The Mode of Action of Cyclic Monoterpenes (−)-Limonene and (+)-α-Pinene on Bacterial Cells
Abstract
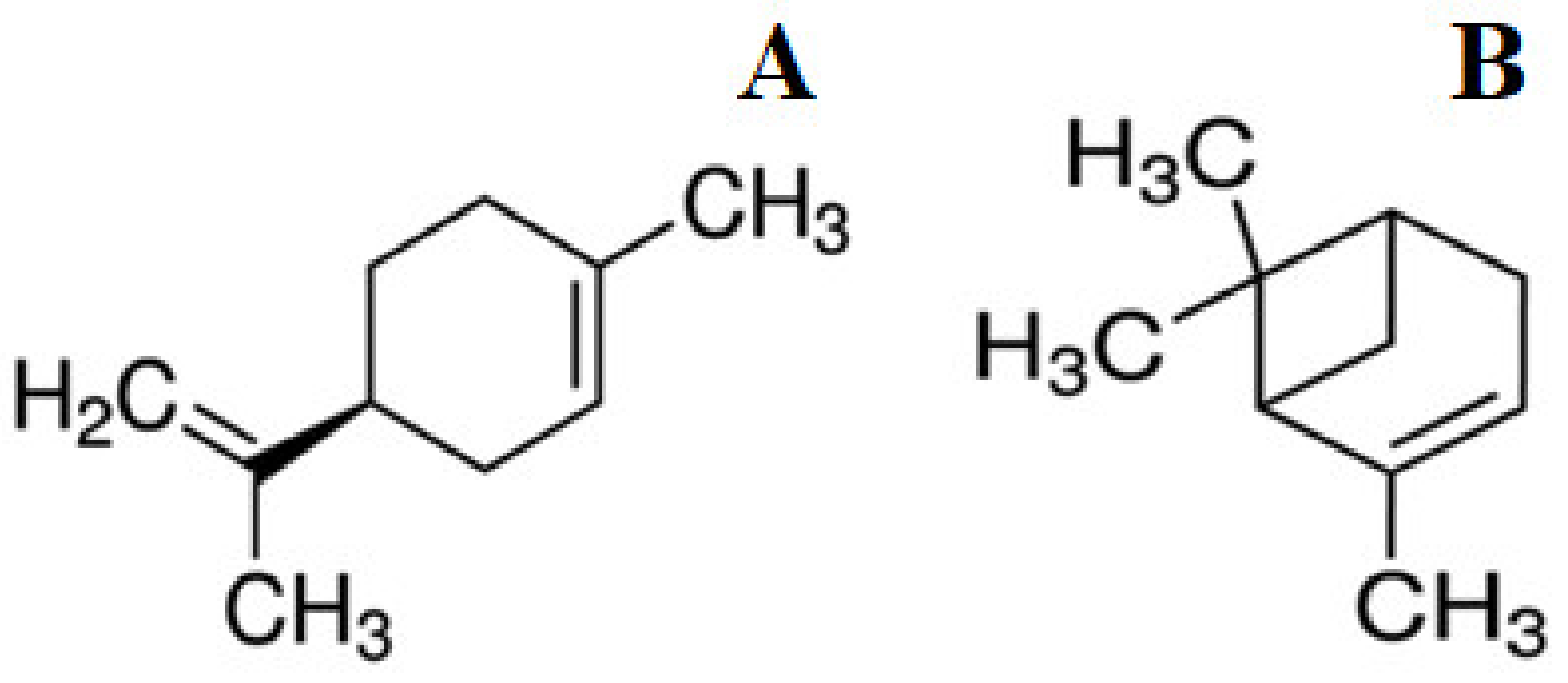
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Plasmids
2.3. Nutrient Media and Growth Conditions
2.4. Enzymes and Chemicals
2.5. Measurement of Production and Activity of Bacterial Luciferase
2.6. Thermal Inactivation and Refolding
2.7. Antibacterial Activity
3. Results
3.1. The Action of (−)-Limonene and (+)-α-Pinene on the LuxBiosensors
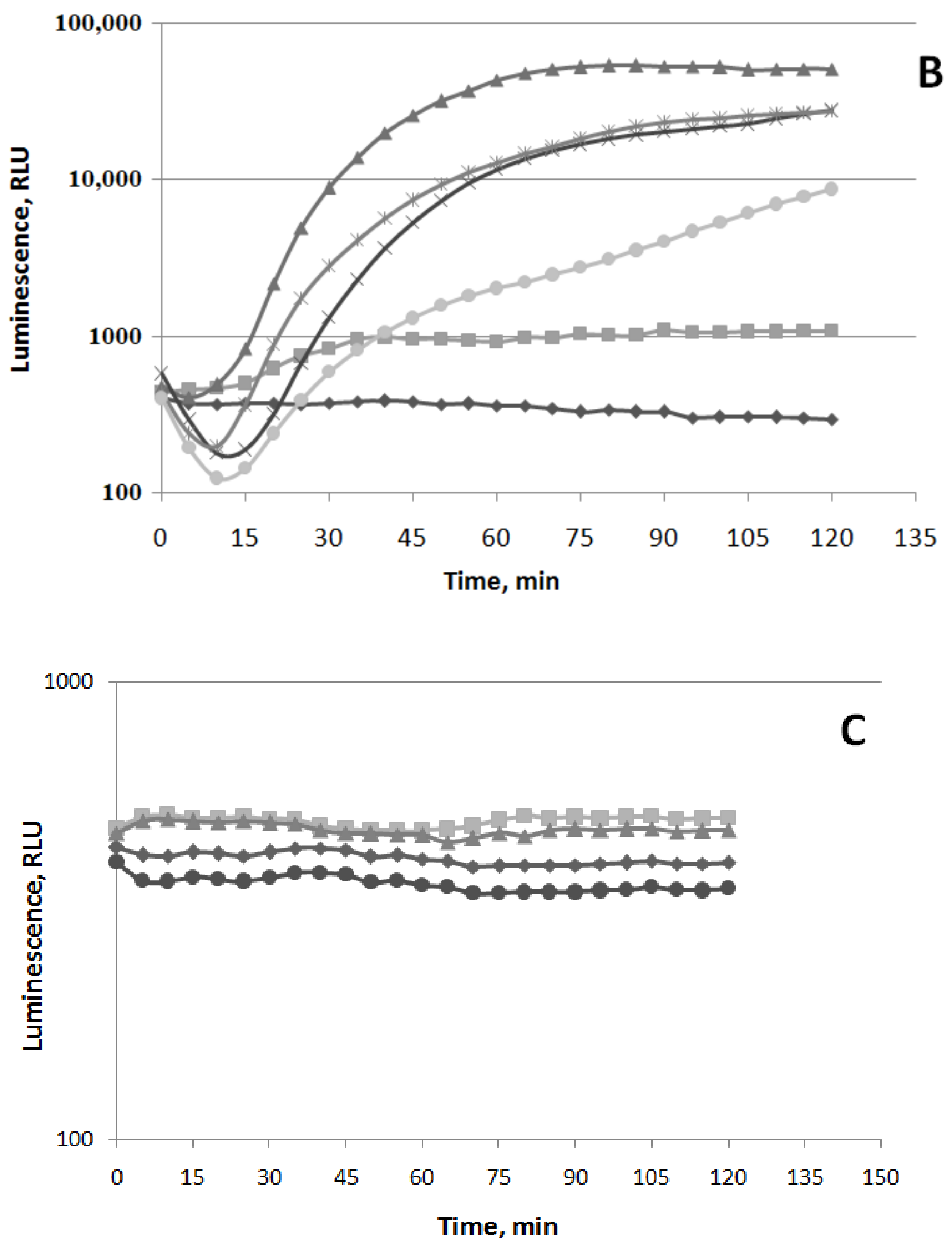
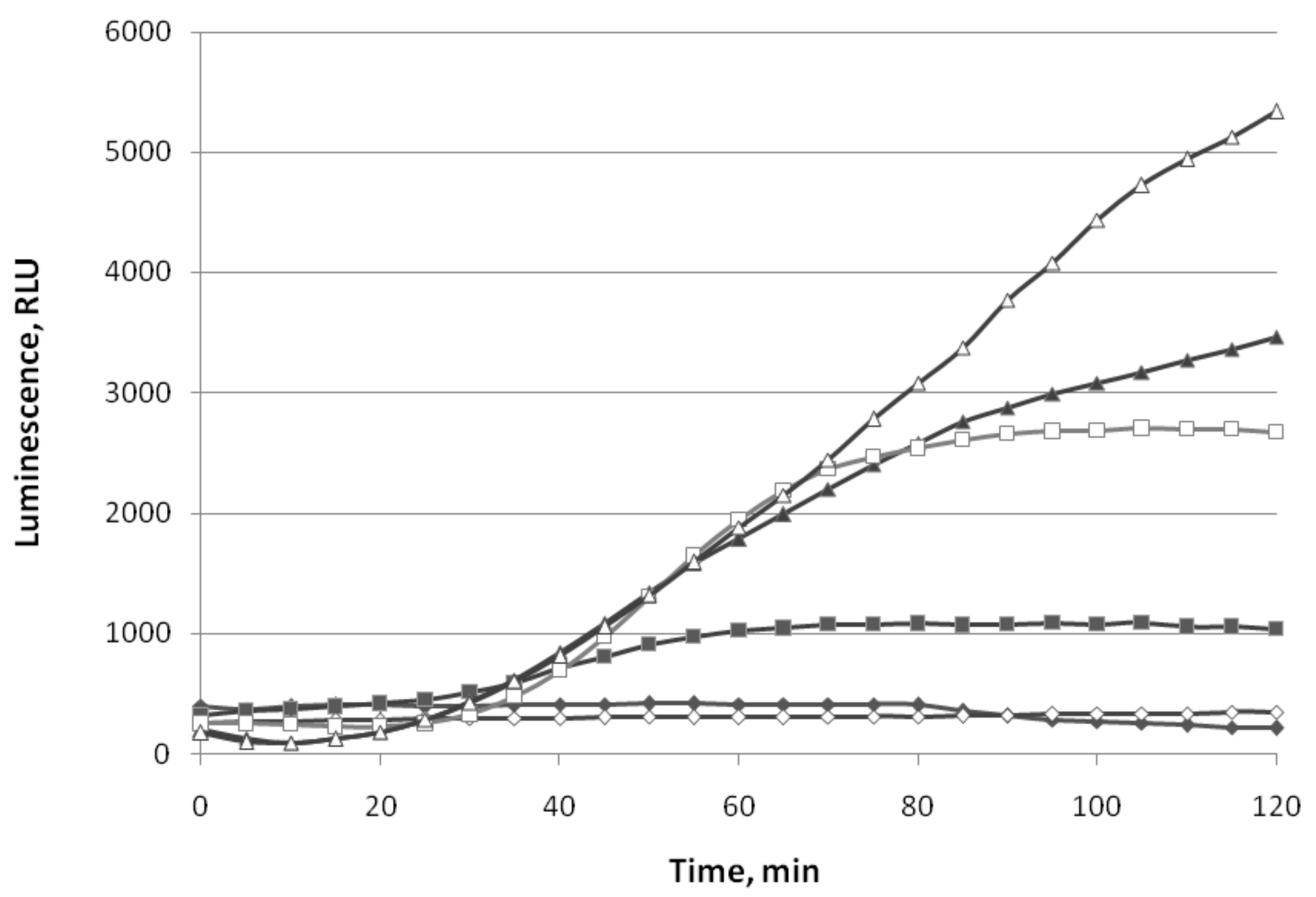
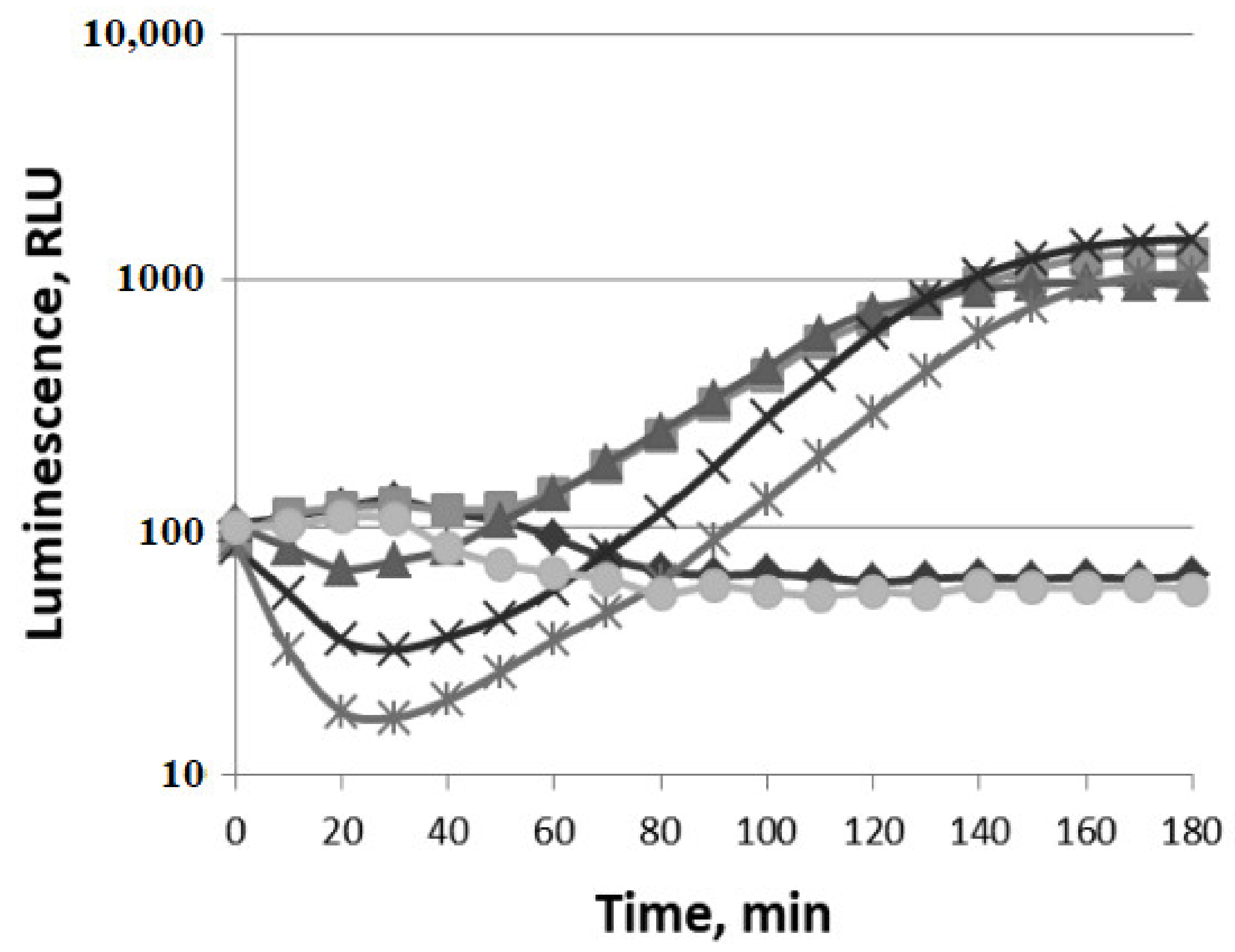
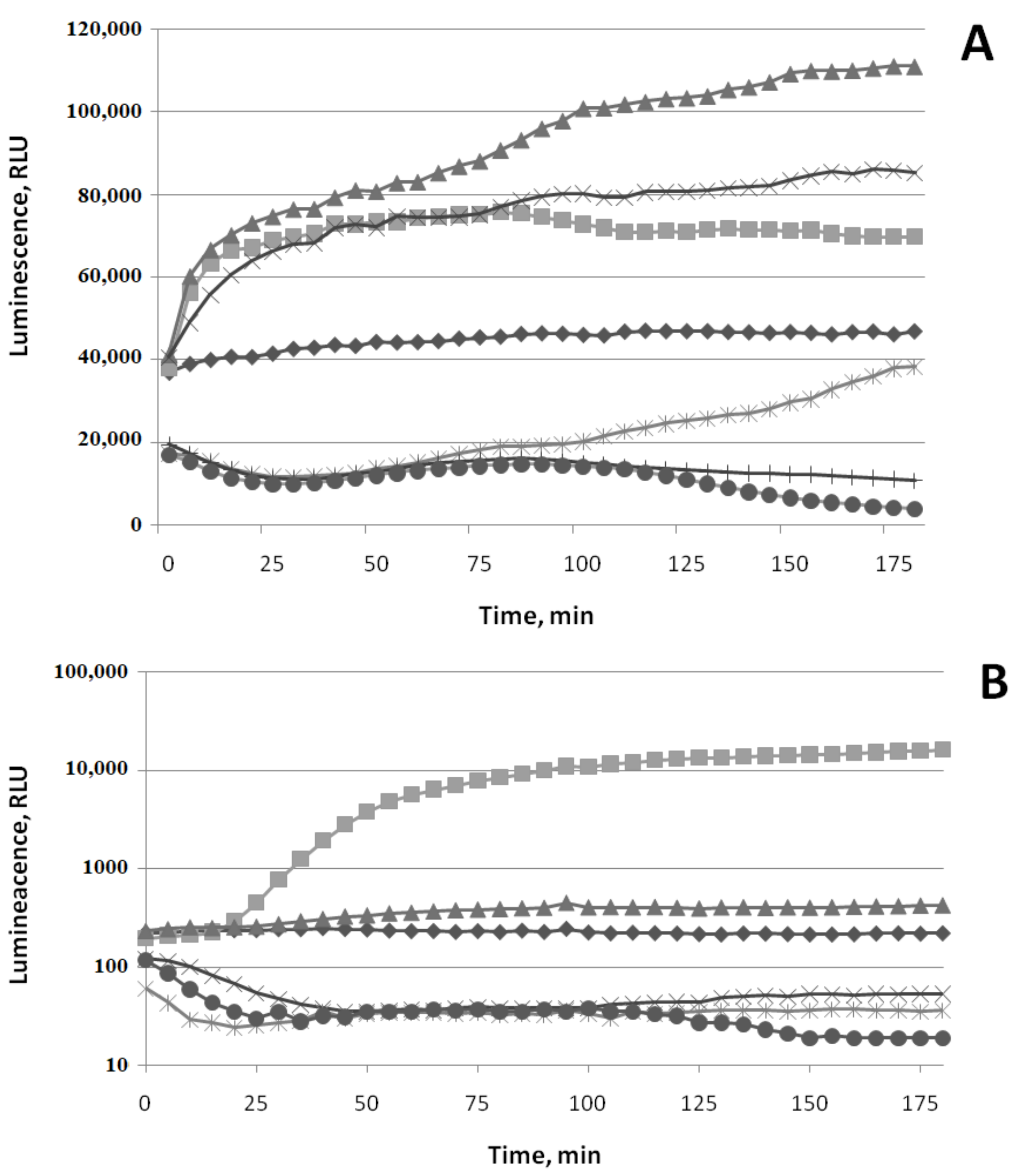
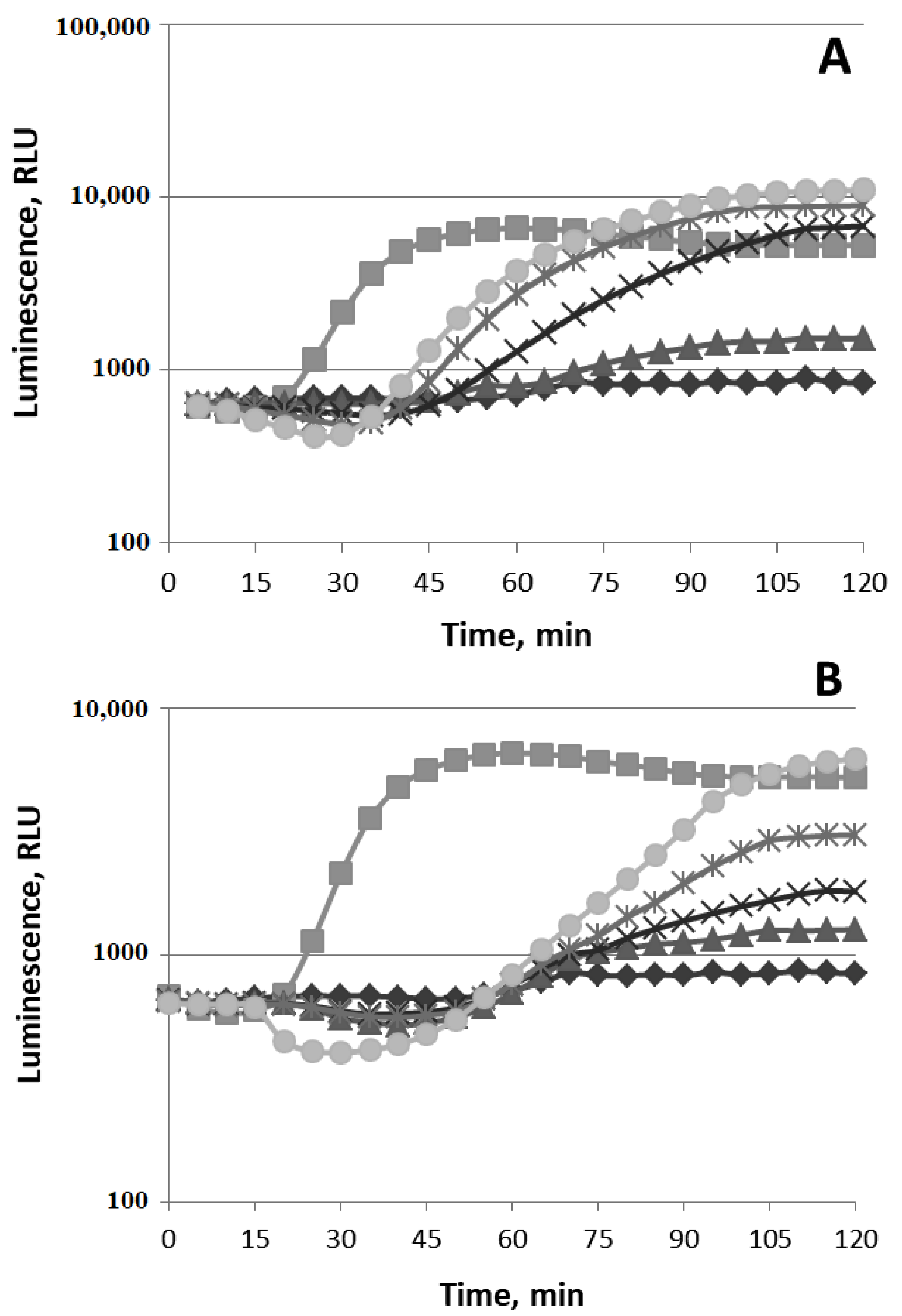
3.1.1. Oxidative Stress
3.1.2. SOSResponse
3.1.3. Cell Membrane Damage
3.1.4. HeatShock
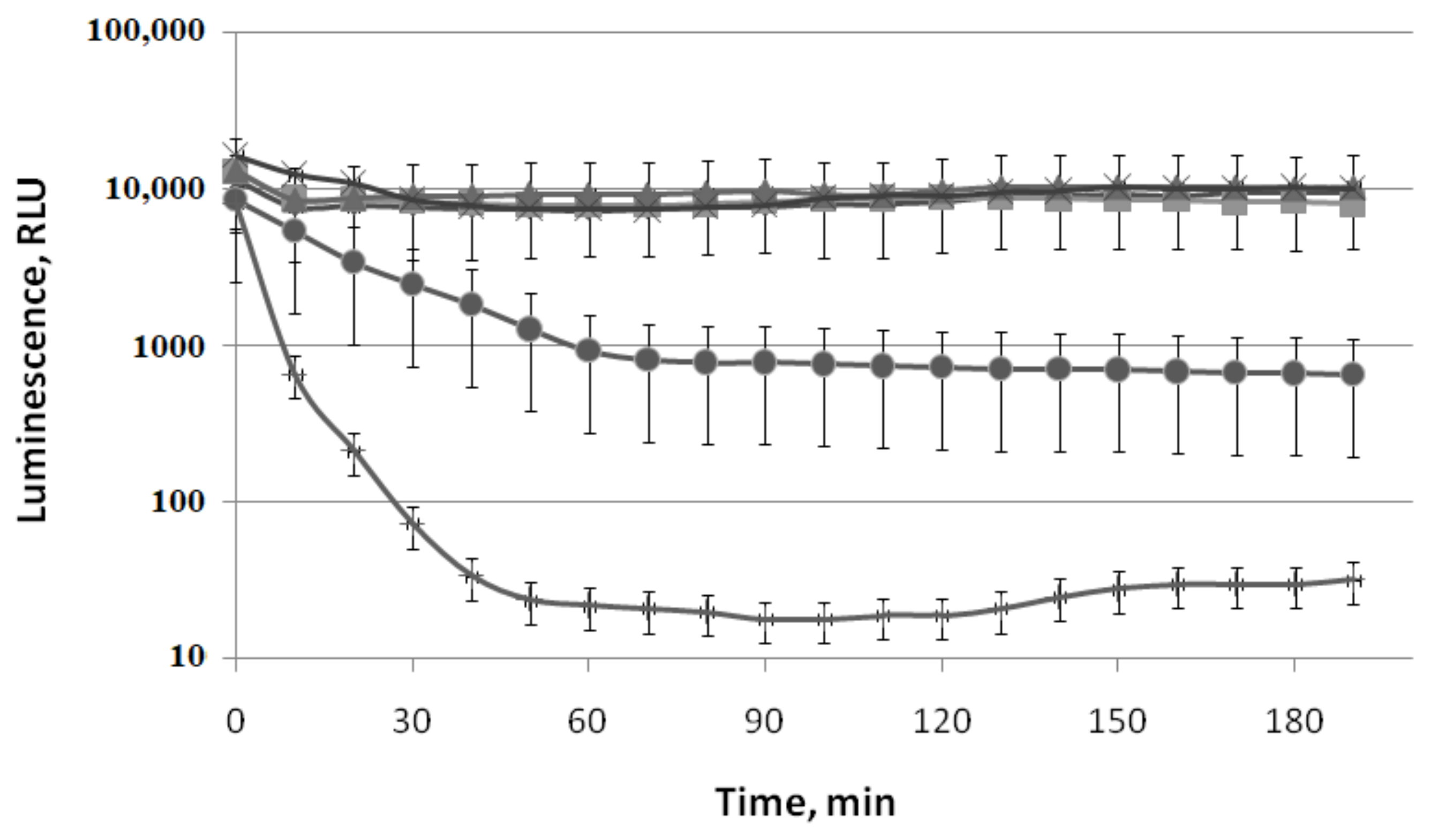
3.1.5. Effect of VOCs on the Enzymatic Activity of Native Bacterial Luciferase P. luminescens
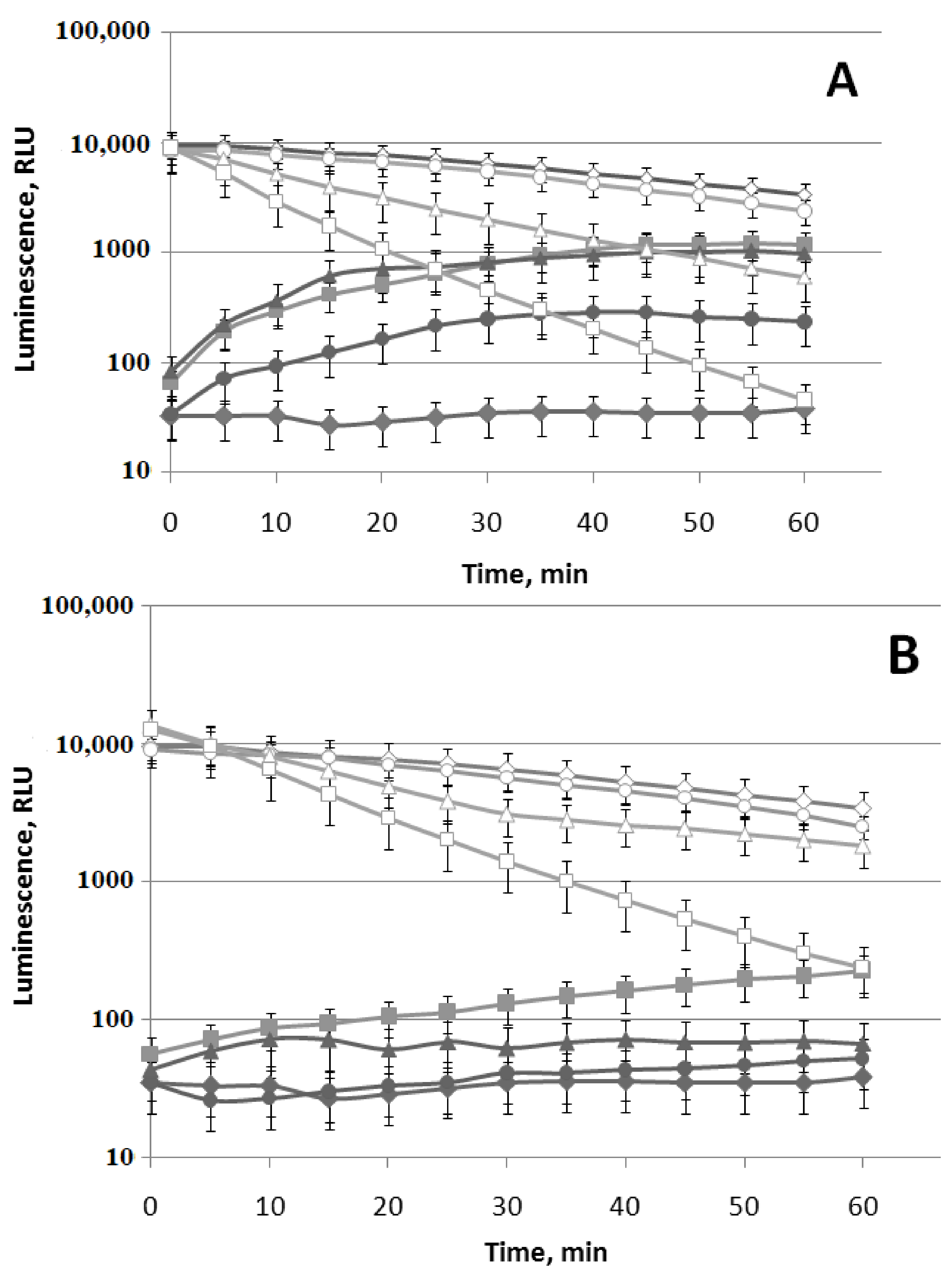
3.2. Effect of (−)-Limonene and (+)-α-Pinene on the DnaKJE-Dependent Refolding of Heat-Inactivated Bacterial Luciferase
3.3. Effect of (−)-Limonene and (+)-α-Pineneon Firefly Luciferase Activity
3.4. Effect of (−)-Limonene and (+)-α-Pinene on the Growth of Bacteria
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Böhme, K.; Barros-Velázquez, J.; Calo-Mata, P.; Aubourg, S.P. Antibacterial, Antiviral and Antifungal Activity of Essential Oils: Mechanisms and Applications. In Antimicrobial Compounds; Springer: Berlin/Heidelberg, Germany, 2014; pp. 51–81. [Google Scholar] [CrossRef]
- Burt, S. Essential Oils: Their Antibacterial Properties and Potential Applications in Foods—A Review. Int. J. Food Microbiol. 2004, 94, 223–253. [Google Scholar] [CrossRef]
- Mercier, B.; Prost, J.; Prost, M. The Essential Oil of Turpentine and Its Major Volatile Fraction (α- and β-Pinenes): A Review. Int. J. Occup. Med. Environ. Health 2009, 22, 331–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bevilacqua, A.; Corbo, M.R.; Sinigaglia, M. In Vitro Evaluation of the Antimicrobial Activity of Eugenol, Limonene, and Citrus Extract against Bacteria and Yeasts, Representative of the Spoiling Microflora of Fruit Juices. J. Food Prot. 2010, 73, 888–894. [Google Scholar] [CrossRef]
