E-Cadherin Expression Distinguishes Mouse from Human Hematopoiesis in the Basophil and Erythroid Lineages
Abstract
:1. Introduction
2. Methods
2.1. Generation of Mice
2.2. Human PBMC Isolation
2.3. LCMV Infections
2.4. Immunohistochemistry
2.5. Flow Cytometry
2.6. Cytospins
2.7. Hematopoietic Progenitor Colony Formation Assay
2.8. Single Cell RNA Sequencing
2.9. Statistics
3. Results
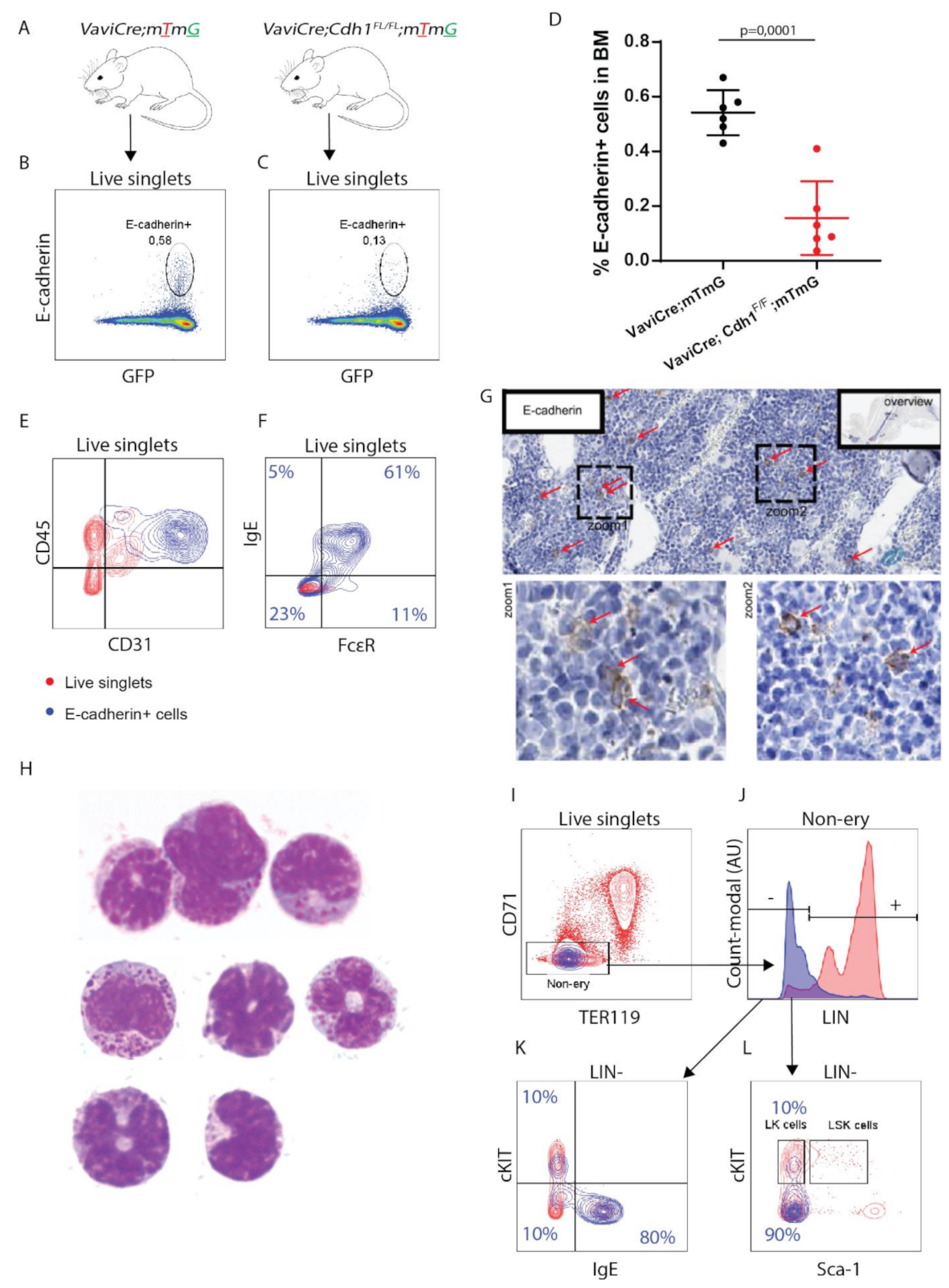
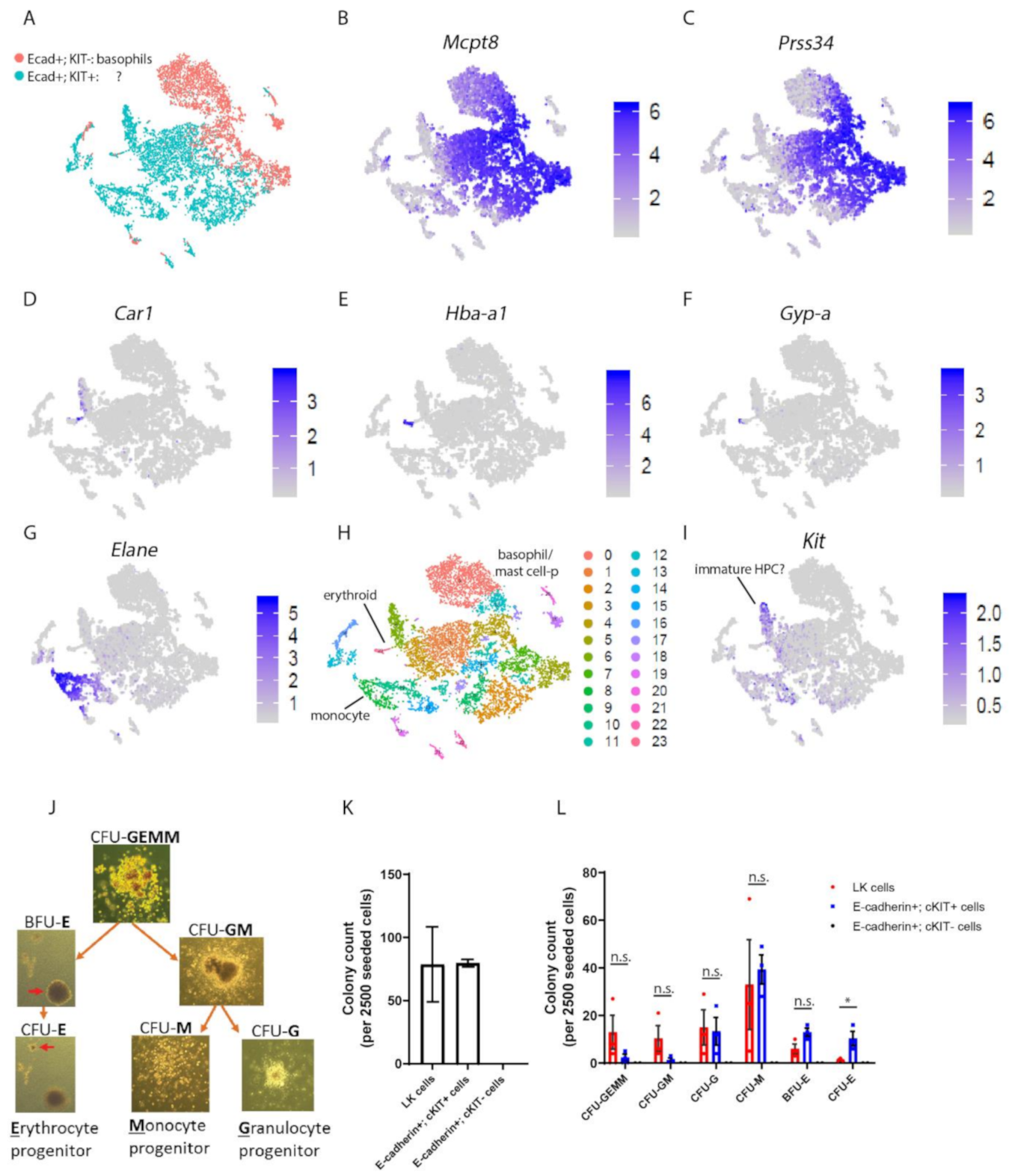
3.1. E-cadherin Is Predominantly Expressed by Basophil/Mast Cell Precursors in Mouse BM
3.2. E-cadherin Is Dispensable for Basophil Formation in Mice
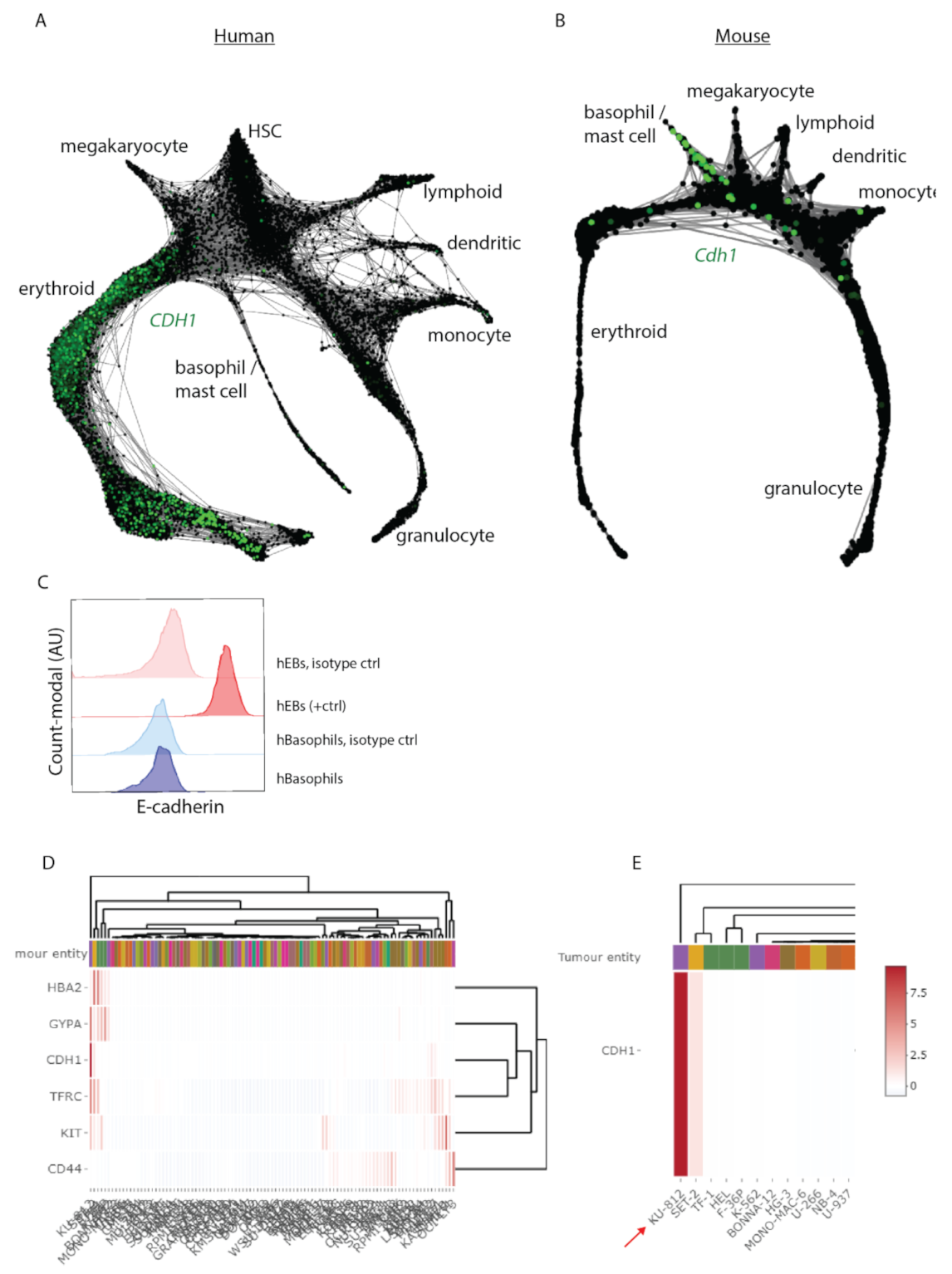
3.3. Cdh1 Expression Distinguishes Human from Mouse Hematopoiesis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koch, A.W.; Bozic, D.; Pertz, O.; Engel, J. Homophilic adhesion by cadherins. Curr. Opin. Struct. Biol. 1999, 9, 275–281. [Google Scholar] [CrossRef]
- Tomschy, A.; Fauser, C.; Landwehr, R.; Engel, J. Homophilic adhesion of E-cadherin occurs by a co-operative two-step interaction of N-terminal domains. EMBO J. 1996, 15, 3507–3514. [Google Scholar] [CrossRef]
- Yamada, S.; Pokutta, S.; Drees, F.; Weis, W.I.; Nelson, W.J. Deconstructing the cadherin-catenin-actin complex. Cell 2005, 123, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Gudipaty, S.A.; Lindblom, J.; Loftus, P.D.; Redd, M.J.; Edes, K.; Davey, C.F.; Krishnegowda, V.; Rosenblatt, J. Mechanical stretch triggers rapid epithelial cell division through Piezo1. Nature 2017, 543, 118–121. [Google Scholar] [CrossRef] [Green Version]
- Lecuit, T.; Yap, A.S. E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat. Cell Biol. 2015, 17, 533–539. [Google Scholar] [CrossRef]
- Benham-Pyle, B.W.; Sim, J.Y.; Hart, K.C.; Pruitt, B.L.; Nelson, W.J. Increasing β-catenin/Wnt3A activity levels drive mechanical strain-induced cell cycle progression through mitosis. eLife 2016, 5, e19799. [Google Scholar] [CrossRef]
- Benham-Pyle, B.W.; Pruitt, B.L.; Nelson, W.J. Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science 2015, 348, 1024–1027. [Google Scholar] [CrossRef] [Green Version]
- Larue, L.; Ohsugi, M.; Hirchenhain, J.; Kemler, R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc. Natl. Acad. Sci. USA 1994, 91, 8263–8267. [Google Scholar] [CrossRef] [Green Version]
