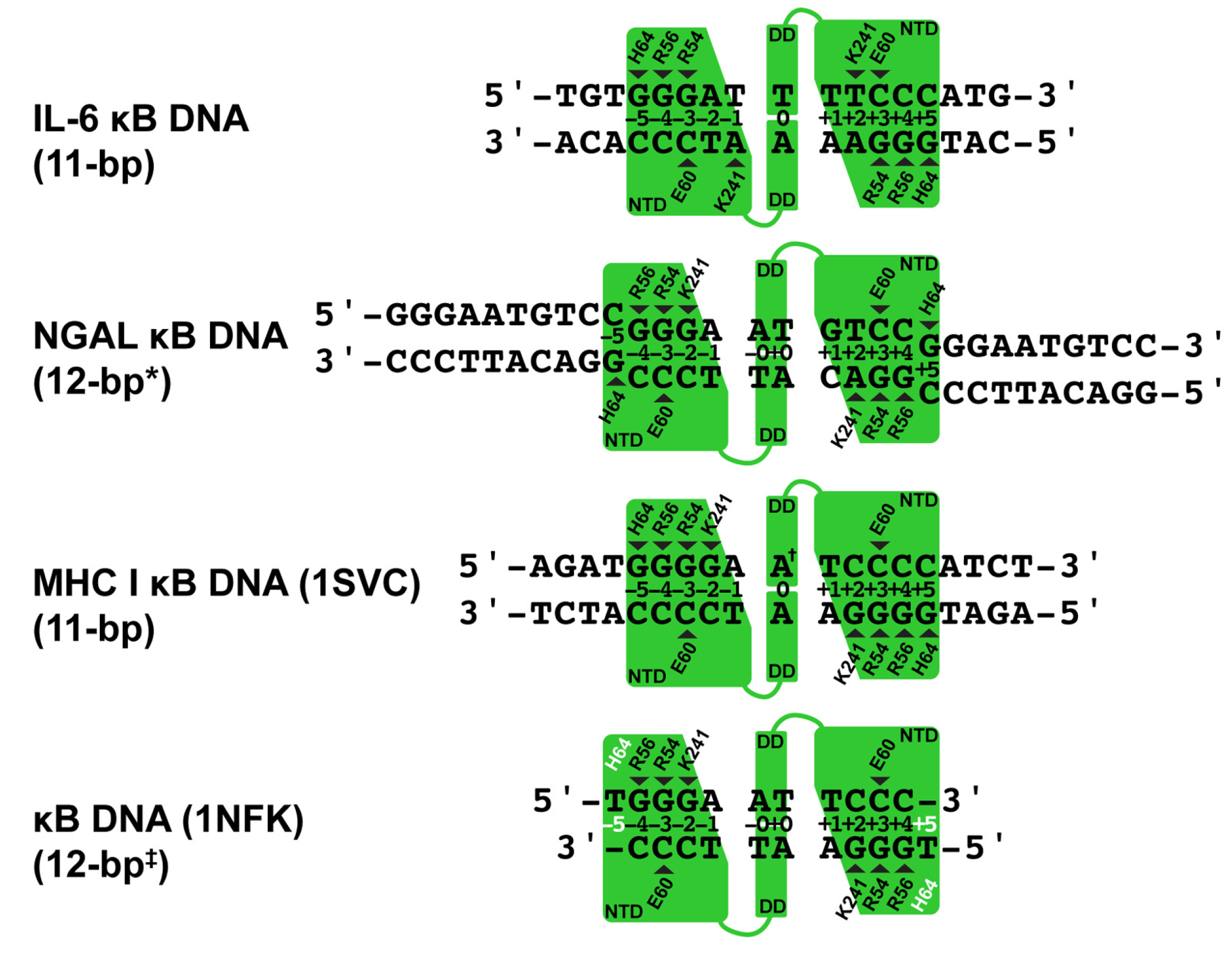
3.3. Comparative Analysis of DNA Base Contacts in p50:DNA Complex Co-Crystal Structures
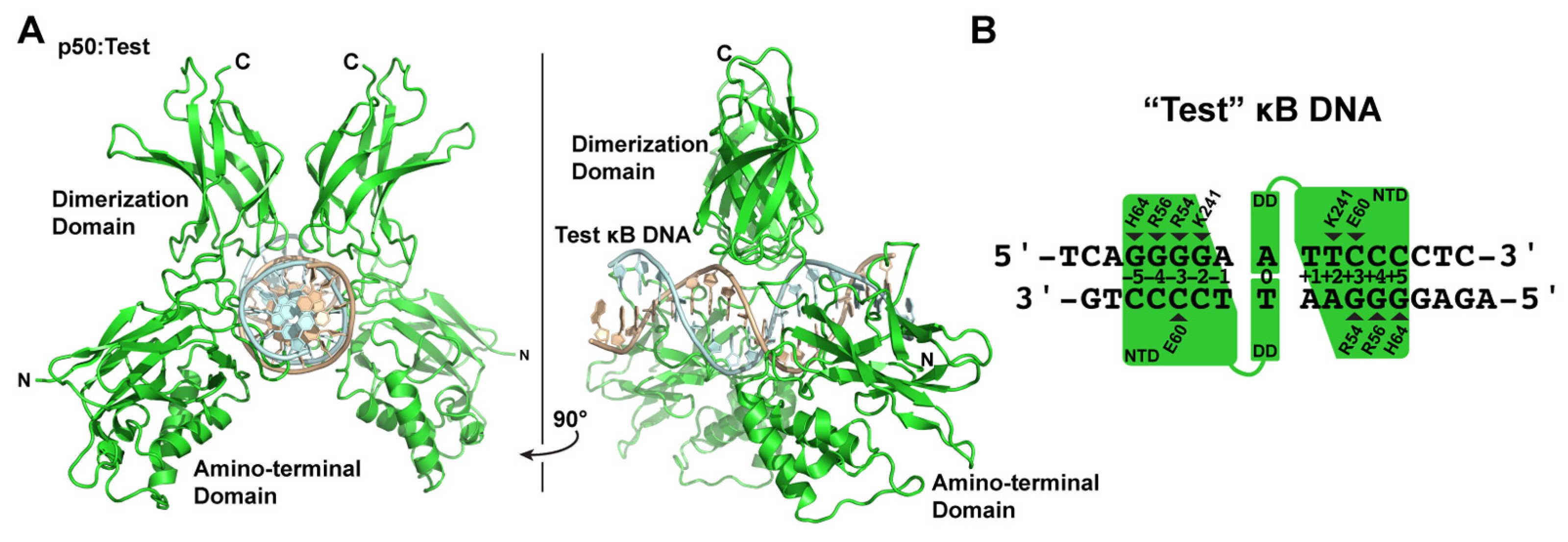
3.4. X-ray Co-Crystal Structure of NF-κB p50 Homodimer in Complex with “Test” κB DNA
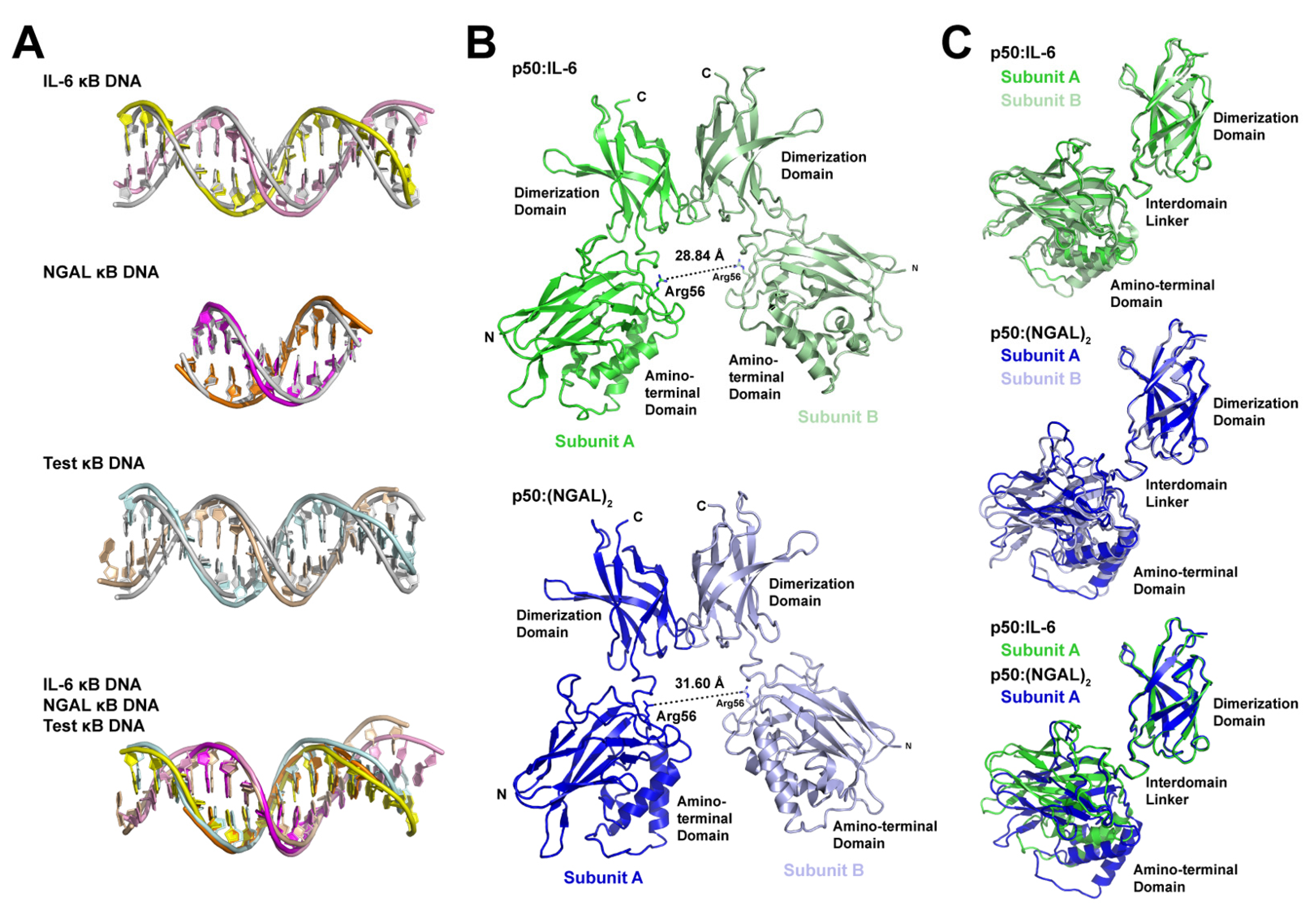
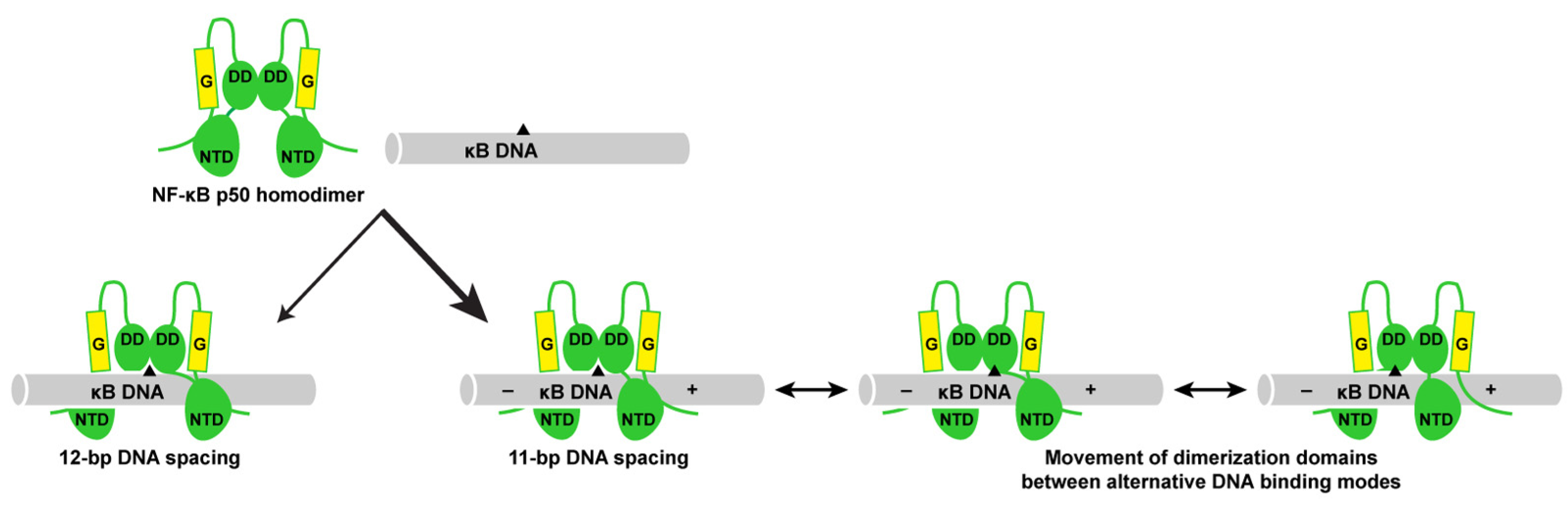
3.5. DNA Is Not Altered Significantly by 11- or 12-Base Pair Spacing
3.6. Individual p50 Subunits Adopt Similar Conformations Regardless of Spacing on Target DNA
3.7. The p50 Homodimer Dimerization Domain Is a Versatile DNA Contacting Unit
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoffmann, A.; Levchenko, A.; Scott, M.L.; Baltimore, D. The IκB-NF-κB signaling module: Temporal control and selective gene activation. Science 2002, 298, 1241–1245. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Ghosh, S. NF-κB, an evolutionarily conserved mediator of immune and inflammatory responses. Adv. Exp. Med. Biol. 2005, 560, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.M.; Gilmore, T.D. Looking Down on NF-κB. Mol. Cell. Biol. 2020, 40, e00104-20. