An Overview of Ovarian Calyx Fluid Proteins of Toxoneuron nigriceps (Viereck) (Hymenoptera: Braconidae): An Integrated Transcriptomic and Proteomic Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Calyx Fluid Collection, Protein Purification, and RNA Extraction
2.3. RNASeq Data Generation and De Novo Transcriptome Assembly
2.4. Protein Separation and Identification
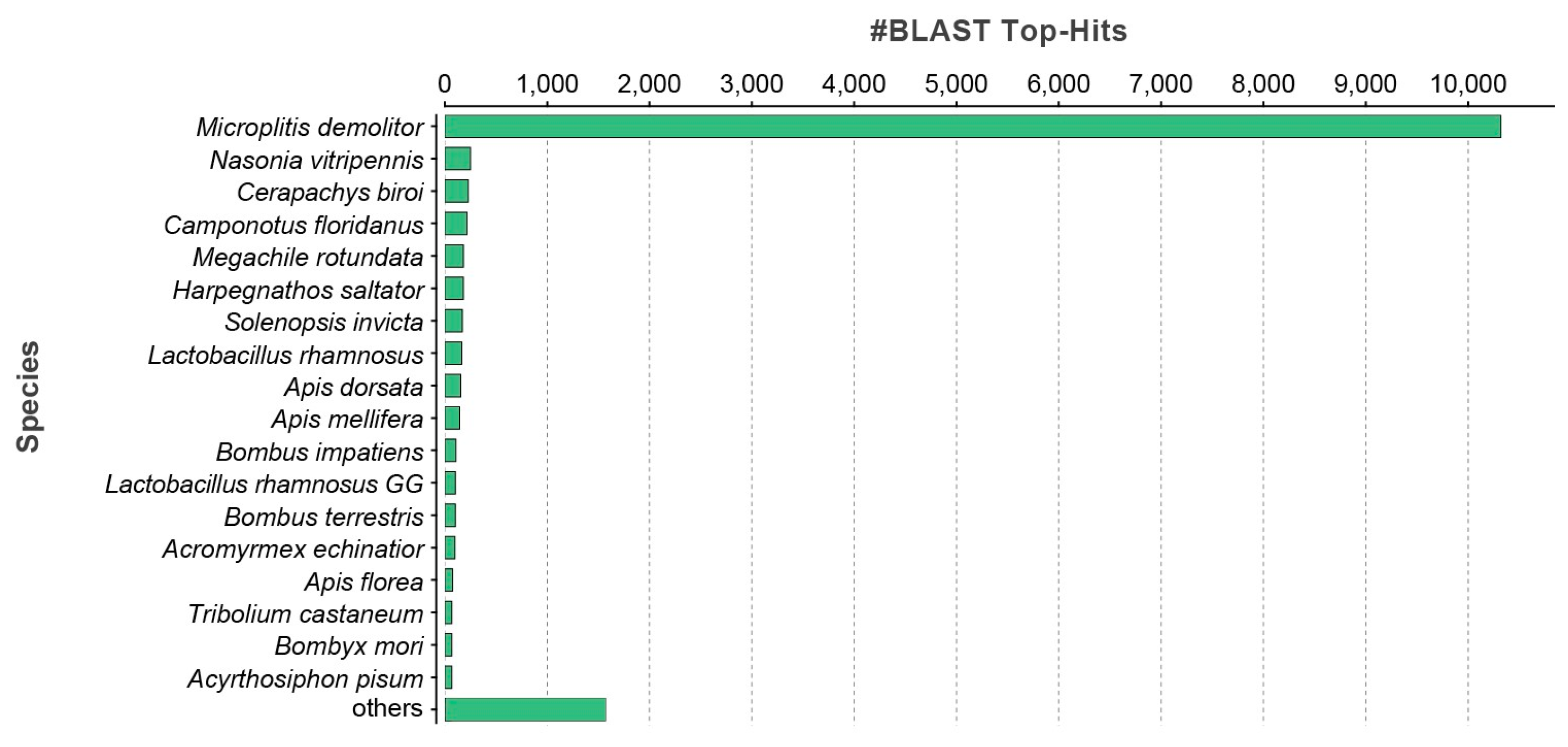
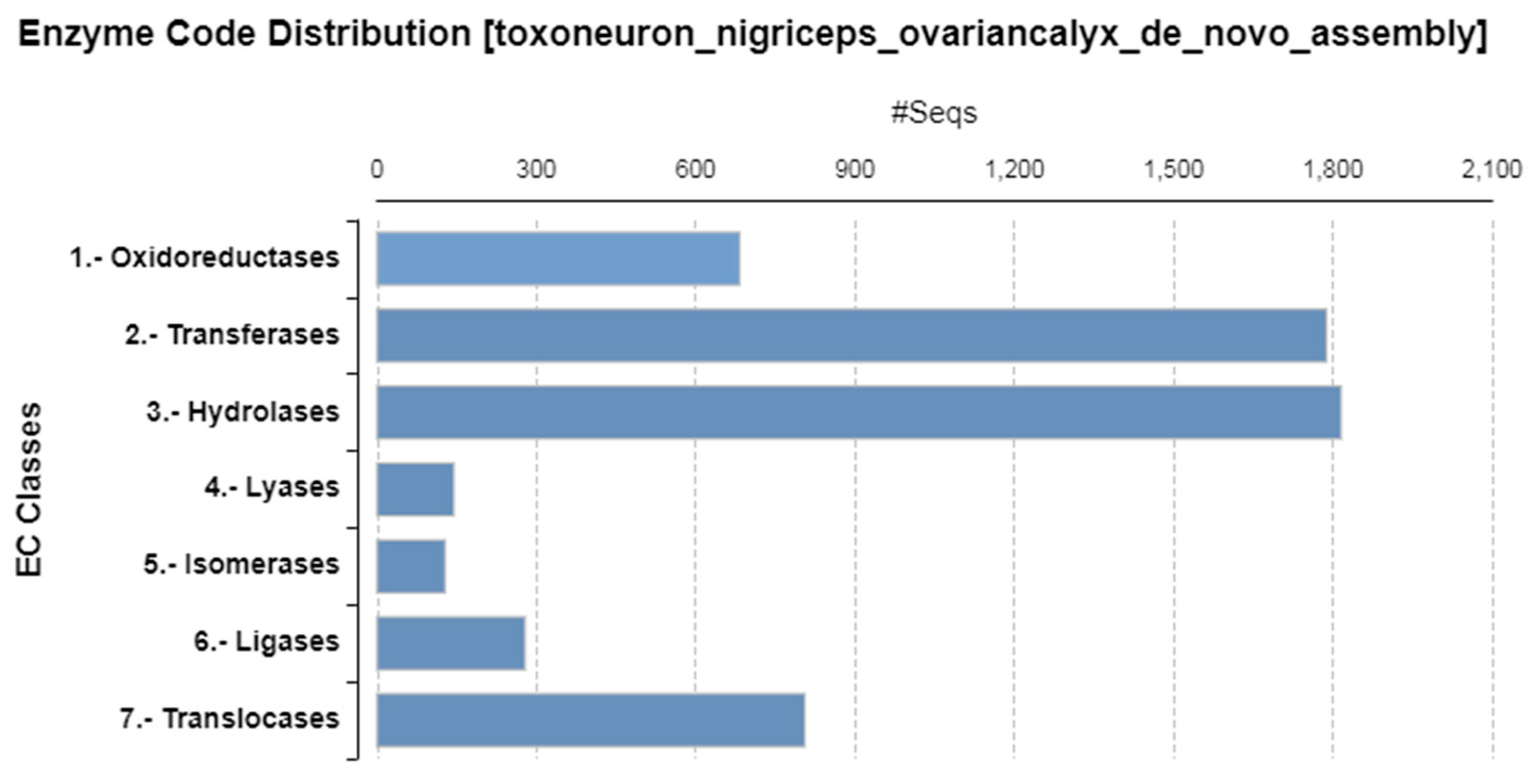
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chapman, A.D. Australian Biodiversity Information Services; Australia. Department of the Environment and Heritage. In Numbers of Living Species in Australia and the World; Australian Government, Department of the Environment and Heritage: Canberra, Australia, 2006; ISBN 9780642568496. [Google Scholar]
- Farrell, B.D.; Mitter, C. Adaptive Radiation in Insects and Plants: Time and Opportunity. Am. Zool. 1994, 34, 57–69. [Google Scholar] [CrossRef]
- Hoy, M.A.; Capinera, J.L.; Paré, P.W.; Farag, M.A.; Trumble, J.T.; Isman, M.B.; Adams, B.J.; Nguyen, K.B.; Panizzi, A.R.; Sánchez, N.E.; et al. Natural Enemies Important in Biological Control. In Encyclopedia of Entomology; Springer: Dordrecht, The Netherlands, 2008; pp. 2555–2567. [Google Scholar]
- Kalyanasundaram, M.; Merlin Kamala, I. Parasitoids. In Ecofriendly Pest Management for Food Security; Academic Press: Cambridge, MA, USA; Elsevier Inc.: Amsterdam, The Netherlands, 2016; pp. 109–138. [Google Scholar]
- Pennacchio, F.; Falabella, P.; Sordetti, R.; Varricchio, P.; Malva, C.; Vinson, B.S. Prothoracic Gland Inactivation in Heliothis virescens (F.) (Lepidoptera: Noctuidae) Larvae Parasitized by Cardiochiles nigriceps Viereck (Hymenoptera: Braconidae). J. Insect Physiol. 1998, 44, 845–857. [Google Scholar] [CrossRef]
- Pennacchio, F. Biochemical and Ultrastructural Alterations in Prothoracic Glands of Heliothis virescens (F.) (Lepidoptera: Noctuidae) Last Instar Larvae Parasitized by Cardiochiles nigriceps Viereck (Hymenoptera: Braconidae). Insect Biochem. Mol. Biol. 1997, 27, 439–450. [Google Scholar] [CrossRef]
- Pennacchio, F.; Tranfaglia, A.; Malva, C. Host-Parasitoid Antagonism in Insects: New Opportunities for Pest Control? Agro. Food Ind. Hi-Tech 2003, 14, 53–56. [Google Scholar]
- Pennacchio, F.; Mancini, D. Aphid Parasitoid Venom and Its Role in Host Regulation. In Parasitoid Viruses; Elsevier Inc.: Amsterdam, The Netherlands, 2012; pp. 247–254. [Google Scholar]
- Asgari, S.; Rivers, D.B. Venom Proteins from Endoparasitoid Wasps and Their Role in Host-Parasite Interactions. Annu. Rev. Entomol. 2011, 56, 313–335. [Google Scholar] [CrossRef]
- Beckage, N.E. Endocrine Interactions Between Endoparasitic Insects and Their Hosts. Annu. Rev. Entomol. 1985, 30, 371–413. [Google Scholar] [CrossRef]