- Boelens, M.C.; Nethe, M.; Klarenbeek, S.; De Ruiter, J.R.; Schut, E.; Bonzanni, N.; Zeeman, A.L.; Wientjens, E.; Van der Burg, E.; Wessels, L.; et al. PTEN Loss in E-Cadherin-Deficient Mouse Mammary Epithelial Cells Rescues Apoptosis and Results in Development of Classical Invasive Lobular Carcinoma. Cell Rep. 2016, 16, 2087–2101. [Google Scholar] [CrossRef] [Green Version]
- Schipper, K.; Seinstra, D.; Paulien Drenth, A.; Van der Burg, E.; Ramovs, V.; Sonnenberg, A.; Van Rheenen, J.; Nethe, M.; Jonkers, J. Rebalancing of actomyosin contractility enables mammary tumor formation upon loss of E-cadherin. Nat. Commun. 2019, 10, 3800. [Google Scholar] [CrossRef]
- Derksen, P.W.; Liu, X.; Saridin, F.; Van der Gulden, H.; Zevenhoven, J.; Evers, B.; Van Beijnum, J.R.; Griffioen, A.W.; Vink, J.; Krimpenfort, P.; et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell 2006, 10, 437–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tinkle, C.L.; Lechler, T.; Pasolli, H.A.; Fuchs, E. Conditional targeting of E-cadherin in skin: Insights into hyperproliferative and degenerative responses. Proc. Natl. Acad. Sci. USA 2004, 101, 552–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bondow, B.J.; Faber, M.L.; Wojta, K.J.; Walker, E.M.; Battle, M.A. E-cadherin is required for intestinal morphogenesis in the mouse. Dev. Biol. 2012, 371, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strumane, K.; Berx, G.; Van Roy, F. Cadherins in cancer. In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2004; Volume 165, pp. 69–103. [Google Scholar] [CrossRef]
- Ohgami, R.S.; Chisholm, K.M.; Ma, L.; Arber, D.A. E-cadherin is a specific marker for erythroid differentiation and has utility, in combination with CD117 and CD34, for enumerating myeloblasts in hematopoietic neoplasms. Am. J. Clin. Pathol. 2014, 141, 656–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldwell, I.; Ruskova, A.; Royle, G.; Liang, J.; Bain, B.J. Pure erythroid leukemia: The value of E-cadherin in making the diagnosis. Am. J. Hematol. 2019, 94, 726–727. [Google Scholar] [CrossRef]
- Murray-Stewart, T.; Woster, P.M.; Casero, R.A., Jr. The re-expression of the epigenetically silenced e-cadherin gene by a polyamine analogue lysine-specific demethylase-1 (LSD1) inhibitor in human acute myeloid leukemia cell lines. Amino Acids 2014, 46, 585–594. [Google Scholar] [CrossRef]