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef]
- Sun, S.C. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017, 17, 545–558. [Google Scholar] [CrossRef]
- Kaltschmidt, C.; Greiner, J.F.W.; Kaltschmidt, B. The transcription factor NF-κB in stem cells and development. Cells 2021, 10, 2042. [Google Scholar] [CrossRef]
- Taniguchi, K.; Karin, M. NF-κB, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.S.; Ghosh, S. Shared principles in NF-κB signaling. Cell 2008, 132, 344–362. [Google Scholar] [CrossRef]
- Liou, H.C.; Nolan, G.P.; Ghosh, S.; Fujita, T.; Baltimore, D. The NF-κB p50 precursor, p105, contains an internal IκB-like inhibitor that preferentially inhibits p50. EMBO J. 1992, 11, 3003–3009. [Google Scholar] [CrossRef]
- Lin, L.; Ghosh, S. A glycine-rich region in NF-κB p105 functions as a processing signal for the generation of the p50 subunit. Mol. Cell. Biol. 1996, 16, 2248–2254. [Google Scholar] [CrossRef]
- Heissmeyer, V.; Krappmann, D.; Wulczyn, F.G.; Scheidereit, C. NF-κB p105 is a target of IκB kinases and controls signal induction of Bcl-3-p50 complexes. EMBO J. 1999, 18, 4766–4778. [Google Scholar] [CrossRef] [PubMed]
- Cartwright, T.; Perkins, N.D.; Caroline, L.W. NFKB1: A suppressor of inflammation, ageing and cancer. FEBS J. 2016, 283, 1812–1822. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.W.; Miyamoto, S. Expanded nuclear roles for IκBs. Sci STKE 2004, 2004, pe48. [Google Scholar] [CrossRef]
- Trinh, D.V.; Zhu, N.; Farhang, G.; Kim, B.J.; Huxford, T. The nuclear IκB protein IκBζ specifically binds NF-κB p50 homodimers and forms a ternary complex on κB DNA. J. Mol. Biol. 2008, 379, 122–135. [Google Scholar] [CrossRef]
- Yamamoto, M.; Yamazaki, S.; Uematsu, S.; Sato, S.; Hemmi, H.; Hoshino, K.; Kaisho, T.; Kuwata, H.; Takeuchi, O.; Takeshige, K.; et al. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IκBζ. Nature 2004, 430, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, S.; Yamazaki, S.; Takeshige, K.; Muta, T. Crucial roles of binding sites for NF-κB and C/EBPs in IκBζ-mediated transcriptional activation. Biochem. J. 2007, 405, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Tartey, S.; Matsushita, K.; Vandenbon, A.; Ori, D.; Imamura, T.; Mino, T.; Standley, D.M.; Hoffmann, J.A.; Reichhart, J.M.; Akira, S.; et al. Akirin2 is critical for inducing inflammatory genes by bridging IκBζ and the SWI/SNF complex. EMBO J. 2014, 33, 2332–2348. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Ghosh, G.; van Duyne, G.; Ghosh, S.; Sigler, P.B. Structure of NF-κB p50 homodimer bound to a κB site. Nature 1995, 373, 303–310. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger, L.; DeLano, W. PyMOL. Available online: http://www.pymol.org/pymol (accessed on 4 July 2023).