- Beckage, N.E.; Gelman, D.B. Wasp Parasitoid Disruption of Host Development: Implications for New Biologically Based Strategies for Insect Control. Annu. Rev. Entomol. 2004, 49, 299–330. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.; Weaver, R. Endocrine Interactions of Insect Parasites and Pathogens; BIOS: Cambridge, UK, 2001. [Google Scholar]
- Lawrence, P.O.; Lanzrein, B. Hormonal Interactions between Insect Endoparasites and Their Host Insects. In Parasites and Pathogens of Insects; Beckage, N.E., Thompson, S.N., Federici, B.A., Eds.; Academic Press: San Diego, CA, USA, 1993; Volume 1, pp. 59–86. [Google Scholar]
- Lawrence, P.O. Host-Parasite Hormonal Interactions: An Overview. J. Insect Physiol. 1986, 32, 295–298. [Google Scholar] [CrossRef]
- Vinson, S.B.; Iwantsch, G.F. Host Suitability for Insect Parasitoids. Annu. Rev. Entomol. 1980, 25, 397–419. [Google Scholar] [CrossRef]
- Pennacchio, F.; Strand, M.R. Evolution of Developmental Strategies in Parasitic Hymenoptera. Annu. Rev. Entomol. 2006, 51, 233–258. [Google Scholar] [CrossRef]
- Schmidt, O.; Theopold, U.; Strand, M. Innate Immunity and Its Evasion and Suppression by Hymenopteran Endoparasitoids. BioEssays 2001, 23, 344–351. [Google Scholar] [CrossRef]
- Webb, B.A.; Strand, M.R.; Dickey, S.E.; Beck, M.H.; Hilgarth, R.S.; Barney, W.E.; Kadash, K.; Kroemer, J.A.; Lindstrom, K.G.; Rattanadechakul, W.; et al. Polydnavirus Genomes Reflect Their Dual Roles as Mutualists and Pathogens. Virology 2006, 347, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Quistad, G.; Nguyen, Q.; Bernasconi, P.; Leisy, D. Purification and Characterization of Insecticidal Toxins from Venom Glands of the Parasitic Wasp, Bracon hebetor. Insect Biochem. Mol. Biol. 1994, 24, 955–961. [Google Scholar] [CrossRef]
- Moreau, S.; Asgari, S. Venom Proteins from Parasitoid Wasps and Their Biological Functions. Toxins 2015, 7, 2385–2412. [Google Scholar] [CrossRef] [PubMed]
- Moreau, S.J.M.; Dingremont, A.; Doury, G.; Giordanengo, P. Effects of Parasitism by Asobara tabida (Hymenoptera: Braconidae) on the Development, Survival and Activity of Drosophila melanogaster Larvae. J. Insect Physiol. 2002, 48, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Falabella, P.; Riviello, L.; De Stradis, M.L.; Stigliano, C.; Varricchio, P.; Grimaldi, A.; de Eguileor, M.; Graziani, F.; Gigliotti, S.; Pennacchio, F. Aphidius ervi Teratocytes Release an Extracellular Enolase. Insect Biochem. Mol. Biol. 2009, 39, 801–813. [Google Scholar] [CrossRef]
- Laurino, S.; Grossi, G.; Pucci, P.; Flagiello, A.; Bufo, S.A.; Bianco, G.; Salvia, R.; Vinson, S.B.; Vogel, H.; Falabella, P. Identification of Major Toxoneuron nigriceps Venom Proteins Using an Integrated Transcriptomic/Proteomic Approach. Insect Biochem. Mol. Biol. 2016, 76, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Falabella, P. The Mechanism Utilized by Toxoneuron nigriceps in Inhibiting the Host Immune System. Invertebr. Surviv. J. 2018, 15, 240–255. [Google Scholar]