- Aggerholm, A.; Holm, M.S.; Guldberg, P.; Olesen, L.H.; Hokland, P. Promoter hypermethylation of p15INK4B, HIC1, CDH1, and ER is frequent in myelodysplastic syndrome and predicts poor prognosis in early-stage patients. Eur. J. Haematol. 2006, 76, 23–32. [Google Scholar] [CrossRef]
- Deneberg, S.; Grövdal, M.; Karimi, M.; Jansson, M.; Nahi, H.; Corbacioglu, A.; Gaidzik, V.; Döhner, K.; Paul, C.; Ekström, T.J.; et al. Gene-specific and global methylation patterns predict outcome in patients with acute myeloid leukemia. Leukemia 2010, 24, 932–941. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Wu, X.; Fossett, N. Drosophila E-cadherin functions in hematopoietic progenitors to maintain multipotency and block differentiation. PLoS ONE 2013, 8, e74684. [Google Scholar] [CrossRef]
- Wanet, A.; Bassal, M.A.; Patel, S.B.; Marchi, F.; Mariani, S.A.; Ahmed, N.; Zhang, H.; Borchiellini, M.; Chen, S.; Zhang, J.; et al. E-cadherin is regulated by GATA-2 and marks the early commitment of mouse hematopoietic progenitors to the basophil and mast cell fates. Sci. Immunol. 2021, 6, eaba0178. [Google Scholar] [CrossRef] [PubMed]
- Armeanu, S.; Bühring, H.J.; Reuss-Borst, M.; Müller, C.A.; Klein, G. E-cadherin is functionally involved in the maturation of the erythroid lineage. J. Cell Biol. 1995, 131, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Voermans, C.; Gerritsen, W.R.; Von dem Borne, A.E.; Van der Schoot, C.E. Increased migration of cord blood-derived CD34+ cells, as compared to bone marrow and mobilized peripheral blood CD34+ cells across uncoated or fibronectin-coated filters. Exp. Hematol. 1999, 27, 1806–1814. [Google Scholar] [CrossRef]
- Siegemund, S.; Shepherd, J.; Xiao, C.; Sauer, K. hCD2-iCre and Vav-iCre mediated gene recombination patterns in murine hematopoietic cells. PLoS ONE 2015, 10, e0124661. [Google Scholar] [CrossRef] [Green Version]
- Muzumdar, M.D.; Tasic, B.; Miyamichi, K.; Li, L.; Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 2007, 45, 593–605. [Google Scholar] [CrossRef]
- Lee, J.J.; McGarry, M.P. When is a mouse basophil not a basophil? Blood 2007, 109, 859–861. [Google Scholar] [CrossRef] [Green Version]