- Li, S.; Olson, W.K.; Lu, X.J. Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures. Nucleic Acids Res. 2019, 47, W26–W34. [Google Scholar] [CrossRef]
- Libermann, T.A.; Baltimore, D. Activation of interleukin-6 gene expression through the NF-κB transcription factor. Mol. Cell. Biol. 1990, 10, 2327–2334. [Google Scholar] [CrossRef]
- Matsusaka, T.; Fujikawa, K.; Nishio, Y.; Mukaida, N.; Matsushima, K.; Kishimoto, T.; Akira, S. Transcription factors NF-IL6 and NF-κB synergistically activate transcription of the inflammatory cytokines, interleukin-6 and interleukin-8. Proc. Natl. Acad. Sci. USA 1993, 90, 10193–10197. [Google Scholar] [CrossRef]
- Kunsch, C.; Ruben, S.M.; Rosen, C.A. Selection of optimal κB/Rel DNA-binding motifs: Interaction of both subunits of NF-κB with DNA is required for transcriptional activation. Mol. Cell. Biol. 1992, 12, 4412–4421. [Google Scholar] [CrossRef]
- Siggers, T.; Chang, A.B.; Teixeira, A.; Wong, D.; Williams, K.J.; Ahmed, B.; Ragoussis, J.; Udalova, I.A.; Smale, S.T.; Bulyk, M.L. Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-κB family DNA binding. Nat. Immunol. 2011, 13, 95–102. [Google Scholar] [CrossRef]
- Mulero, M.C.; Wang, V.Y.; Huxford, T.; Ghosh, G. Genome reading by the NF-κB transcription factors. Nucleic Acids Res. 2019, 47, 9967–9989. [Google Scholar] [CrossRef]
- Huxford, T.; Huang, D.B.; Malek, S.; Ghosh, G. The crystal structure of the IκBα/NF-κB complex reveals mechanisms of NF-κB inactivation. Cell 1998, 95, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, M.D.; Harrison, S.C. Structure of an IκBα/NF-κB complex. Cell 1998, 95, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Michel, F.; Soler-Lopez, M.; Petosa, C.; Cramer, P.; Siebenlist, U.; Müller, C.W. Crystal structure of the ankyrin repeat domain of Bcl-3: A unique member of the IκB protein family. EMBO J. 2001, 20, 6180–6190. [Google Scholar] [CrossRef]
- Malek, S.; Huang, D.B.; Huxford, T.; Ghosh, S.; Ghosh, G. X-ray crystal structure of an IκBβ:NF-κB p65 homodimer complex. J. Biol. Chem. 2003, 278, 23094–23100. [Google Scholar] [CrossRef]
- Karlsen, J.R.; Borregaard, N.; Cowland, J.B. Induction of neutrophil gelatinase-associated lipocalin expression by co-stimulation with interleukin-17 and tumor necrosis factor-α is controlled by IκBζ but neither by C/EBP-β nor C/EBP-δ. J. Biol. Chem. 2010, 285, 14088–14100. [Google Scholar] [CrossRef]
- Cowland, J.B.; Sorensen, O.E.; Sehested, M.; Borregaard, N. Neutrophil gelatinase-associated lipocalin is up-regulated in human epithelial cells by IL-1β, but not by TNF-α. J. Immunol. 2003, 171, 6630–6639. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.B.; Vu, D.; Cassiday, L.A.; Zimmerman, J.M.; Maher, L.J., 3rd; Ghosh, G. Crystal structure of NF-κB (p50)2 complexed to a high-affinity RNA aptamer. Proc. Natl. Acad. Sci. USA 2003, 100, 9268–9273. [Google Scholar] [CrossRef]
- Ghosh, G.; Huang, D.B.; Huxford, T. Molecular mimicry of the NF-κB DNA target site by a selected RNA aptamer. Curr. Opin. Struct. Biol. 2004, 14, 21–27. [Google Scholar] [CrossRef]
- Müller, C.W.; Rey, F.A.; Sodeoka, M.; Verdine, G.L.; Harrison, S.C. Structure of the NF-κB p50 homodimer bound to DNA. Nature 1995, 373, 311–317. [Google Scholar] [CrossRef]
- Müller, C.W.; Rey, F.A.; Harrison, S.C. Comparison of two different DNA-binding modes of the NF-κB p50 homodimer. Nat. Struct. Biol. 1996, 3, 224–227. [Google Scholar] [CrossRef]