- Teng, Z.; Wu, H.; Ye, X.; Xiong, S.; Xu, G.; Wang, F.; Fang, Q.; Ye, G. An Ovarian Protein Involved in Passive Avoidance of an Endoparasitoid to Evade Its Host Immune Response. J. Proteome Res. 2019, 18, 2695–2705. [Google Scholar] [CrossRef]
- Salvia, R.; Grimaldi, A.; Girardello, R.; Scieuzo, C.; Scala, A.; Bufo, S.A.; Vogel, H.; Falabella, P. Aphidius ervi Teratocytes Release Enolase and Fatty Acid Binding Protein Through Exosomal Vesicles. Front. Physiol. 2019, 10, 715. [Google Scholar] [CrossRef]
- Lapointe, R.; Wilson, R.; Vilaplana, L.; O’Reilly, D.R.; Falabella, P.; Douris, V.; Bernier-Cardou, M.; Pennacchio, F.; Iatrou, K.; Malva, C.; et al. Expression of a Toxoneuron nigriceps Polydnavirus-Encoded Protein Causes Apoptosis-like Programmed Cell Death in Lepidopteran Insect Cells. J. Gen. Virol. 2005, 86, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Falabella, P.; Varricchio, P.; Gigliotti, S.; Tranfaglia, A.; Pennacchio, F.; Malva, C. Toxoneuron nigriceps Polydnavirus Encodes a Putative Aspartyl Protease Highly Expressed in Parasitized Host Larvae. Insect Mol. Biol. 2003, 12, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Falabella, P.; Caccialupi, P.; Varricchio, P.; Malva, C.; Pennacchio, F. Protein Tyrosine Phosphatases of Toxoneuron nigriceps Bracovirus as Potential Disrupters of Host Prothoracic Gland Function. Arch. Insect Biochem. Physiol. 2006, 61, 157–169. [Google Scholar] [CrossRef]
- Falabella, P.; Varricchio, P.; Provost, B.; Espagne, E.; Ferrarese, R.; Grimaldi, A.; de Eguileor, M.; Fimiani, G.; Ursini, M.V.; Malva, C.; et al. Characterization of the IκB-like Gene Family in Polydnaviruses Associated with Wasps Belonging to Different Braconid Subfamilies. J. Gen. Virol. 2007, 88, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Provost, B.; Varricchio, P.; Arana, E.; Espagne, E.; Falabella, P.; Huguet, E.; La Scaleia, R.; Cattolico, L.; Poirié, M.; Malva, C.; et al. Bracoviruses Contain a Large Multigene Family Coding for Protein Tyrosine Phosphatases. J. Virol. 2004, 78, 13090–13103. [Google Scholar] [CrossRef] [PubMed]
- Salvia, R.; Grossi, G.; Amoresano, A.; Scieuzo, C.; Nardiello, M.; Giangrande, C.; Laurenzana, I.; Ruggieri, V.; Bufo, S.A.; Vinson, S.B.; et al. The Multifunctional Polydnavirus TnBVANK1 Protein: Impact on Host Apoptotic Pathway. Sci. Rep. 2017, 7, 11775. [Google Scholar] [CrossRef]
- Malva, C.; Varricchio, P.; Falabella, P.; La Scaleia, R.; Graziani, F.; Pennacchio, F. Physiological and Molecular Interaction in the Host–Parasitoid System Heliothis virescens–Toxoneuron nigriceps: Current Status and Future Perspectives. Insect Biochem. Mol. Biol. 2004, 34, 177–183. [Google Scholar] [CrossRef]
- Varricchio, P.; Falabella, P.; Sordetti, R.; Graziani, F.; Malva, C.; Pennacchio, F. Cardiochiles Nigriceps Polydnavirus: Molecular Characterization and Gene Expression in Parasitized Heliothis virescens Larvae. Insect Biochem. Mol. Biol. 1999, 29, 1087–1096. [Google Scholar] [CrossRef]