- Avalos, M.; van Wezel, G.P.; Raaijmakers, J.M.; Garbeva, P. Healthy Scents: Microbial Volatiles as New Frontier in Antibiotic Research? Curr. Opin. Microbiol. 2018, 45, 84–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giorgio, A.; de Stradis, A.; Lo Cantore, P.; Iacobellis, N.S. Biocide Effects of Volatile Organic Compounds Produced by Potential BiocontrolRhizobacteria on Sclerotinia sclerotiorum. Front. Microbiol. 2015, 6, 1056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tyc, O.; Song, C.; Dickschat, J.S.; Vos, M.; Garbeva, P. The Ecological Role of Volatile and Soluble Secondary Metabolites Produced by Soil Bacteria. Trends Microbiol. 2017, 25, 280–292. [Google Scholar] [CrossRef]
- Cai, R.; Hu, M.; Zhang, Y.; Niu, C.; Yue, T.; Yuan, Y.; Wang, Z. Antifungal Activity and Mechanism of Citral, Limonene and Eugenol against Zygosaccharomyces rouxii. LWT 2019, 106, 50–56. [Google Scholar] [CrossRef]
- Lo Cantore, P.; Shanmugaiah, V.; Iacobellis, N.S. Antibacterial Activity of Essential Oil Components and Their Potential Use in Seed Disinfection. J. Agric. Food Chem. 2009, 57, 9454–9461. [Google Scholar] [CrossRef]
- Silva, A.C.R.D.; Lopes, P.M.; Azevedo, M.M.B.D.; Costa, D.C.M.; Alviano, C.S.; Alviano, D.S. Biological Activities of a-Pinene and β-Pinene Enantiomers. Molecules 2012, 17, 6305. [Google Scholar] [CrossRef] [Green Version]
- Van Vuuren, S.F.; Viljoen, A.M. Antimicrobial Activity of Limonene Enantiomers and 1,8-Cineole Alone and in Combination. Flavour Fragr. J. 2007, 22, 540–544. [Google Scholar] [CrossRef]
- Salehi, B.; Upadhyay, S.; ErdoganOrhan, I.; Kumar Jugran, A.; Jayaweera, S.L.D.; Dias, D.A.; Sharopov, F.; Taheri, Y.; Martins, N.; Baghalpour, N.; et al. Therapeutic Potential of α- and β-Pinene: A Miracle Gift of Nature. Biomolecules 2019, 9, 738. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.-H.; Cai, M.; Liu, Y.-S.; Sun, P.-L.; Luo, S.-L. Antibacterial Activity and Mechanisms of Essential Oil from Citrus Medica L. Var. Sarcodactylis. Molecules 2019, 24, 1577. [Google Scholar] [CrossRef] [Green Version]