- Huang, D.B.; Huxford, T.; Chen, Y.Q.; Ghosh, G. The role of DNA in the mechanism of NF-κB dimer formation: Crystal structures of the dimerization domains of the p50 and p65 subunits. Structure 1997, 5, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.W.; Harrison, S.C. The structure of the NF-κB p50:DNA-complex: A starting point for analyzing the Rel family. FEBS Lett. 1995, 369, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Q.; Ghosh, S.; Ghosh, G. A novel DNA recognition mode by the NF-κB p65 homodimer. Nat. Struct. Biol. 1998, 5, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Biancalana, M.; Natan, E.; Lenardo, M.J.; Fersht, A.R. NF-κB Rel subunit exchange on a physiological timescale. Protein Sci. 2021, 30, 1818–1832. [Google Scholar] [CrossRef]
- Phelps, C.B.; Sengchanthalangsy, L.L.; Huxford, T.; Ghosh, G. Mechanism of IκBα binding to NF-κB dimers. J. Biol. Chem. 2000, 275, 29840–29846. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Barrera, L.A.; Ersing, I.; Willox, B.; Schmidt, S.C.; Greenfeld, H.; Zhou, H.; Mollo, S.B.; Shi, T.T.; Takasaki, K.; et al. The NF-κB genomic landscape in lymphoblastoid B cells. Cell Rep. 2014, 8, 1595–1606. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Zhang, X.; Edwards, J.P.; Mosser, D.M. NF-κB1 (p50) homodimers differentially regulate pro- and anti-inflammatory cytokines in macrophages. J. Biol. Chem. 2006, 281, 26041–26050. [Google Scholar] [CrossRef]
- Cheng, C.S.; Feldman, K.E.; Lee, J.; Verma, S.; Huang, D.B.; Huynh, K.; Chang, M.; Ponomarenko, J.V.; Sun, S.C.; Benedict, C.A.; et al. The specificity of innate immune responses is enforced by repression of interferon response elements by NF-κB p50. Sci. Signal. 2011, 4, ra11. [Google Scholar] [CrossRef]
- Chen, Y.Q.; Sengchanthalangsy, L.L.; Hackett, A.; Ghosh, G. NF-κB p65 (RelA) homodimer uses distinct mechanisms to recognize DNA targets. Structure 2000, 8, 419–428. [Google Scholar] [CrossRef]
- Hou, S.; Guan, H.; Ricciardi, R.P. Phosphorylation of serine 337 of NF-κB p50 is critical for DNA binding. J. Biol. Chem. 2003, 278, 45994–45998. [Google Scholar] [CrossRef]
- Mulero, M.C.; Shahabi, S.; Ko, M.S.; Schiffer, J.M.; Huang, D.B.; Wang, V.Y.; Amaro, R.E.; Huxford, T.; Ghosh, G. Protein cofactors are essential for high-affinity DNA binding by the nuclear factor κB RelA subunit. Biochemistry 2018, 57, 2943–2957. [Google Scholar] [CrossRef] [PubMed]
| p50:DNA Complex | Spacing on DNA | Distance (Å) |
|---|---|---|
| p50:IL-6 | 11-bp | 28.84 1 |
| p50:(NGAL)2 | 12-bp | 31.60 |
| p50:Test | 11-bp | 28.57 |
| 1SVC | 11-bp | 28.71 |
| 1NFK | 12-bp | 32.36 |
| IL-6 A | IL-6 B | NGAL A | NGAL B | Test A | |
|---|---|---|---|---|---|
| IL-6 A | - | - | - | - | - |
| IL-6 B | 1.056 1 | - | - | - | - |
| NGAL A | 2.246 | 2.111 | - | - | - |
| NGAL B | 3.262 | 3.524 | 2.073 | - | - |
| Test A | 1.487 | 0.671 | 2.154 | 3.627 | - |
| Test B | 0.805 | 0.913 | 1.732 | 2.853 | 1.145 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, N.; Mealka, M.; Mitchel, S.; Milani, C.; Acuña, L.M.; Rogers, E.; Lahana, A.N.; Huxford, T. X-ray Crystallographic Study of Preferred Spacing by the NF-κB p50 Homodimer on κB DNA. Biomolecules 2023, 13, 1310. https://doi.org/10.3390/biom13091310
Zhu N, Mealka M, Mitchel S, Milani C, Acuña LM, Rogers E, Lahana AN, Huxford T. X-ray Crystallographic Study of Preferred Spacing by the NF-κB p50 Homodimer on κB DNA. Biomolecules. 2023; 13(9):1310. https://doi.org/10.3390/biom13091310
Chicago/Turabian StyleZhu, Norman, Matthew Mealka, Shane Mitchel, Christy Milani, Lisa M. Acuña, Eric Rogers, Ashlee N. Lahana, and Tom Huxford. 2023. "X-ray Crystallographic Study of Preferred Spacing by the NF-κB p50 Homodimer on κB DNA" Biomolecules 13, no. 9: 1310. https://doi.org/10.3390/biom13091310





