DPH1 Gene Mutations Identify a Candidate SAM Pocket in Radical Enzyme Dph1•Dph2 for Diphthamide Synthesis on EF2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Media, Growth Conditions and Assays
2.2. Sequence Alignments and Protein Modeling Based on the Archaeal CmnDph2 Structure
2.3. Assaying Diphthamide-Modified EF2 and ADP Ribosylation (ADPR) of EF2 with DT
3. Results and Discussion
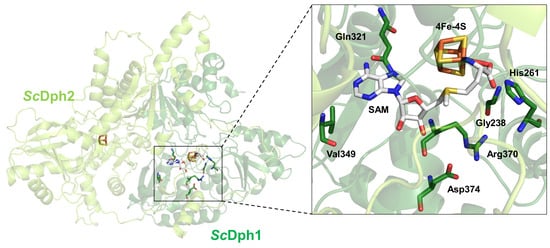
3.1. In Search for a Potential SAM Pocket in the Yeast Dph1•Dph2 Heterodimer
3.2. Conserved Residues in Dph1 Qualify for a SAM Pocket Relevant to Diphthamide Synthesis
3.3. Unmodified EF2 from the SAM Pocket Mutants Escapes ADP Ribosylation by DT
4. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frey, P.A.; Hegeman, A.D.; Ruzicka, F.J. The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 63–88. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, B.J.; McCarthy, E.L.; Booker, S.J. Radical S-adenosylmethionine enzymes in human health and disease. Annu. Rev. Biochem. 2016, 85, 485–514. [Google Scholar] [CrossRef] [PubMed]
- Broderick, J.B.; Duffus, B.R.; Duschene, K.S.; Shepard, E.M. Radical S-adenosylmethionine enzymes. Chem. Rev. 2014, 114, 4229–4317. [Google Scholar] [CrossRef]
- Yokoyama, K.; Lilla, E.A. C-C bond forming radical SAM enzymes involved in the construction of carbon skeletons of cofactors and natural products. Nat. Prod. Rep. 2018, 35, 660–694. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, B.M.; Broderick, W.E.; Broderick, J.B. Mechanism of radical initiation in the radical SAM enzyme superfamily. Annu. Rev. Biochem. 2023, 92, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Broderick, J.B. Biochemistry: A radically different enzyme. Nature. 2010, 465, 877–878. [Google Scholar] [CrossRef]
- Zhu, X.; Dzikovski, B.; Su, X.; Torelli, A.T.; Zhang, Y.; Ealick, S.E.; Freed, J.H.; Lin, H. Mechanistic understanding of Pyrococcus horikoshii Dph2, a [4Fe-4S] enzyme required for diphthamide biosynthesis. Mol. Biosyst. 2011, 7, 74–81. [Google Scholar] [CrossRef]
- Dong, M.; Dando, E.E.; Kotliar, I.; Su, X.; Dzikovski, B.; Freed, J.H.; Lin, H. The asymmetric function of Dph1-Dph2 heterodimer in diphthamide biosynthesis. J. Biol. Inorg. Chem. 2019, 24, 777–782. [Google Scholar] [CrossRef]
- Zhang, H.; Quintana, J.; Ütkür, K.; Adrian, L.; Hawer, H.; Mayer, K.; Gong, X.; Castanedo, L.; Schulten, A.; Janina, N.; et al. Translational fidelity and growth of Arabidopsis require stress-sensitive diphthamide biosynthesis. Nat. Commun. 2022, 13, 4009. [Google Scholar] [CrossRef]
- Ütkür, K.; Mayer, K.; Khan, M.; Manivannan, T.; Schaffrath, R.; Brinkmann, U. DPH1 and DPH2 variants that confer susceptibility to diphthamide deficiency syndrome in human cells and yeast models. Dis. Model. Mech. 2023, 16, dmm050207. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, X.; Torelli, A.T.; Lee, M.; Dzikovski, B.; Koralewski, R.M.; Wang, E.; Freed, J.; Krebs, C.; Ealick, S.E.; et al. Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature 2010, 465, 891–896. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Kim, J.; Su, X.; Lin, H. Reconstitution of diphthine synthase activity in vitro. Biochemistry 2010, 49, 9649–9657. [Google Scholar] [CrossRef] [PubMed]
