Defining the Protease and Protease Inhibitor (P/PI) Proteomes of Healthy and Diseased Human Skin by Modified Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
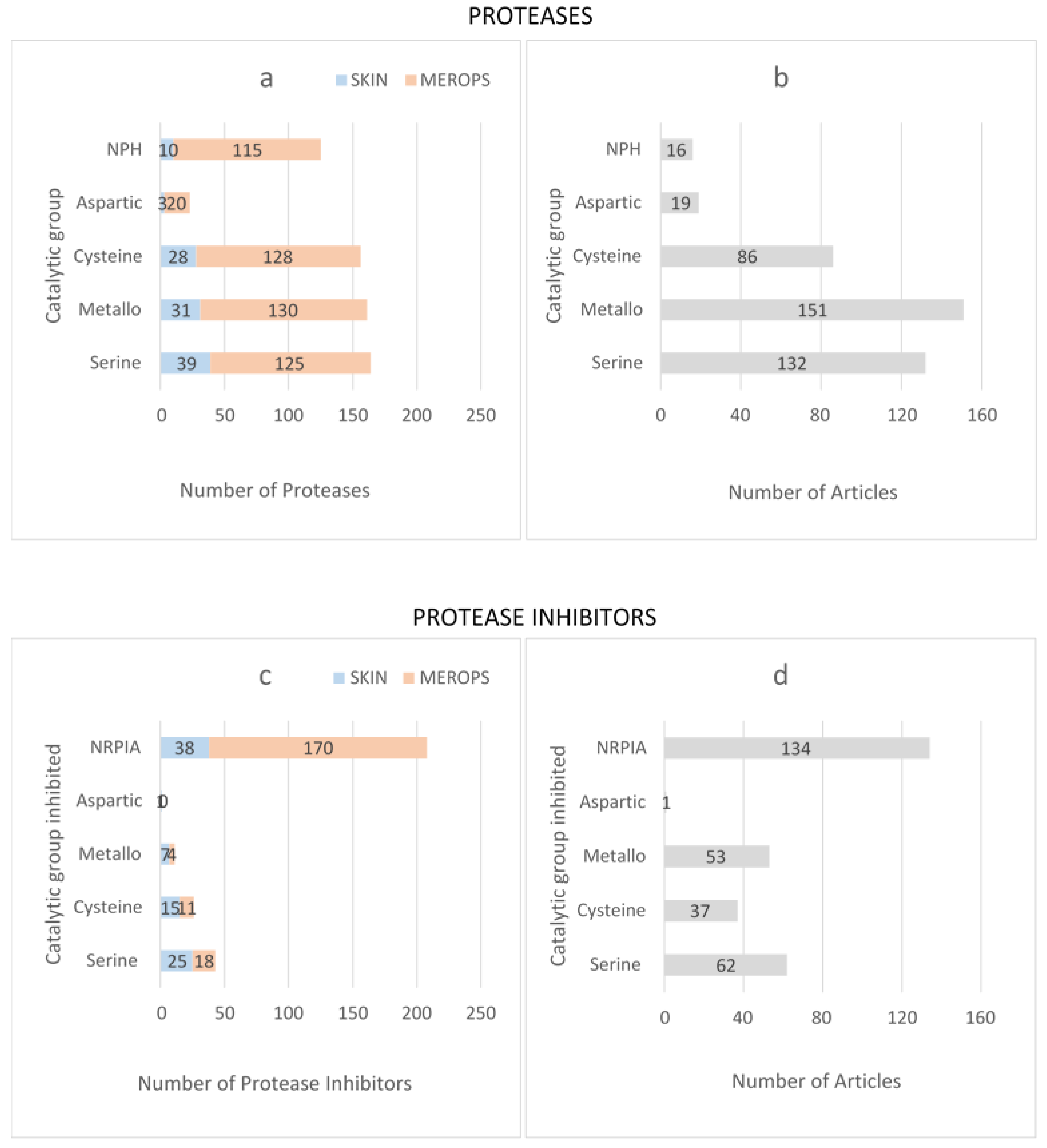
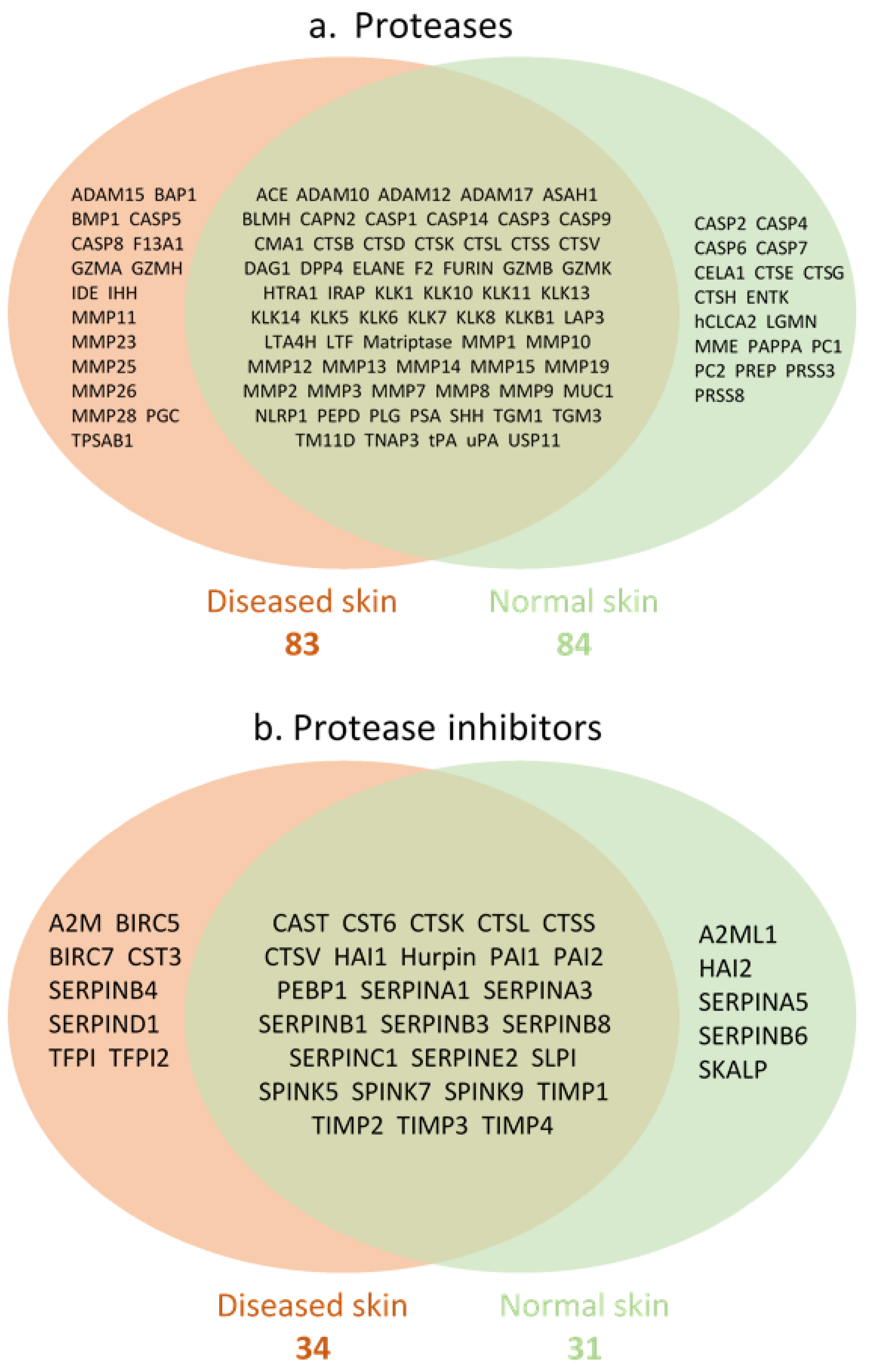
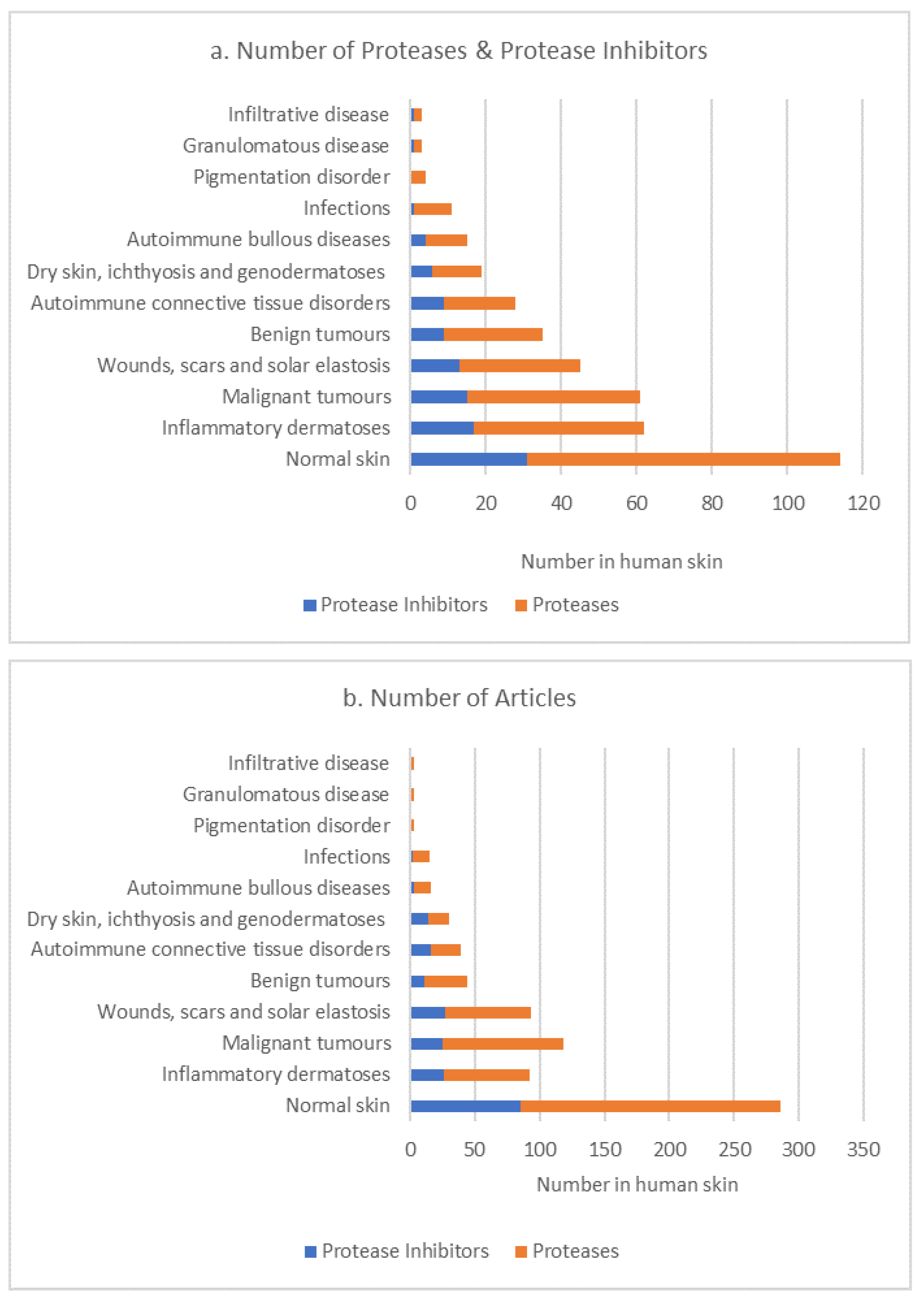
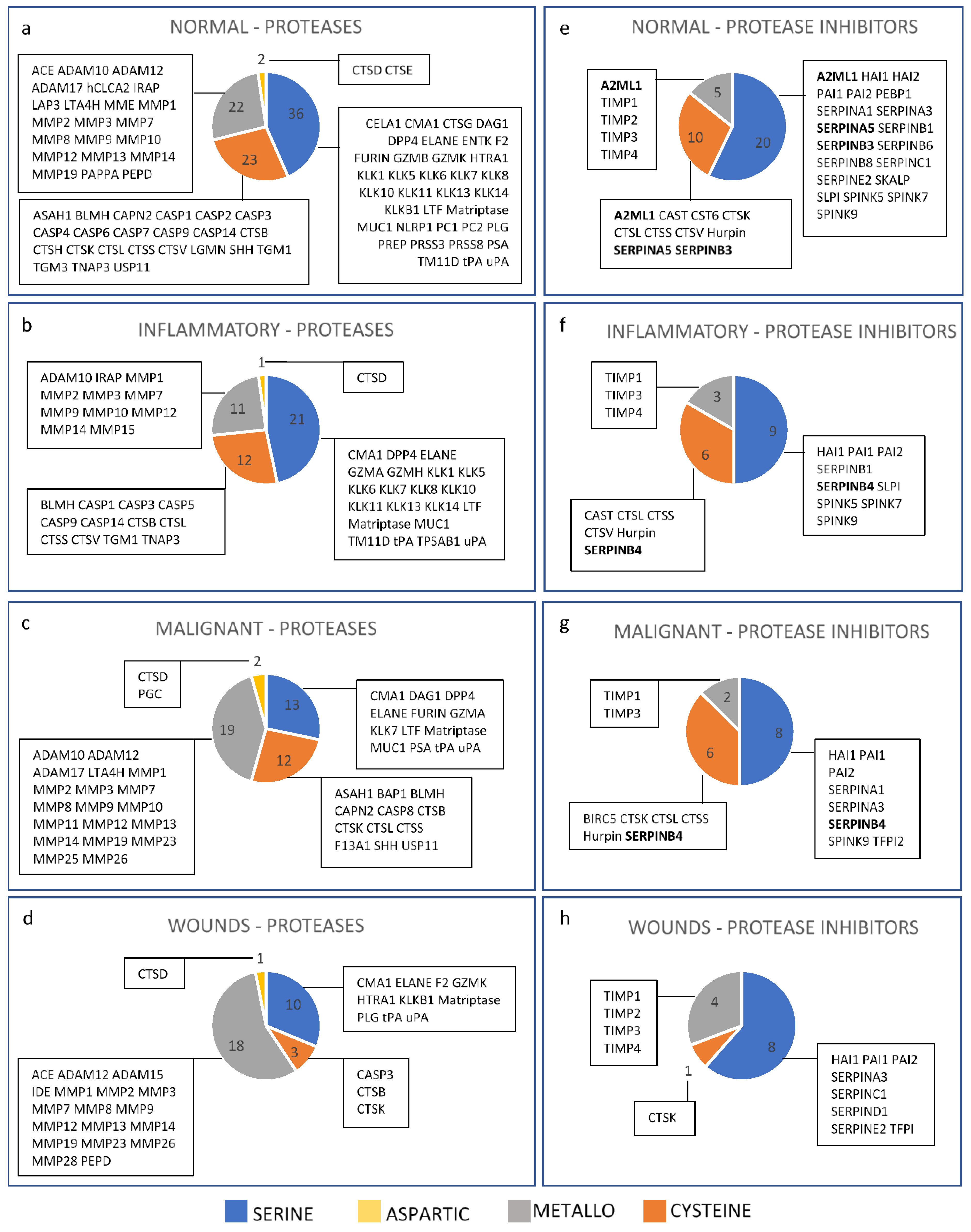
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cabral-Pacheco, G.A.; Garza-Veloz, I.; La Rosa, C.C.-D.; Ramirez-Acuña, J.M.; Perez-Romero, B.A.; Guerrero-Rodriguez, J.F.; Martinez-Avila, N.; Martinez-Fierro, M.L. The Roles of Matrix Metalloproteinases and Their Inhibitors in Human Diseases. Int. J. Mol. Sci. 2020, 21, 9739. [Google Scholar] [CrossRef]
- Nauroy, P.; Nyström, A. Kallikreins: Essential epidermal messengers for regulation of the skin microenvironment during homeostasis, repair and disease. Matrix Biol. Plus 2019, 6–7, 100019. [Google Scholar] [CrossRef]
- Gutierrez-Fernandez, A.; Inada, M.; Balbín, M.; Fueyo-Silva, A.; Pitiot, A.; Astudillo, A.; Hirose, K.; Hirata, M.; Shapiro, S.D.; Noel, A.; et al. Increased inflammation delays wound healing in mice deficient in collagenase-2 (MMP-8). FASEB J. 2007, 21, 2580–2591. [Google Scholar] [CrossRef]
- Hattori, N.; Mochizuki, S.; Kishi, K.; Nakajima, T.; Takaishi, H.; D’Armiento, J.; Okada, Y. MMP-13 Plays a Role in Keratinocyte Migration, Angiogenesis, and Contraction in Mouse Skin Wound Healing. Am. J. Pathol. 2009, 175, 533–546. [Google Scholar] [CrossRef] [Green Version]
- Hiroyasu, S.; Hiroyasu, A.; Granville, D.J.; Tsuruta, D. Pathological functions of granzyme B in inflammatory skin diseases. J. Dermatol. Sci. 2021, 104, 76–82. [Google Scholar] [CrossRef]