- Vespermann, K.A.C.; Paulino, B.N.; Barcelos, M.C.S.; Pessôa, M.G.; Pastore, G.M.; Molina, G. Biotransformation of α- and β-Pinene into Flavor Compounds. Appl. Microbiol. Biotechnol. 2017, 101, 1805–1817. [Google Scholar] [CrossRef]
- Van Dyk, T.K.; Majarian, W.R.; Konstantinov, K.B.; Young, R.M.; Dhurjati, P.S.; LaRossa, R.A. Rapid and Sensitive Pollutant Detection by Induction of Heat Shock Gene-Bioluminescence Gene Fusions. Appl. Environ. Microbiol. 1994, 60, 1414–1420. [Google Scholar] [CrossRef] [Green Version]
- Ben-Israel, O.; Ben-Israel, H.; Ulitzur, S. Identification and Quantification of Toxic Chemicals by Use of Escherichia coli Carryinglux Genes Fused to Stress Promoters. Appl. Environ. Microbiol. 1998, 64, 4346–4352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bechor, O.; Smulski, D.R.; van Dyk, T.K.; LaRossa, R.A.; Belkin, S. Recombinant Microorganisms as Environmental Biosensors: Pollutants Detection by Escherichia coli Bearing FabA′::Lux Fusions. J. Biotechnol. 2002, 94, 125–132. [Google Scholar] [CrossRef]
- Vollmer, C.A.; van Dyk, T.K. Stress Responsive Bacteria: Biosensors as Environmental Monitors. Adv. Microb. Physiol. 2004, 49, 131–174. [Google Scholar] [CrossRef]
- Zavilgelsky, G.B.; Kotova, V.Y.; Manukhov, I.V. Action of 1,1-Dimethylhydrazine on Bacterial Cells Is Determined by Hydrogen Peroxide. Mutat. Res. Toxicol. Environ. Mutagen. 2007, 634, 172–176. [Google Scholar] [CrossRef]
- Kotova, V.Y.; Manukhov, I.V.; Zavilgelskii, G.B. Lux-Biosensors for Detection of SOS-Response, Heat Shock, and Oxidative Stress. Appl. Biochem. Microbiol. 2010, 46, 781–788. [Google Scholar] [CrossRef]
- Woutersen, M.; Belkin, S.; Brouwer, B.; van Wezel, A.P.; Heringa, M.B. Are Luminescent Bacteria Suitable for Online Detection and Monitoring of Toxic Compounds in Drinking Water and Its Sources? Anal. Bioanal. Chem. 2011, 400, 915–929. [Google Scholar] [CrossRef] [Green Version]