- Salvia, R.; Cozzolino, F.; Scieuzo, C.; Grimaldi, A.; Franco, A.; Vinson, S.B.; Monti, M.; Falabella, P. Identification and Functional Characterization of Toxoneuron nigriceps Ovarian Proteins Involved in the Early Suppression of Host Immune Response. Insects 2022, 13, 144. [Google Scholar] [CrossRef]
- Salvia, R.; Scieuzo, C.; Grimaldi, A.; Fanti, P.; Moretta, A.; Franco, A.; Varricchio, P.; Vinson, S.B.; Falabella, P. Role of Ovarian Proteins Secreted by Toxoneuron nigriceps (Viereck) (Hymenoptera, Braconidae) in the Early Suppression of Host Immune Response. Insects 2021, 12, 33. [Google Scholar] [CrossRef]
- Vinson, S.B.; Guillot, F.S.; Hays, D.B. Rearing of Cardiochiles nigriceps in the Laboratory, with Heliothis virescens as Hosts. Ann. Entomol. Soc. Am. 1973, 66, 1170–1172. [Google Scholar] [CrossRef]
- Vanderzant, E.S.; Richardson, C.D.; Fort, S.W. Rearing of the Bollworm on Artificial Diet. J. Econ. Entomol. 1962, 55, 140. [Google Scholar] [CrossRef]
- Vogel, H.; Badapanda, C.; Knorr, E.; Vilcinskas, A. RNA-Sequencing Analysis Reveals Abundant Developmental Stage-Specific and Immunity-Related Genes in the Pollen Beetle Meligethes aeneus. Insect Mol. Biol. 2014, 23, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-Throughput Functional Annotation and Data Mining with the Blast2GO Suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef]
- Butturini, E.; Gotte, G.; Dell’Orco, D.; Chiavegato, G.; Marino, V.; Canetti, D.; Cozzolino, F.; Monti, M.; Pucci, P.; Mariotto, S. Intermolecular Disulfide Bond Influences Unphosphorylated STAT3 Dimerization and Function. Biochem. J. 2016, 473, 3205–3219. [Google Scholar] [CrossRef]
- Bertini, L.; Cozzolino, F.; Proietti, S.; Falconieri, G.S.; Iacobucci, I.; Salvia, R.; Falabella, P.; Monti, M.; Caruso, C. What Antarctic Plants Can Tell Us about Climate Changes: Temperature as a Driver for Metabolic Reprogramming. Biomolecules 2021, 11, 1094. [Google Scholar] [CrossRef]
- Moreau, S.J.M.; Guillot, S. Advances and Prospects on Biosynthesis, Structures and Functions of Venom Proteins from Parasitic Wasps. Insect Biochem. Mol. Biol. 2005, 35, 1209–1223. [Google Scholar] [CrossRef] [PubMed]
- Felföldi, G.; Marokhazi, J.; Képiro, M.; Venekei, I. Identification of natural target proteins indicates functions of a serralysin-type metalloprotease, PrtA, in anti-immune mechanisms. Appl. Environ. Microbiol. 2009, 75, 3120–3126. [Google Scholar] [CrossRef]
- Price, D.R.G.; Bell, H.A.; Hinchliffe, G.; Fitches, E.; Weaver, R.; Gatehouse, J.A. A venom metalloproteinase from the parasitic wasp Eulophus pennicornis is toxic towards its host, tomato moth (Lacanobia oleracae). Insect Mol. Biol. 2009, 18, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-T.; Hsieh, C.; Wu, L.-C.; Chang, H.C.; Kao, S.-S.; Meng, M.; Hsieh, F.-C. Purification and Properties of an Insecticidal Metalloprotease Produced by Photorhabdus luminescens Strain 0805-P5G, the Entomopathogenic Nematode Symbiont. Int. J. Mol. Sci. 2013, 14, 308–321. [Google Scholar] [CrossRef] [PubMed]