- Davis, G.E.; Saunders, W.B. Molecular Balance of Capillary Tube Formation versus Regression in Wound Repair: Role of Matrix Metalloproteinases and Their Inhibitors. J. Investig. Dermatol. Symp. Proc. 2006, 11, 44–56. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Lim, J.; Oh, J.-H.; Cho, S.; Chung, J.H. IGF-1 Upregulates Biglycan and Decorin by Increasing Translation and Reducing ADAMTS5 Expression. Int. J. Mol. Sci. 2021, 22, 1403. [Google Scholar] [CrossRef]
- Han, Y.-P.; Yan, C.; Garner, W.L. Proteolytic Activation of Matrix Metalloproteinase-9 in Skin Wound Healing Is Inhibited by α-1-Antichymotrypsin. J. Investig. Dermatol. 2008, 128, 2334–2342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emami, N.; Diamandis, E.P. Human kallikrein-related peptidase 14 (KLK14) is a new activator component of the KLK proteolytic cascade-Possible function in seminal plasma and skin. J. Biol. Chem. 2008, 283, 3031–3041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hovnanian, A. Netherton syndrome: Skin inflammation and allergy by loss of protease inhibition. Cell Tissue Res. 2013, 351, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Müller, F.; Hausser, I.; Berg, D.; Casper, C.; Maiwald, R.; Jung, A.; Jung, H.; Korge, B. Genetic analysis of a severe case of Netherton syndrome and application for prenatal testing. Br. J. Dermatol. 2002, 146, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, K.; Kanada, K.; Macleod, D.T.; Borkowski, A.W.; Morizane, S.; Nakatsuji, T.; Cogen, A.L.; Gallo, R. TLR2 Expression Is Increased in Rosacea and Stimulates Enhanced Serine Protease Production by Keratinocytes. J. Investig. Dermatol. 2011, 131, 688–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bullen, E.C.; Longaker, M.T.; Updike, D.L.; Benton, R.; Ladin, D.; Hou, Z.; Howard, E.W. Tissue Inhibitor of Metalloproteinases-1 Is Decreased and Activated Gelatinases Are Increased in Chronic Wounds. J. Investig. Dermatol. 