- Guyer, M.S.; Reed, R.R.; Steitz, J.A.; Low, K.B. Identification of a Sex-Factor-Affinity Site in E. coli as Γδ. Cold Spring Harb. Symp. Quant. Biol. 1981, 45, 135–140. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-Step Inactivation of Chromosomal Genes in Escherichia coli K-12 Using PCR Products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-Frame, Single-Gene Knockout Mutants: The Keio Collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlioz, A.; Touati, D. Isolation of Superoxide Dismutase Mutants in Escherichia Coli: Is Superoxide Dismutase Necessary for Aerobic Life? EMBO J. 1986, 5, 623–630. [Google Scholar] [CrossRef]
- Riether, K.; Dollard, M.-A.; Billard, P. Assessment of Heavy Metal Bioavailability Using Escherichia coli ZntAp::Lux and CopAp::Lux-Based Biosensors. Appl. Microbiol. Biotechnol. 2001, 57, 712–716. [Google Scholar] [CrossRef] [PubMed]
- Zavilgelsky, G.B.; Kotova, V.Y.; Manukhov, I.V. Sensor bioluminescent systems with lux operons for detection of toxical compounds. Russ. J. Phys. Chem. 2012, 31, 15–20. [Google Scholar]
- Van Dyk, T.K.; Rosson, R.A. Photorhabdus Luminescens LuxCDABE Promoter Probe Vectors. In Bioluminescence Methods and Protocols; Humana Press: Totowa, NJ, USA, 1998; Volume 102, pp. 85–96. [Google Scholar] [CrossRef]
- Zavilgelsky, G.B.; Zarubina, A.P.; Manukhov, I.V. Sequencing and Comparative Analysis of the Lux Operon of Photorhabdusluminescens Strain ZM1: ERIC Elements as Putative Recombination Spots. Mol. Biol. 2002, 36, 792–804. [Google Scholar] [CrossRef]
- Zavilgelsky, G.B.; Kotova, V.Y.; Manukhov, I.V. The Effects of the Regulatory Proteins RcsA and RcsB on the Expression of the Vibrio fischeri Lux Operon in Escherichia coli. Mol. Biol. 2003, 37, 704–711. [Google Scholar] [CrossRef]
- Lundovskich, I.A.; Leontieva, O.V.; Dementieva, E.I.; Ugarova, N.N. Bioluminescence and Chemiluminescence: Perspectives for 21st Century; Roda, A., Pazzagli, M., Kricka, L.J., Stanley, P.E., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 1999; pp. 420–424. [Google Scholar]