- de Graaf, D.C.; Aerts, M.; Brunain, M.; Desjardins, C.A.; Jacobs, F.J.; Werren, J.H.; Devreese, B. Insights into the Venom Composition of the Ectoparasitoid Wasp Nasonia vitripennis from Bioinformatic and Proteomic Studies. Insect Mol. Biol. 2010, 19, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Scieuzo, C.; Salvia, R.; Franco, A.; Pezzi, M.; Cozzolino, F.; Chicca, M.; Scapoli, C.; Vogel, H.; Monti, M.; Ferracini, C.; et al. An Integrated Transcriptomic and Proteomic Approach to Identify the Main Torymus sinensis Venom Components. Sci. Rep. 2021, 11, 5032. [Google Scholar] [CrossRef]
- Zhang, G.; Lu, Z.-Q.; Jiang, H.; Asgari, S. Negative Regulation of Prophenoloxidase (ProPO) Activation by a Clip-Domain Serine Proteinase Homolog (SPH) from Endoparasitoid Venom. Insect Biochem. Mol. Biol. 2004, 34, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Asgari, S.; Zhan, G.; Zareie, R.; Schmidt, O. A serine proteinase homolog venom protein from an endoparasitoid wasp inhibits melanization of the host hemolymph. Insect Biochem. Mol. Biol. 2003, 33, 1017–1024. [Google Scholar] [CrossRef]
- Choo, Y.M.; Lee, K.S.; Yoon, H.J.; Kim, B.Y.; Sohn, M.R.; Roh, J.Y.; Je, Y.H.; Kim, N.J.; Kim, I.; Woo, S.D.; et al. Dual function of a bee venom serine protease: Prophenoloxidase-activating factor in arthropods and fibrin(ogen)olytic enzyme in mammals. PLoS ONE 2010, 5, e10393. [Google Scholar] [CrossRef]
- Beck, M.; Theopold, U.; Schmidt, O. Evidence for serine protease inhibitor activity in the ovarian calyx fluid of the endoparasitoid Venturia canescens. J. Insect Physiol. 2000, 46, 1275–1283. [Google Scholar] [CrossRef]
- Colinet, H.; Renault, D.; Charoy-Guével, B.; Com, E. Metabolic and Proteomic Profiling of Diapause in the Aphid Parasitoid Praon volucre. PLoS ONE 2012, 7, e32606. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, D.; Feng, B.; Gou, M.; Liu, X.; Li, Q. Identification and Characterization of Aldehyde Dehydrogenase 9 from Lampetra japonica and Its Protective Role against Cytotoxicity. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2015, 187, 102–109. [Google Scholar] [CrossRef]
- Martinson, E.O.; Martinson, V.G.; Edwards, R.; Mrinalini; Werren, J.H. Laterally Transferred Gene Recruited as a Venom in Parasitoid Wasps. Mol. Biol. Evol. 2016, 33, 1042–1052. [Google Scholar] [CrossRef]
- Song, Y.; Gu, F.; Zhou, W.; Li, P.; Wu, F.; Sheng, S. Parasitoid Wasps Can Manipulate Host Trehalase to the Benefit of Their Offspring. Insects 2022, 13, 833. [Google Scholar] [CrossRef]
- Grossi, G.; Grimaldi, A.; Cardone, R.A.; Monné, M.; Reshkin, S.J.; Girardello, R.; Greco, M.R.; Coviello, E.; Laurino, S.; Falabella, P. Extracellular Matrix Degradation via Enolase/Plasminogen Interaction: Evidence for a Mechanism Conserved in Metazoa. Biol. Cell 2016, 108, 161–178. [Google Scholar] [CrossRef]