- Ugajin, T.; Kojima, T.; Mukai, K.; Obata, K.; Kawano, Y.; Minegishi, Y.; Eishi, Y.; Yokozeki, H.; Karasuyama, H. Basophils preferentially express mouse Mast Cell Protease 11 among the mast cell tryptase family in contrast to mast cells. J. Leukoc. Biol. 2009, 86, 1417–1425. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, B.M.; Liang, H.E.; Bando, J.K.; Wu, D.; Cheng, L.E.; McKerrow, J.K.; Allen, C.D.; Locksley, R.M. Genetic analysis of basophil function in vivo. Nat. Immunol. 2011, 12, 527–535. [Google Scholar] [CrossRef]
- Wada, T.; Ishiwata, K.; Koseki, H.; Ishikura, T.; Ugajin, T.; Ohnuma, N.; Obata, K.; Ishikawa, R.; Yoshikawa, S.; Mukai, K.; et al. Selective ablation of basophils in mice reveals their nonredundant role in acquired immunity against ticks. J. Clin. Investig. 2010, 120, 2867–2875. [Google Scholar] [CrossRef] [Green Version]
- Poorafshar, M.; Helmby, H.; Troye-Blomberg, M.; Hellman, L. MMCP-8, the first lineage-specific differentiation marker for mouse basophils. Elevated numbers of potent IL-4-producing and MMCP-8-positive cells in spleens of malaria-infected mice. Eur. J. Immunol. 2000, 30, 2660–2668. [Google Scholar] [CrossRef]
- El Hachem, C.; Hener, P.; Kirstetter, P.; Li, J.; Chan, S.; Li, M. Treatment of MCPT8(DTR) mice with high- or low-dose diphtheria toxin leads to differential depletion of basophils and granulocyte-macrophage progenitors. Eur. J. Immunol. 2018, 48, 861–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koblitz, J.; Dirks, W.G.; Eberth, S.; Nagel, S.; Steenpass, L.; Pommerenke, C. DSMZCellDive: Diving into high-throughput cell line data. F1000Research 2022, 11, 420. [Google Scholar] [CrossRef] [PubMed]
- Stellrecht-Broomhall, K.A. Evidence for immune-mediated destruction as mechanism for LCMV-induced anemia in persistently infected mice. Viral Immunol. 1991, 4, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Libregts, S.F.; Gutiérrez, L.; De Bruin, A.M.; Wensveen, F.M.; Papadopoulos, P.; Van Ijcken, W.; Ozgür, Z.; Philipsen, S.; Nolte, M.A. Chronic IFN-γ production in mice induces anemia by reducing erythrocyte life span and inhibiting erythropoiesis through an IRF-1/PU.1 axis. Blood 2011, 118, 2578–2588. [Google Scholar] [CrossRef] [Green Version]