- Sambrook, J.; Russel, D.W. Molecular Cloning, 3-Volume Set: A Laboratory Manual; Cold Spring Harboc Laboratory Press: New York, NY, USA, 2001; p. 999. [Google Scholar]
- Lundin, A.; Thore, A. Comparison of Methods for Extraction of Bacterial Adenine Nucleotides Determined by Firefly Assay. Appl. Microbiol. 1975, 30, 713–721. [Google Scholar] [CrossRef]
- Nunoshiba, T.; Nishioka, H. ‘Rec-Lac Test’ for Detecting SOS-Inducing Activity of Environmental Genotoxic Substances. Mutat. Res. Repair 1991, 254, 71–77. [Google Scholar] [CrossRef]
- Goñi, P.; López, P.; Sánchez, C.; Gómez-Lus, R.; Becerril, R.; Nerín, C. Antimicrobial Activity in the Vapour Phase of a Combination of Cinnamon and Clove Essential Oils. Food Chem. 2009, 116, 982–989. [Google Scholar] [CrossRef]
- Melkina, O.E.; Khmel, I.A.; Plyuta, V.A.; Koksharova, O.A.; Zavilgelsky, G.B. Ketones 2-Heptanone, 2-Nonanone, and 2-Undecanone Inhibit DnaK-Dependent Refolding of Heat-Inactivated Bacterial Luciferases in Escherichia coli Cells Lacking Small Chaperon IbpB. Appl. Microbiol. Biotechnol. 2017, 101, 5765–5771. [Google Scholar] [CrossRef] [PubMed]
- Matuszewska, M.; Kuczyńska-Wiśnik, D.; Laskowska, E.; Liberek, K. The Small Heat Shock Protein IbpA of Escherichia coli Cooperates with IbpB in Stabilization of Thermally Aggregated Proteins in a Disaggregation Competent State. J. Biol. Chem. 2005, 280, 12292–12298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mogk, A.; Deuerling, E.; Vorderwülbecke, S.; Vierling, E.; Bukau, B. Small Heat Shock Proteins, ClpB and the DnaK System Form a Functional Triade in Reversing Protein Aggregation. Mol. Microbiol. 2003, 50, 585–595. [Google Scholar] [CrossRef]