- Lin, H. S-Adenosylmethionine-dependent alkylation reactions: When are radical reactions used? Bioorg. Chem. 2011, 39, 161–170. [Google Scholar] [CrossRef]
- Dong, M.; Kathiresan, V.; Fenwick, M.K.; Torelli, A.T.; Zhang, Y.; Caranto, J.D.; Dzikovski, B.; Sharma, A.; Lancaster, K.M.; Freed, J.H.; et al. Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis. Science 2018, 359, 1247–1250. [Google Scholar] [CrossRef] [PubMed]
- Van Ness, B.G.; Howard, J.B.; Bodley, J.W. ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. J. Biol. Chem. 1980, 255, 10717–10720. [Google Scholar] [CrossRef]
- Uthman, S.; Liu, S.; Giorgini, F.; Stark, M.J.R.; Costanzo, M.; Schaffrath, R. Diphtheria disease and genes involved in formation of diphthamide, key effector of the diphtheria toxin. In Insight and Control of Infectious Disease in Global Scenario; Kumar, R., Ed.; INTECH OAP: Rijeka, Croatia, 2012; pp. 333–356. [Google Scholar] [CrossRef]
- Jørgensen, R.; Merrill, A.R.; Andersen, G.R. The life and death of translation elongation factor 2. Biochem. Soc. Trans. 2006, 34, 1–6. [Google Scholar] [CrossRef]
- Liu, S.; Milne, G.T.; Kuremsky, J.G.; Fink, G.R.; Leppla, S.H. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol. Cell. Biol. 2004, 24, 9487–9497. [Google Scholar] [CrossRef]
- Uthman, S.; Bär, C.; Scheidt, V.; Liu, S.; ten Have, S.; Giorgini, F.; Stark, M.J.R.; Schaffrath, R. The amidation step of diphthamide biosynthesis in yeast requires DPH6, a gene identified through mining the DPH1-DPH5 interaction network. PLoS Genet. 2013, 9, e1003334. [Google Scholar] [CrossRef]
- Arend, M.; Ütkür, K.; Hawer, H.; Mayer, K.; Ranjan, N.; Adrian, L.; Brinkmann, U.; Schaffrath, R. Yeast gene KTI13 (alias DPH8) operates in the initiation step of diphthamide synthesis on elongation factor 2. Microb. Cell 2023, 10, 195–203. [Google Scholar] [CrossRef]
- Hawer, H.; Ütkür, K.; Arend, M.; Mayer, K.; Adrian, L.; Brinkmann, U.; Schaffrath, R. Importance of diphthamide modified EF2 for translational accuracy and competitive cell growth in yeast. PLoS ONE 2018, 13, e0205870. [Google Scholar] [CrossRef]
- Pellegrino, S.; Demeshkina, N.; Mancera-Martinez, E.; Melnikov, S.; Simonetti, A.; Myasnikov, A.; Yusupov, M.; Yusupova, G.; Hashem, Y. Structural insights into the role of diphthamide on elongation factor 2 in mRNA reading-frame maintenance. J. Mol. Biol. 2018, 430, 2677–2687. [Google Scholar] [CrossRef] [PubMed]
- Djumagulov, M.; Demeshkina, N.; Jenner, L.; Rozov, A.; Yusupov, M.; Yusupova, G. Accuracy mechanism of eukaryotic ribosome translocation. Nature 2021, 600, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Shin, B.-S.; Ivanov, I.P.; Kim, J.-R.; Cao, C.; Kinzy, T.G.; Dever, T.E. eEF2 diphthamide modification restrains spurious frameshifting to maintain translational fidelity. Nucleic Acids Res. 2023, 51, 6899–6913. [Google Scholar] [CrossRef]
- Loucks, C.M.; Parboosingh, J.S.; Shaheen, R.; Bernier, F.P.; Mcleod, D.R.; Seidahmed, M.Z.; Puffenberger, E.G.; Ober, C.; Hegele, R.A.; Boycott, K.M.; et al. Matching two independent cohorts validates DPH1 as a gene responsible for autosomal recessive intellectual disability with short stature, craniofacial, and ectodermalanomalies. Hum. Mutat. 2015, 36, 1015–1019. [Google Scholar] [CrossRef] [PubMed]