- Kramer, K.J.; Koga, D. Insect Chitin: Physical State, Synthesis, Degradation and Metabolic Regulation. Insect Biochem. 1986, 16, 851–877. [Google Scholar] [CrossRef]
- Cônsoli, F.L.; Lewis, D.; Keeley, L.; Vinson, S.B. Characterization of a CDNA Encoding a Putative Chitinase from Teratocytes of the Endoparasitoid Toxoneuron nigriceps. Entomol. Exp. Appl. 2007, 122, 271–278. [Google Scholar] [CrossRef]
- Rossi, G.D.; Labate, M.T.V.; Labate Carlos, A.; Vinson, S.B.; Cônsoli Fernando, L. Characterization of a Toxoneuron nigriceps (Viereck) (Hymenoptera: Braconidae)—Derived chitinase and its potential for pest control. Pestic. Biochem. Physiol. 2012, 104, 96–102. [Google Scholar] [CrossRef]
- An, X.; Yu, L.; Wang, S.; Ao, Y.; Zhan, X.; Liu, Q.; Zhao, Y.; Li, M.; Shu, X.; Li, F.; et al. Kinetic Characterization and Inhibitor Screening of Pyruvate Kinase I from Babesia Microti. Front. Microbiol. 2021, 12, 710678. [Google Scholar] [CrossRef]
- Labella, C.; Kanawati, B.; Vogel, H.; Schmitt-Kopplin, P.; Laurino, S.; Bianco, G.; Falabella, P. Identification of Two Arginine Kinase Forms of Endoparasitoid Leptomastix dactylopii Venom by Bottom up-Sequence Tag Approach. J. Mass Spectrom. 2015, 50, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, A.; UlAbdin, Z.; Webb, B.A.; Arif, M.J.; Jamil, A. De Novo Sequencing and Transcriptome Analysis of Female Venom Glands of Ectoparasitoid Bracon hebetor (Say.) (Hymenoptera: Braconidae). Comp. Biochem. Physiol. Part D Genom. Proteom. 2016, 20, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.S.; Kim, B.Y.; Kim, H.J.; Seo, S.J.; Yoon, H.J.; Choi, Y.S.; Kim, I.; Han, Y.S.; Je, Y.H.; Lee, S.M.; et al. Transferrin Inhibits Stress-Induced Apoptosis in a Beetle. Free Radic. Biol. Med. 2006, 41, 1151–1161. [Google Scholar] [CrossRef]
- Chang, C.L.; Liang, G.H.; Geib, S.M. Proteomic Interactions between the Parasitoid Diachasmimorpha longicaudata and the Oriental Fruit Fly, Bactrocera dorsalis during Host Parasitism. J. Asia Pac. Entomol. 2018, 21, 335–344. [Google Scholar] [CrossRef]
- Kurama, T.; Kurata, S.; Natori, S. Molecular Characterization of an Insect Transferrin and Its Selective Incorporation into Eggs During Oogenesis. Eur. J. Biochem. 1995, 228, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Kayukawa, T.; Murata, M.; Kobayashi, I.; Muramatsu, D.; Okada, C.; Uchino, K.; Sezutsu, H.; Kiuchi, M.; Tamura, T.; Hiruma, K.; et al. Hormonal Regulation and Developmental Role of Krüppel Homolog 1, a Repressor of Metamorphosis, in the Silkworm Bombyx mori. Dev. Biol. 2014, 388, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Li, X.; Zhao, J.; Zhu-Salzman, K. Cloning and Characterization of Methoprene-tolerant (Met) and Krüppel Homolog 1 (Kr-h1) Genes in the Wheat Blossom Midge, Sitodiplosis mosellana. Insect Sci. 2020, 27, 292–303. [Google Scholar] [CrossRef]