- Mogk, A.; Schlieker, C.; Friedrich, K.L.; Schönfeld, H.-J.; Vierling, E.; Bukau, B. Refolding of Substrates Bound to Small Hsps Relies on a Disaggregation Reaction Mediated Most Efficiently by ClpB/DnaK. J. Biol. Chem. 2003, 278, 31033–31042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palomares, A.J.; DeLuca, M.A.; Helinski, D.R. Firefly Luciferase as a Reporter Enzyme for Measuring Gene Expression in Vegetative and Symbiotic Rhizobium meliloti and Other Gram-Negative Bacteria. Gene 1989, 81, 55–64. [Google Scholar] [CrossRef]
- Subramani, S.; DeLuca, M. Applications of the Firefly Luciferase as a Reporter Gene. In Genetic Engineering; Springer US: Boston, MA, USA, 1988; pp. 75–89. [Google Scholar] [CrossRef]
- Wood, K.V.; DeLuca, M. Photographic Detection of Luminescence in Escherichia coli Containing the Gene for Firefly Luciferase. Anal. Biochem. 1987, 161, 501–507. [Google Scholar] [CrossRef]
- Dwyer, D.J.; Kohanski, M.A.; Hayete, B.; Collins, J.J. Gyrase Inhibitors Induce an Oxidative Damage Cellular Death Pathway in Escherichia coli. Mol. Syst. Biol. 2007, 3, 91. [Google Scholar] [CrossRef]
- Dwyer, D.J.; Belenky, P.A.; Yang, J.H.; MacDonald, I.C.; Martell, J.D.; Takahashi, N.; Chan, C.T.Y.; Lobritz, M.A.; Braff, D.; Schwarz, E.G.; et al. Antibiotics Induce Redox-Related Physiological Alterations as Part of Their Lethality. Proc. Natl. Acad. Sci. USA 2014, 111, E2100–E2109. [Google Scholar] [CrossRef] [Green Version]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How Antibiotics Kill Bacteria: From Targets to Networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef] [Green Version]
- Julian, W.T.; Vasilchenko, A.V.; Shpindyuk, D.D.; Poshvina, D.V.; Vasilchenko, A.S. Bacterial-Derived Plant Protection Metabolite 2,4-Diacetylphloroglucinol: Effects on Bacterial Cells at Inhibitory and Subinhibitory Concentrations. Biomolecules 2020, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Makemson, J.C.; Hastings, J.W. Poising of the Arginine Pool and Control of Bioluminescence in BeneckeaHarveyi. J. Bacteriol. 1979, 140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holzman, T.F.; Baldwin, T.O. Binding of 2,2-Diphenylpropylamine at the Aldehyde Site of Bacterial Luciferase Increases the Affinity of the Reduced Riboflavin 5′-Phosphate Site. Biochemistry 1981, 20, 5524–5528. [Google Scholar] [CrossRef]