1995, 104, 236–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, G.J.; Wang, Z.Q.; Datta, S.C.; Varani, J.; Kang, S.; Voorhees, J.J. Pathophysiology of Premature Skin Aging Induced by Ultraviolet Light. N. Engl. J. Med. 1997, 337, 1419–1429. [Google Scholar] [CrossRef] [PubMed]
- Furio, L.; Hovnanian, A. Netherton syndrome: Defective kallikrein inhibition in the skin leads to skin inflammation and allergy. Biol. Chem. 2014, 395, 945–958. [Google Scholar] [CrossRef] [PubMed]
- Ainscough, J.S.; Macleod, T.; McGonagle, D.; Brakefield, R.; Baron, J.M.; Alase, A.; Wittmann, M.; Stacey, M. Cathepsin S is the major activator of the psoriasis-associated proinflammatory cytokine IL-36 gamma. Proc. Natl. Acad. Sci. USA 2017, 114, E2748–E2757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Veer, S.J.; Furio, L.; Harris, J.M.; Hovnanian, A. Proteases: Common culprits in human skin disorders. Trends Mol. Med. 2014, 20, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Hibbert, S.A.; Ozols, M.; Griffiths, C.E.M.; Watson, R.E.B.; Bell, M.; Sherratt, M.J. Defining tissue proteomes by systematic literature review. Sci. Rep. 2018, 8, 546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozols, M.; Eckersley, A.; Mellody, K.T.; Mallikarjun, V.; Warwood, S.; O’Cualain, R.; Knight, D.; Watson, R.; Griffiths, C.; Swift, J.; et al. Peptide location fingerprinting reveals modification-associated biomarker candidates of ageing in human tissue proteomes. Aging Cell 2021, 20, e13355. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R. Sensitivity and specificity: Twin goals of proteomics assays. Can they be combined? Expert Rev. Proteom. 