- Liu, S.; Li, K.; Gao, Y.; Liu, X.; Chen, W.; Ge, W.; Feng, Q.; Palli, S.R.; Li, S. Antagonistic Actions of Juvenile Hormone and 20-Hydroxyecdysone within the Ring Gland Determine Developmental Transitions in Drosophila. Proc. Natl. Acad. Sci. USA 2018, 115, 139–144. [Google Scholar] [CrossRef]
- Zhang, T.; Song, W.; Li, Z.; Qian, W.; Wei, L.; Yang, Y.; Wang, W.; Zhou, X.; Meng, M.; Peng, J.; et al. Krüppel Homolog 1 Represses Insect Ecdysone Biosynthesis by Directly Inhibiting the Transcription of Steroidogenic Enzymes. Proc. Natl. Acad. Sci. USA 2018, 115, 3960–3965. [Google Scholar] [CrossRef] [PubMed]
- Salvia, R.; Nardiello, M.; Scieuzo, C.; Scala, A.; Bufo, S.A.; Rao, A.; Vogel, H.; Falabella, P. Novel Factors of Viral Origin Inhibit TOR Pathway Gene Expression. Front. Physiol. 2018, 9, 1678. [Google Scholar] [CrossRef]
- Scieuzo, C.; Nardiello, M.; Salvia, R.; Pezzi, M.; Chicca, M.; Leis, M.; Bufo, S.A.; Vinson, S.B.; Rao, A.; Vogel, H.; et al. Ecdysteroidogenesis and Development in Heliothis virescens (Lepidoptera: Noctuidae): Focus on PTTH-Stimulated Pathways. J. Insect Physiol. 2018, 107, 57–67. [Google Scholar] [CrossRef]
- Jiang, J.; Xu, Y.; Lin, X. Role of Broad-Complex (Br) and Krüppel Homolog 1 (Kr-H1) in the Ovary Development of Nilaparvata lugens. Front. Physiol. 2017, 8. [Google Scholar] [CrossRef]
- Ojani, R.; Fu, X.; Ahmed, T.; Liu, P.; Zhu, J. Krüppel Homologue 1 Acts as a Repressor and an Activator in the Transcriptional Response to Juvenile Hormone in Adult Mosquitoes. Insect Mol. Biol. 2018, 27, 268–278. [Google Scholar] [CrossRef]
- Yue, Y.; Yang, R.-L.; Wang, W.-P.; Zhou, Q.-H.; Chen, E.-H.; Yuan, G.-R.; Wang, J.-J.; Dou, W. Involvement of Met and Kr-H1 in JH-Mediated Reproduction of Female Bactrocera dorsalis (Hendel). Front. Physiol. 2018, 9, 482. [Google Scholar] [CrossRef]
- Zhang, W.-N.; Ma, L.; Liu, C.; Chen, L.; Xiao, H.-J.; Liang, G.-M. Dissecting the Role of Krüppel Homolog 1 in the Metamorphosis and Female Reproduction of the Cotton Bollworm, Helicoverpa armigera. Insect Mol. Biol. 2018, 27, 492–504. [Google Scholar] [CrossRef]
- Saha, T.T.; Roy, S.; Pei, G.; Dou, W.; Zou, Z.; Raikhel, A.S. Synergistic Action of the Transcription Factors Krüppel Homolog 1 and Hairy in Juvenile Hormone/Methoprene-Tolerant-Mediated Gene-Repression in the Mosquito Aedes aegypti. PLoS Genet. 2019, 15, e1008443. [Google Scholar] [CrossRef]
- Tang, Y.; He, H.; Qu, X.; Cai, Y.; Ding, W.; Qiu, L.; Li, Y. RNA Interference-mediated Knockdown of the Transcription Factor Krüppel Homologue 1 Suppresses Vitellogenesis in Chilo suppressalis. Insect Mol. Biol. 2020, 29, 183–192. [Google Scholar] [CrossRef]