- Hawer, H.; Mendelsohn, B.A.; Mayer, K.; Kung, A.; Malhotra, A.; Tuupanen, S.; Schleit, J.; Brinkmann, U.; Schaffrath, R. Diphthamide-deficiency syndrome: A novel human developmental disorder and ribosomopathy. Eur. J. Hum. Genet. 2020, 28, 1497–1508. [Google Scholar] [CrossRef]
- Shankar, S.P.; Grimsrud, K.; Lanoue, L.; Egense, A.; Willis, B.; Hörberg, J.; Al Abdi, L.; Mayer, K.; Ütkür, K.; Monaghan, K.G.; et al. Undiagnosed Diseases Network. A novel DPH5-related diphthamide-deficiency syndrome causing embryonic lethality or profound neurodevelopmental disorder. Genet. Med. 2022, 24, 1567–1582. [Google Scholar] [CrossRef]
- Liu, S.; Wiggins, J.F.; Sreenath, T.; Kulkarni, A.B.; Ward, J.M.; Leppla, S.H. Dph3, a small protein required for diphthamide biosynthesis, is essential in mouse development. Mol. Cell. Biol. 2006, 26, 3835–3841. [Google Scholar] [CrossRef]
- Webb, T.R.; Cross, S.H.; McKie, L.; Edgar, R.; Vizor, L.; Harrison, J.; Peters, J.; Jackson, I.J. Diphthamide modification of eEF2 requires a J-domain protein and is essential for normal development. J. Cell Sci. 2008, 121, 3140–3145. [Google Scholar] [CrossRef]
- Liu, S.; Bachran, C.; Gupta, P.; Miller-Randolph, S.; Wang, H.; Crown, D.; Zhang, Y.; Wein, A.N.; Singh, R.; Fattah, R.; et al. Diphthamide modification on eukaryotic elongation factor 2 is needed to assure fidelity of mRNA translation and mouse development. Proc. Natl. Acad. Sci. USA 2012, 109, 13817–13822. [Google Scholar] [CrossRef]
- Toulmay, A.; Schneiter, R. A two-step method for the introduction of single or multiple defined point mutations into the genome of Saccharomyces cerevisiae. Yeast 2006, 23, 825–831. [Google Scholar] [CrossRef]
- Janke, C.; Magiera, M.M.; Rathfelder, N.; Taxis, C.; Reber, S.; Maekawa, H.; Moreno-Borchart, A.; Doenges, G.; Schwob, E.; Schiebel, E.; et al. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast 2004, 21, 947–962. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Sherman, F. Getting started with yeast. Methods Enzymol. 2002, 350, 3–41. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Stahl, S.; da Silva Mateus Seidl, A.R.; Ducret, A.; Kux van Geijtenbeek, S.; Michel, S.; Racek, T.; Birzele, F.; Haas, A.K.; Rueger, R.; Gerg, M.; et al. Loss of diphthamide pre-activates NF-kappaB and death receptor pathways and renders MCF7 cells hypersensitive to tumor necrosis factor. Proc. Natl. Acad. Sci. USA 2015, 112, 10732–10737. [Google Scholar] [CrossRef]
- Zachariae, W.; Shin, T.H.; Galova, M.; Obermaier, B.; Nasmyth, K. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science 1996, 274, 201–1204. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Chem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Mayer, K.; Schröder, A.; Schnitger, J.; Stahl, S.; Brinkmann, U. Influence of DPH1 and DPH5 protein variants on the synthesis of diphthamide, the target of ADPRibosylating toxins. Toxins 2017, 9, 78. [Google Scholar] [CrossRef]
- Fenwick, M.K.; Dong, M.; Lin, H.; Ealick, S.E. The crystal structure of Dph2 in complex with elongation factor 2 reveals the structural basis for the first step of diphthamide biosynthesis. Biochemistry 2019, 58, 4343–4351. [Google Scholar] [CrossRef]
- Shao, Y.; Molestak, E.; Su, W.; Stankevič, M.; Tchórzewski, M. Sordarin—An anti-fungal antibiotic with a unique modus operandi. Br. J. Pharmacol. 2022, 179, 1125–1145. [Google Scholar] [CrossRef] [PubMed]