- Tusi, B.K.; Wolock, S.L.; Weinreb, C.; Hwang, Y.; Hidalgo, D.; Zilionis, R.; Waisman, A.; Huh, J.R.; Klein, A.M.; Socolovsky, M. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature 2018, 555, 54–60. [Google Scholar] [CrossRef]
- Pellin, D.; Loperfido, M.; Baricordi, C.; Wolock, S.L.; Montepeloso, A.; Weinberg, O.K.; Biffi, A.; Klein, A.M.; Biasco, L. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat. Commun. 2019, 10, 2395. [Google Scholar] [CrossRef] [Green Version]
- Chirumbolo, S.; Vella, A.; Ortolani, R.; De Gironcoli, M.; Solero, P.; Tridente, G.; Bellavite, P. Differential response of human basophil activation markers: A multi-parameter flow cytometry approach. Clin. Mol. Allergy CMA 2008, 6, 12. [Google Scholar] [CrossRef] [Green Version]
- Blom, T.; Huang, R.; Aveskogh, M.; Nilsson, K.; Hellman, L. Phenotypic characterization of KU812, a cell line identified as an immature human basophilic leukocyte. Eur. J. Immunol. 1992, 22, 2025–2032. [Google Scholar] [CrossRef]
- Klewer, T.; Bakic, L.; Müller-Reichert, T.; Kiewisz, R.; Jessberger, G.; Kiessling, N.; Roers, A.; Jessberger, R. E-Cadherin restricts mast cell degranulation in mice. Eur. J. Immunol. 2022, 52, 44–53. [Google Scholar] [CrossRef]
- Gambardella, A.R.; Poto, R.; Tirelli, V.; Schroeder, J.T.; Marone, G.; Mattei, F.; Varricchi, G.; Schiavoni, G. Differential Effects of Alarmins on Human and Mouse Basophils. Front. Immunol. 2022, 13, 894163. [Google Scholar] [CrossRef]
- Liu, B.; Staron, M.; Li, Z. Murine but not human basophil undergoes cell-specific proteolysis of a major endoplasmic reticulum chaperone. PLoS ONE 2012, 7, e39442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pecaric-Petkovic, T.; Didichenko, S.A.; Kaempfer, S.; Spiegl, N.; Dahinden, C.A. Human basophils and eosinophils are the direct target leukocytes of the novel IL-1 family member IL-33. Blood 2009, 113, 1526–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banh, C.; Fugere, C.; Brossay, L. Immunoregulatory functions of KLRG1 cadherin interactions are dependent on forward and reverse signaling. Blood 2009, 114, 5299–5306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, M.; Maruyama, T.; Saito, N.; Koganei, S.; Yamamoto, K.; Matsumoto, N. Killer cell lectin-like receptor G1 binds three members of the classical cadherin family to inhibit NK cell cytotoxicity. J. Exp. Med. 2006, 203, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Grundemann, C.; Bauer, M.; Schweier, O.; Von Oppen, N.; Lassing, U.; Saudan, P.; Becker, K.F.; Karp, K.; Hanke, T.; Bachmann, M.F.; et al. Cutting edge: Identification of E-cadherin as a ligand for the murine killer cell lectin-like receptor G1. J. Immunol. 