2013, 10, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Chakraborti, S.; Mandal, M.; Das, S.; Mandal, A.; Chakraborti, T. Regulation of matrix metalloproteinases: An overview. Mol. Cell. Biochem. 2003, 253, 269–285. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Waller, M.; Barrett, A.J.; Bateman, A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014, 42, D503–D509. [Google Scholar] [CrossRef] [Green Version]
- Rawlings, N.D. Twenty-five years of nomenclature and classification of proteolytic enzymes. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2020, 1868, 140345. [Google Scholar] [CrossRef]
- Rawlings, N.D. Peptidase inhibitors in the MEROPS database. Biochimie 2010, 92, 1463–1483. [Google Scholar] [CrossRef]
- Szél, E.; Bozó, R.; Hunyadi-Gulyás, É.; Manczinger, M.; Szabó, K.; Kemény, L.; Bata-Csörgő, Z.; Groma, G. Comprehensive Proteomic Analysis Reveals Intermediate Stage of Non-Lesional Psoriatic Skin and Points out the Importance of Proteins Outside this Trend. Sci. Rep. 2019, 9, 11382. [Google Scholar] [CrossRef]
- Wang, W.; Xia, Z.; Farré, J.-C.; Subramani, S. TRIM37 deficiency induces autophagy through deregulating the MTORC1-TFEB axis. Autophagy 2018, 14, 1574–1585. [Google Scholar] [CrossRef] [Green Version]
- Niizuma, H.; Cheng, E.H.; Hsieh, J.J. Taspase 1: A protease with many biological surprises. Mol. Cell. Oncol. 2015, 2, e999513. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Abreu, J.G.; Yokota, C.; MacDonald, B.T.; Singh, S.; Coburn, K.L.A.; Cheong, S.-M.; Zhang, M.M.; Ye, Q.-Z.; Hang, H.C.; et al. Tiki1 Is Required for Head Formation via Wnt Cleavage-Oxidation and Inactivation. Cell 2012, 149, 1565–1577. [Google Scholar] [CrossRef] [Green Version]