- Moné, Y.; Monnin, D.; Kremer, N. The Oxidative Environment: A Mediator of Interspecies Communication That Drives Symbiosis Evolution. Proc. R. Soc. B Biol. Sci. 2014, 281, 20133112. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-D.; Shen, Z.-J.; Liu, X.-M.; Li, Z.; Zhang, Q.-W.; Liu, X.-X. Molecular Identification of Three Novel Glutaredoxin Genes That Play Important Roles in Antioxidant Defense in Helicoverpa armigera. Insect Biochem. Mol. Biol. 2016, 75, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Wrońska, A.K.; Boguś, M.I. Heat Shock Proteins (HSP 90, 70, 60, and 27) in Galleria mellonella (Lepidoptera) Hemolymph Are Affected by Infection with Conidiobolus coronatus (Entomophthorales). PLoS ONE 2020, 15, e0228556. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Li, G.; Ji, X.; Yang, Y.; Guo, X.; Sun, Q. Identification of an Inositol-3-Phosphate Synthase 1-B Gene (AccIPS1-B) from Apis cerana cerana and Its Role in Abiotic Stress. Cell Stress Chaperones 2019, 24, 1101–1113. [Google Scholar] [CrossRef]
- Harrison, M.C.; Niño, L.M.J.; Rodrigues, M.A.; Ryll, J.; Flatt, T.; Oettler, J.; Bornberg-Bauer, E. Gene Coexpression Network Reveals Highly Conserved, Well-Regulated Anti-Ageing Mechanisms in Old Ant Queens. Genome Biol. Evol. 2021, 13, evab093. [Google Scholar] [CrossRef]
- Federici, B.A.; Bigot, Y. Origin and evolution of polydnaviruses by symbiogenesis on insect DNA viruses in endoparasitic wasps. J. Insect Phys. 2003, 49, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Strand, M.R.; Burke, G.R. Polydnaviruses: From discovery to current insights. Virology 2015, 479, 393–402. [Google Scholar] [CrossRef]
- Pinto, C.P.G.; Walker, A.A.; King, G.F.; Rossi, G.D. Immunosuppressive, antimicrobial and insecticidal activities of inhibitor cystine knot peptides produced by teratocytes of the endoparasitoid wasp Cotesia flavipes (Hymenoptera: Braconidae). Insect Sci. 2022, 30, 4. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salvia, R.; Scieuzo, C.; Boschi, A.; Pezzi, M.; Mistri, M.; Munari, C.; Chicca, M.; Vogel, H.; Cozzolino, F.; Monaco, V.; et al. An Overview of Ovarian Calyx Fluid Proteins of Toxoneuron nigriceps (Viereck) (Hymenoptera: Braconidae): An Integrated Transcriptomic and Proteomic Approach. Biomolecules 2023, 13, 1547. https://doi.org/10.3390/biom13101547
Salvia R, Scieuzo C, Boschi A, Pezzi M, Mistri M, Munari C, Chicca M, Vogel H, Cozzolino F, Monaco V, et al. An Overview of Ovarian Calyx Fluid Proteins of Toxoneuron nigriceps (Viereck) (Hymenoptera: Braconidae): An Integrated Transcriptomic and Proteomic Approach. Biomolecules. 2023; 13(10):1547. https://doi.org/10.3390/biom13101547
Chicago/Turabian StyleSalvia, Rosanna, Carmen Scieuzo, Andrea Boschi, Marco Pezzi, Michele Mistri, Cristina Munari, Milvia Chicca, Heiko Vogel, Flora Cozzolino, Vittoria Monaco, and et al. 2023. "An Overview of Ovarian Calyx Fluid Proteins of Toxoneuron nigriceps (Viereck) (Hymenoptera: Braconidae): An Integrated Transcriptomic and Proteomic Approach" Biomolecules 13, no. 10: 1547. https://doi.org/10.3390/biom13101547