2006, 176, 1311–1315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiraishi, K.; Tsuzaka, K.; Yoshimoto, K.; Kumazawa, C.; Nozaki, K.; Abe, T.; Tsubota, K.; Takeuchi, T. Critical role of the fifth domain of E-cadherin for heterophilic adhesion with alpha E beta 7, but not for homophilic adhesion. J. Immunol. 2005, 175, 1014–1021. [Google Scholar] [CrossRef] [Green Version]
- Karecla, P.I.; Bowden, S.J.; Green, S.J.; Kilshaw, P.J. Recognition of E-cadherin on epithelial cells by the mucosal T cell integrin alpha M290 beta 7 (alpha E beta 7). Eur. J. Immunol. 1995, 25, 852–856. [Google Scholar] [CrossRef]
- Cepek, K.L.; Shaw, S.K.; Parker, C.M.; Russell, G.J.; Morrow, J.S.; Rimm, D.L.; Brenner, M.B. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the alpha E beta 7 integrin. Nature 1994, 372, 190–193. [Google Scholar] [CrossRef]
- Shields, B.D.; Koss, B.; Taylor, E.M.; Storey, A.J.; West, K.L.; Byrum, S.D.; Mackintosh, S.G.; Edmondson, R.; Mahmoud, F.; Shalin, S.C.; et al. Loss of E-Cadherin Inhibits CD103 Antitumor Activity and Reduces Checkpoint Blockade Responsiveness in Melanoma. Cancer Res. 2019, 79, 1113–1123. [Google Scholar] [CrossRef] [Green Version]
- Le Floc’h, A.; Jalil, A.; Vergnon, I.; Le Maux Chansac, B.; Lazar, V.; Bismuth, G.; Chouaib, S.; Mami-Chouaib, F. Alpha E beta 7 integrin interaction with E-cadherin promotes antitumor CTL activity by triggering lytic granule polarization and exocytosis. J. Exp. Med. 2007, 204, 559–570. [Google Scholar] [CrossRef]
- Higgins, J.M.; Mandlebrot, D.A.; Shaw, S.K.; Russell, G.J.; Murphy, E.A.; Chen, Y.T.; Nelson, W.J.; Parker, C.M.; Brenner, M.B. Direct and regulated interaction of integrin alphaEbeta7 with E-cadherin. J. Cell Biol. 1998, 140, 197–210. [Google Scholar] [CrossRef] [Green Version]
- Henson, S.M.; Franzese, O.; Macaulay, R.; Libri, V.; Azevedo, R.I.; Kiani-Alikhan, S.; Plunkett, F.J.; Masters, J.E.; Jackson, S.; Griffiths, S.J.; et al. KLRG1 signaling induces defective Akt (ser473) phosphorylation and proliferative dysfunction of highly differentiated CD8+ T cells. Blood 2009, 113, 6619–6628. [Google Scholar] [CrossRef]