- Rijken, F.; Bruijnzeel, P.L.; van Weelden, H.; Kiekens, R.C. Responses of Black and White Skin to Solar-Simulating Radiation: Differences in DNA Photodamage, Infiltrating Neutrophils, Proteolytic Enzymes Induced, Keratinocyte Activation, and IL-10 Expression. J. Investig. Dermatol. 2004, 122, 1448–1455. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.-S.; Song, J.H.; Youn, U.J.; Hyun, J.W.; Jeong, W.S.; Lee, M.Y.; Choi, H.J.; Lee, H.-K.; Chae, S. Inhibition of UVB-induced wrinkle formation and MMP-9 expression by mangiferin isolated from Anemarrhena asphodeloides. Eur. J. Pharmacol. 2012, 689, 38–44. [Google Scholar] [CrossRef]
- Dong, T.; Santos, S.; Yang, Z.; Yang, S.; Kirkhus, N.E. Sputum and salivary protein biomarkers and point-of-care biosensors for the management of COPD. Analyst 2020, 145, 1583–1604. [Google Scholar] [CrossRef] [PubMed]
- Olejarz, W.; Łacheta, D.; Kubiak-Tomaszewska, G. Matrix Metalloproteinases as Biomarkers of Atherosclerotic Plaque Instability. Int. J. Mol. Sci. 2020, 21, 3946. [Google Scholar] [CrossRef] [PubMed]
- Vafadari, B.; Salamian, A.; Kaczmarek, L. MMP-9 in translation: From molecule to brain physiology, pathology, and therapy. J. Neurochem. 2015, 139, 91–114. [Google Scholar] [CrossRef] [Green Version]
- Raymond, A.-A.; Méchin, M.-C.; Nachat, R.; Toulza, E.; Tazi-Ahnini, R.; Serre, G.; Simon, M. Nine procaspases are expressed in normal human epidermis, but only caspase-14 is fully processed. Br. J. Dermatol. 2007, 156, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Vesela, B.; Svandova, E.; Berghe, T.V.; Tucker, A.; Vandenabeele, P.; Matalova, E. Non-apoptotic role for caspase-7 in hair follicles and the surrounding tissue. Histochem. J. 2015, 46, 443–455. [Google Scholar] [CrossRef]