- Holzman, T.; Baldwin, T. Corrections—Reversible Inhibition of the Bacterial Luciferase Catalyzed Bioluminescence Reaction by Aldehyde Substrate: Kinetic Mechanism and Ligand Effects. Biochemistry 1983, 22, 5464. [Google Scholar] [CrossRef]
- Virta, M.; Åkerman, K.E.O.; Saviranta, P.; Oker-Blom, C.; Karp, M.T. Real-Time Measurement of Cell Permeabilization with Low-Molecular-Weight Membranolytic Agents. J. Antimicrob. Chemother. 1995, 36, 303–315. [Google Scholar] [CrossRef]
- Gamer, J.; Bujard, H.; Bukau, B. Physical Interaction between Heat Shock Proteins DnaK, DnaJ, and GrpE and the Bacterial Heat Shock Transcription Factor Σ32. Cell 1992, 69, 833–842. [Google Scholar] [CrossRef]
- Liberek, K.; Galitski, T.P.; Zylicz, M.; Georgopoulos, C. The DnaK Chaperone Modulates the Heat Shock Response of Escherichia coli by Binding to the Sigma 32 Transcription Factor. Proc. Natl. Acad. Sci. USA 1992, 89, 3516–3520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomoyasu, T.; Gamer, J.; Bukau, B.; Kanemori, M.; Mori, H.; Rutman, A.J.; Oppenheim, A.B.; Yura, T.; Yamanaka, K.; Niki, H. Escherichia coli FtsH Is a Membrane-Bound, ATP-Dependent Protease Which Degrades the Heat-Shock Transcription Factor Sigma 32. EMBO J. 1995, 14, 2551–2560. [Google Scholar] [CrossRef]
- Arsène, F.; Tomoyasu, T.; Bukau, B. The Heat Shock Response of Escherichia coli. Int. J. Food Microbiol. 2000, 55, 3–9. [Google Scholar] [CrossRef]
- Cassel, J.A.; Ilyin, S.; McDonnell, M.E.; Reitz, A.B. Novel Inhibitors of Heat Shock Protein Hsp70-Mediated Luciferase Refolding That Bind to DnaJ. Bioorg. Med. Chem. 2012, 20, 3609–3614. [Google Scholar] [CrossRef]
- Moses, M.A.; Zuehlke, A.D.; Neckers, L. Molecular Chaperone Inhibitors. In Heat Shock Proteins in the Immune System; Springer International Publishing: Cham, Switzerland, 2018; pp. 21–40. [Google Scholar] [CrossRef]