- Tessmer, M.S.; Fugere, C.; Stevenaert, F.; Naidenko, O.V.; Chong, H.J.; Leclercq, G.; Brossay, L. KLRG1 binds cadherins and preferentially associates with SHIP-1. Int. Immunol. 2007, 19, 391–400. [Google Scholar] [CrossRef] [Green Version]
- Sarfati, M.; Wakahara, K.; Chapuy, L.; Delespesse, G. Mutual Interaction of Basophils and T Cells in Chronic Inflammatory Diseases. Front. Immunol. 2015, 6, 399. [Google Scholar] [CrossRef] [Green Version]
- Sicklinger, F.; Meyer, I.S.; Li, X.; Radtke, D.; Dicks, S.; Kornadt, M.P.; Mertens, C.; Meier, J.K.; Lavine, K.J.; Zhang, Y.; et al. Basophils balance healing after myocardial infarction via IL-4/IL-13. J. Clin. Investig. 2021, 131, e136778. [Google Scholar] [CrossRef]
- Gomez, M.R.; Talke, Y.; Hofmann, C.; Ketelsen, I.; Hermann, F.; Reich, B.; Goebel, N.; Schmidbauer, K.; Dunger, N.; Brühl, H.; et al. Basophils control T-cell responses and limit disease activity in experimental murine colitis. Mucosal Immunol. 2014, 7, 188–199. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Liu, Y.; Han, X.; Mei, Y.; Yang, J.; Zhang, Z.J.; Lu, X.; Ji, P. Rats provide a superior model of human stress erythropoiesis. Exp. Hematol. 2019, 78, 21–34.e23. [Google Scholar] [CrossRef]
- Paulson, R.F.; Shi, L.; Wu, D.C. Stress erythropoiesis: New signals and new stress progenitor cells. Curr. Opin. Hematol. 2011, 18, 139–145. [Google Scholar] [CrossRef] [Green Version]
- Liao, C.; Prabhu, K.S.; Paulson, R.F. Monocyte-derived macrophages expand the murine stress erythropoietic niche during the recovery from anemia. Blood 2018, 132, 2580–2593. [Google Scholar] [CrossRef] [Green Version]
- Trompouki, E.; Bowman, T.V.; Lawton, L.N.; Fan, Z.P.; Wu, D.C.; DiBiase, A.; Martin, C.S.; Cech, J.N.; Sessa, A.K.; Leblanc, J.L.; et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 2011, 147, 577–589. [Google Scholar] [CrossRef]
- Paulson, R.F.; Hariharan, S.; Little, J.A. Stress erythropoiesis: Definitions and models for its study. Exp. Hematol. 2020, 89, 43–54.e42. [Google Scholar] [CrossRef]