- A Hitchon, C.; Danning, C.L.; Illei, G.G.; El-Gabalawy, H.S.; Boumpas, D.T. Gelatinase expression and activity in the synovium and skin of patients with erosive psoriatic arthritis. J. Rheumatol. 2002, 29, 107–117. [Google Scholar]
- D’ortho, M.P.; Will, H.; Atkinson, S.; Butler, G.; Messent, A.; Gavrilovic, J.; Smith, B.; Timpl, R.; Zardi, L.; Murphy, G. Membrane-type matrix metalloproteinases 1 and 2 exhibit broad-spectrum proteolytic capacities comparable to many matrix metalloproteinases. Eur. J. Biochem. 1997, 250, 751–757. [Google Scholar] [CrossRef] [Green Version]
- Rodrıguez-Manzaneque, J.C.; Milchanowski, A.B.; Dufour, E.K.; Leduc, R.; Iruela-Arispe, M.L. Characterization of METH-1/ADAMTS1 processing reveals two distinct active forms. J. Biol. Chem. 2000, 275, 33471–33479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roh, M.R.; Zheng, Z.; Kim, H.S.; Kwon, J.E.; Jeung, H.-C.; Rha, S.Y.; Chung, K.Y. Differential expression patterns of MMPs and their role in the invasion of epithelial premalignant tumors and invasive cutaneous squamous cell carcinoma. Exp. Mol. Pathol. 2012, 92, 236–242. [Google Scholar] [CrossRef]
- Nwomeh, B.C.; Liang, H.-X.; Diegelmann, R.F.; Cohen, I.K.; Yager, D.R. Dynamics of the matrix metalloproteinases MMP-1 and MMP-8 in acute open human dermal wounds. Wound Repair Regen. 1998, 6, 127–134. [Google Scholar] [CrossRef]
- Arif, S.; Attiogbe, E.; Moulin, V.J. Granulation tissue myofibroblasts during normal and pathological skin healing: The interaction between their secretome and the microenvironment. Wound Repair Regen. 2021, 29, 563–572. [Google Scholar] [CrossRef]
- Jiang, L.; Dai, Y.; Cui, F.; Pan, Y.; Zhang, H.; Xiao, J.; Xiaobing, F.U. Expression of cytokines, growth factors and apoptosis-related signal molecules in chronic pressure ulcer wounds healing. Spinal Cord 2013, 52, 145–151. [Google Scholar] [CrossRef] [Green Version]