- Lazarev, V.F.; Sverchinsky, D.V.; Mikhaylova, E.R.; Semenyuk, P.I.; Komarova, E.Y.; Niskanen, S.A.; Nikotina, A.D.; Burakov, A.V.; Kartsev, V.G.; Guzhova, I.V.; et al. Sensitizing Tumor Cells to Conventional Drugs: HSP70 Chaperone Inhibitors, Their Selection and Application in Cancer Models. Cell Death Dis. 2018, 9, 41. [Google Scholar] [CrossRef] [PubMed]




| Strain | CGSC ** ID | Genotype | Reference | Comments |
|---|---|---|---|---|
| MG1655 | 6300 | F-ilvG rfb-50 rph-1 | [22] | Prototroph |
| BW25113 | 7636 | F−, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), л−, rph-1, Δ(rhaD-rhaB)568, hsdR514 | [23] | (This is) The parent strain for the Keio Collection of single-gene knockouts. |
| JW3914-1 | 10827 | ΔkatG729::kan | [24] | Keio Collection strain |
| JW3663 | 10689 | ibpB::kan | [24] | Keio Collection strain |
| JW3933-3 | 12039 | ΔoxyR749::kan | [24] | Keio Collection strain |
| QC868 | no | F−leu6 thrA1 proA2 thi-1 lacY1 tonA1 rpsL31 supE44 hsdR−sodA+sodB+Smr | [25] | The parent strain for QC871 |
| QC871 | no | F−leu6 thrA1 proA2 thi-1 lacY1 tonA1 rpsL31 supE44 hsdR−sodA25 sodB2CmrKanrSmr | [25] | The double mutant (sodAB−) without the manganese- and iron-cofactored superoxide dismutases |
| E. coli Strain | DIZ, mm * |
|---|---|
| MG1655 (wt) | 7 ± 0.5 ** |
| QC871 sodAB- | 8 ± 0.5 |
| JW3933-3 ΔoxyR749::kan | 16 ± 0.5 |
| JW3914-1 ΔkatG729::kan | 10 ± 0.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melkina, O.E.; Plyuta, V.A.; Khmel, I.A.; Zavilgelsky, G.B. The Mode of Action of Cyclic Monoterpenes (−)-Limonene and (+)-α-Pinene on Bacterial Cells. Biomolecules 2021, 11, 806. https://doi.org/10.3390/biom11060806
Melkina OE, Plyuta VA, Khmel IA, Zavilgelsky GB. The Mode of Action of Cyclic Monoterpenes (−)-Limonene and (+)-α-Pinene on Bacterial Cells. Biomolecules. 2021; 11(6):806. https://doi.org/10.3390/biom11060806
Chicago/Turabian StyleMelkina, Olga E., Vladimir A. Plyuta, Inessa A. Khmel, and Gennadii B. Zavilgelsky. 2021. "The Mode of Action of Cyclic Monoterpenes (−)-Limonene and (+)-α-Pinene on Bacterial Cells" Biomolecules 11, no. 6: 806. https://doi.org/10.3390/biom11060806