- Perry, J.M.; Harandi, O.F.; Porayette, P.; Hegde, S.; Kannan, A.K.; Paulson, R.F. Maintenance of the BMP4-dependent stress erythropoiesis pathway in the murine spleen requires hedgehog signaling. Blood 2009, 113, 911–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, X.; Schulz, V.P.; Li, J.; Wu, K.; Liu, J.; Xue, F.; Hu, J.; Mohandas, N.; Gallagher, P.G. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood 2014, 123, 3466–3477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pishesha, N.; Thiru, P.; Shi, J.; Eng, J.C.; Sankaran, V.G.; Lodish, H.F. Transcriptional divergence and conservation of human and mouse erythropoiesis. Proc. Natl. Acad. Sci. USA 2014, 111, 4103–4108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ulirsch, J.C.; Lacy, J.N.; An, X.; Mohandas, N.; Mikkelsen, T.S.; Sankaran, V.G. Altered chromatin occupancy of master regulators underlies evolutionary divergence in the transcriptional landscape of erythroid differentiation. PLoS Genet. 2014, 10, e1004890. [Google Scholar] [CrossRef] [Green Version]
- Grady, W.M.; Willis, J.; Guilford, P.J.; Dunbier, A.K.; Toro, T.T.; Lynch, H.; Wiesner, G.; Ferguson, K.; Eng, C.; Park, J.G.; et al. Methylation of the CDH1 promoter as the second genetic hit in hereditary diffuse gastric cancer. Nat. Genet. 2000, 26, 16–17. [Google Scholar] [CrossRef]
- Droufakou, S.; Deshmane, V.; Roylance, R.; Hanby, A.; Tomlinson, I.; Hart, I.R. Multiple ways of silencing E-cadherin gene expression in lobular carcinoma of the breast. Int. J. Cancer 2001, 92, 404–408. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krimpenfort, R.A.; Behr, F.M.; Nieuwland, M.; Rink, I.d.; Kerkhoven, R.; Lindern, M.v.; Nethe, M. E-Cadherin Expression Distinguishes Mouse from Human Hematopoiesis in the Basophil and Erythroid Lineages. Biomolecules 2022, 12, 1706. https://doi.org/10.3390/biom12111706
Krimpenfort RA, Behr FM, Nieuwland M, Rink Id, Kerkhoven R, Lindern Mv, Nethe M. E-Cadherin Expression Distinguishes Mouse from Human Hematopoiesis in the Basophil and Erythroid Lineages. Biomolecules. 2022; 12(11):1706. https://doi.org/10.3390/biom12111706
Chicago/Turabian StyleKrimpenfort, Rosa A., Felix M. Behr, Marja Nieuwland, Iris de Rink, Ron Kerkhoven, Marieke von Lindern, and Micha Nethe. 2022. "E-Cadherin Expression Distinguishes Mouse from Human Hematopoiesis in the Basophil and Erythroid Lineages" Biomolecules 12, no. 11: 1706. https://doi.org/10.3390/biom12111706