- Bär, C.; Zabel, R.; Liu, S.; Stark, M.J.; Schaffrath, R. A versatile partner of eukaryotic protein complexes that is involved in multiple biological processes: Kti11/Dph3. Mol. Microbiol. 2008, 69, 1221–1233. [Google Scholar] [CrossRef] [PubMed]
- Botet, J.; Rodriguez-Mateos, M.; Ballesta, J.P.; Revuelta, J.L.; Remacha, M. A chemical genomic screen in Saccharomyces cerevisiae reveals a role for diphthamidation of translation elongation factor 2 in inhibition of protein synthesis by sordarin. Antimicrob. Agents Chemother. 2008, 52, 1623–1629. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Dong, M.; Zhang, Y.; Lee, E.A.; Lin, H. Cbr1 is a Dph3 reductase required for the tRNA wobble uridine modification. Nat. Chem. Biol. 2016, 12, 995–997. [Google Scholar] [CrossRef]
- Dong, M.; Su, X.; Dzikovski, B.; Dando, E.E.; Zhu, X.; Du, J.; Freed, J.H.; Lin, H. Dph3 is an electron donor for Dph1-Dph2 in the first step of eukaryotic diphthamide biosynthesis. J. Am. Chem. Soc. 2014, 136, 1754–1757. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, D.; Dzikovski, B.; Majer, S.H.; Coleman, R.; Chandrasekaran, S.; Fenwick, M.K.; Crane, B.R.; Lancaster, K.M.; Freed, J.H.; et al. Dph3 enables aerobic diphthamide biosynthesis by donating one iron atom to transform a [3Fe-4S] to a [4Fe-4S] cluster in Dph1-Dph2. J. Am. Chem. Soc. 2021, 143, 9314–9319. [Google Scholar] [CrossRef]
- Glatt, S.; Zabel, R.; Vonkova, I.; Kumar, A.; Netz, D.J.; Pierik, A.J.; Rybin, V.; Lill, R.; Gavin, A.C.; Balbach, J.; et al. Structure of the Kti11/Kti13 heterodimer and its double role in modification of tRNA and eukaryotic elongation factor 2. Structure 2015, 23, 149–160. [Google Scholar] [CrossRef]
- Jaciuk, M.; Scherf, D.; Kaszuba, K.; Gaik, M.; Rau, A.; Kościelniak, A.; Krutyhołowa, R.; Rawski, M.; Indyka, P.; Graziadei, A.; et al. Cryo-EM structure of the fully assembled Elongator complex. Nucleic Acids Res. 2023, 51, 2011–2032. [Google Scholar] [CrossRef]
- Schaffrath, R.; Abdel-Fattah, W.; Klassen, R.; Stark, M.J. The diphthamide modification pathway from Saccharomyces cerevisiae—Revisited. Mol. Microbiol. 2014, 94, 1213–1226. [Google Scholar] [CrossRef]
- Tsuda-Sakurai, K.; Miura, M. The hidden nature of protein translational control by diphthamide: The secrets under the leather. J. Biochem. 2019, 165, 1–8. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ütkür, K.; Schmidt, S.; Mayer, K.; Klassen, R.; Brinkmann, U.; Schaffrath, R. DPH1 Gene Mutations Identify a Candidate SAM Pocket in Radical Enzyme Dph1•Dph2 for Diphthamide Synthesis on EF2. Biomolecules 2023, 13, 1655. https://doi.org/10.3390/biom13111655
Ütkür K, Schmidt S, Mayer K, Klassen R, Brinkmann U, Schaffrath R. DPH1 Gene Mutations Identify a Candidate SAM Pocket in Radical Enzyme Dph1•Dph2 for Diphthamide Synthesis on EF2. Biomolecules. 2023; 13(11):1655. https://doi.org/10.3390/biom13111655
Chicago/Turabian StyleÜtkür, Koray, Sarina Schmidt, Klaus Mayer, Roland Klassen, Ulrich Brinkmann, and Raffael Schaffrath. 2023. "DPH1 Gene Mutations Identify a Candidate SAM Pocket in Radical Enzyme Dph1•Dph2 for Diphthamide Synthesis on EF2" Biomolecules 13, no. 11: 1655. https://doi.org/10.3390/biom13111655