- Velasco, G.; Pendás, A.M.; Fueyo, A.; Knäuper, V.; Murphy, G.; López-Otín, C. Cloning and Characterization of Human MMP-23, a New Matrix Metalloproteinase Predominantly Expressed in Reproductive Tissues and Lacking Conserved Domains in Other Family Members. J. Biol. Chem. 1999, 274, 4570–4576. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.M.; Galea, C.A.; Schmunk, G.; Smith, B.; Edwards, R.A.; Norton, R.S.; Chandy, K.G. Intracellular Trafficking of the KV1.3 Potassium Channel Is Regulated by the Prodomain of a Matrix Metalloprotease. J. Biol. Chem. 2013, 288, 6451–6464. [Google Scholar] [CrossRef] [Green Version]
- The Manchester Proteome. Available online: https://www.manchesterproteome.manchester.ac.uk/#/Protease_Proteome (accessed on 17 March 2022).

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stewart-McGuinness, C.; Platt, C.I.; Ozols, M.; Goh, B.; Griffiths, T.W.; Sherratt, M.J. Defining the Protease and Protease Inhibitor (P/PI) Proteomes of Healthy and Diseased Human Skin by Modified Systematic Review. Biomolecules 2022, 12, 475. https://doi.org/10.3390/biom12030475
Stewart-McGuinness C, Platt CI, Ozols M, Goh B, Griffiths TW, Sherratt MJ. Defining the Protease and Protease Inhibitor (P/PI) Proteomes of Healthy and Diseased Human Skin by Modified Systematic Review. Biomolecules. 2022; 12(3):475. https://doi.org/10.3390/biom12030475
Chicago/Turabian StyleStewart-McGuinness, Callum, Christopher I. Platt, Matiss Ozols, Brian Goh, Tamara W. Griffiths, and Michael J. Sherratt. 2022. "Defining the Protease and Protease Inhibitor (P/PI) Proteomes of Healthy and Diseased Human Skin by Modified Systematic Review" Biomolecules 12, no. 3: 475. https://doi.org/10.3390/biom12030475





