Mapping the Salt Stress-Induced Changes in the Root miRNome in Pokkali Rice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Small RNA Library Construction and Sequencing
2.3. mRNA and Degradome Sequencing and Analysis
2.4. Expression Analysis of miRs and Target mRNA
2.5. TaqMan PCR Assay
2.6. miR-eQTL Search and Mapping
3. Results
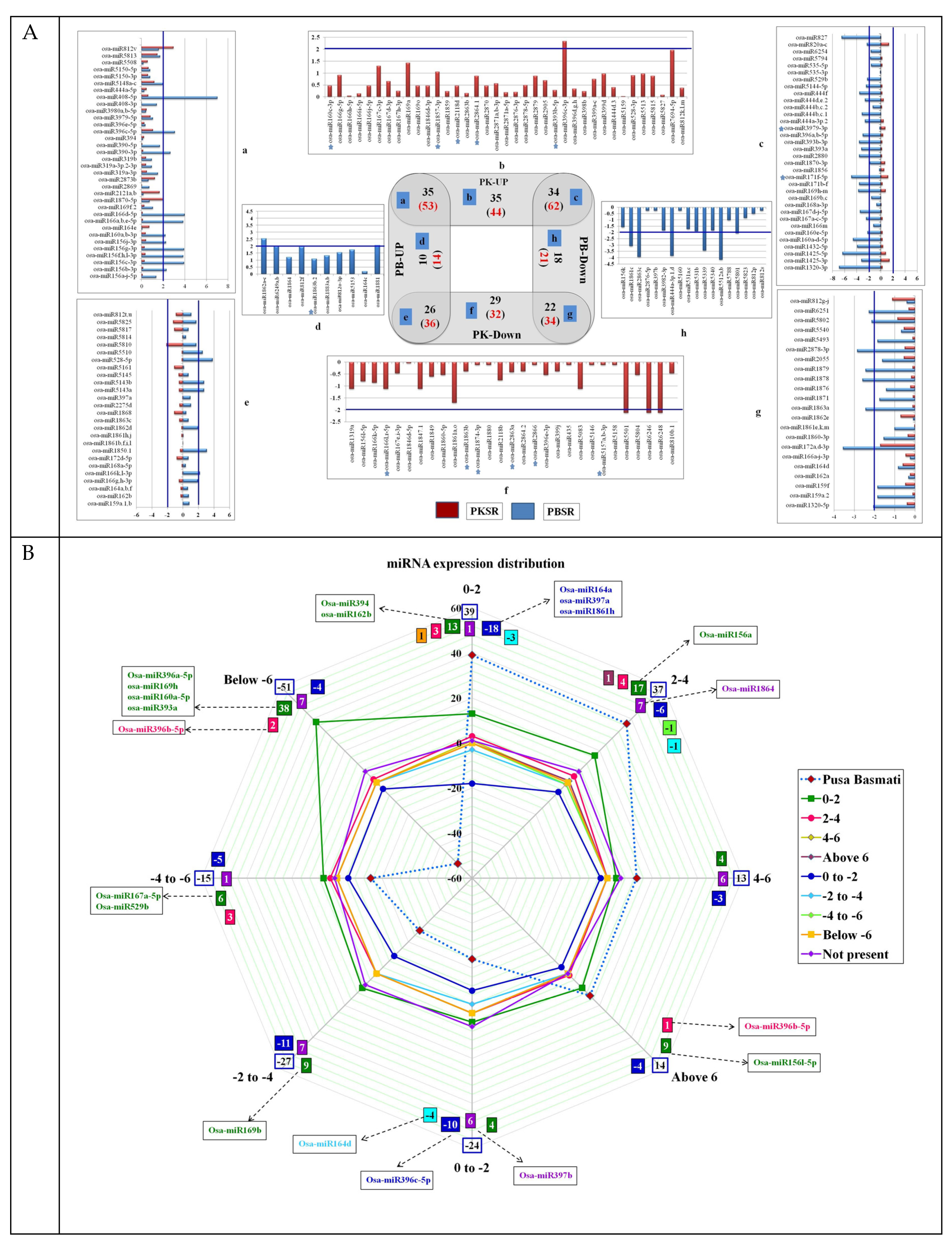
3.1. Expression Profiling of miRs in Roots
3.2. Validation of Salt-Regulated miRs Using the Taqman PCR Assay
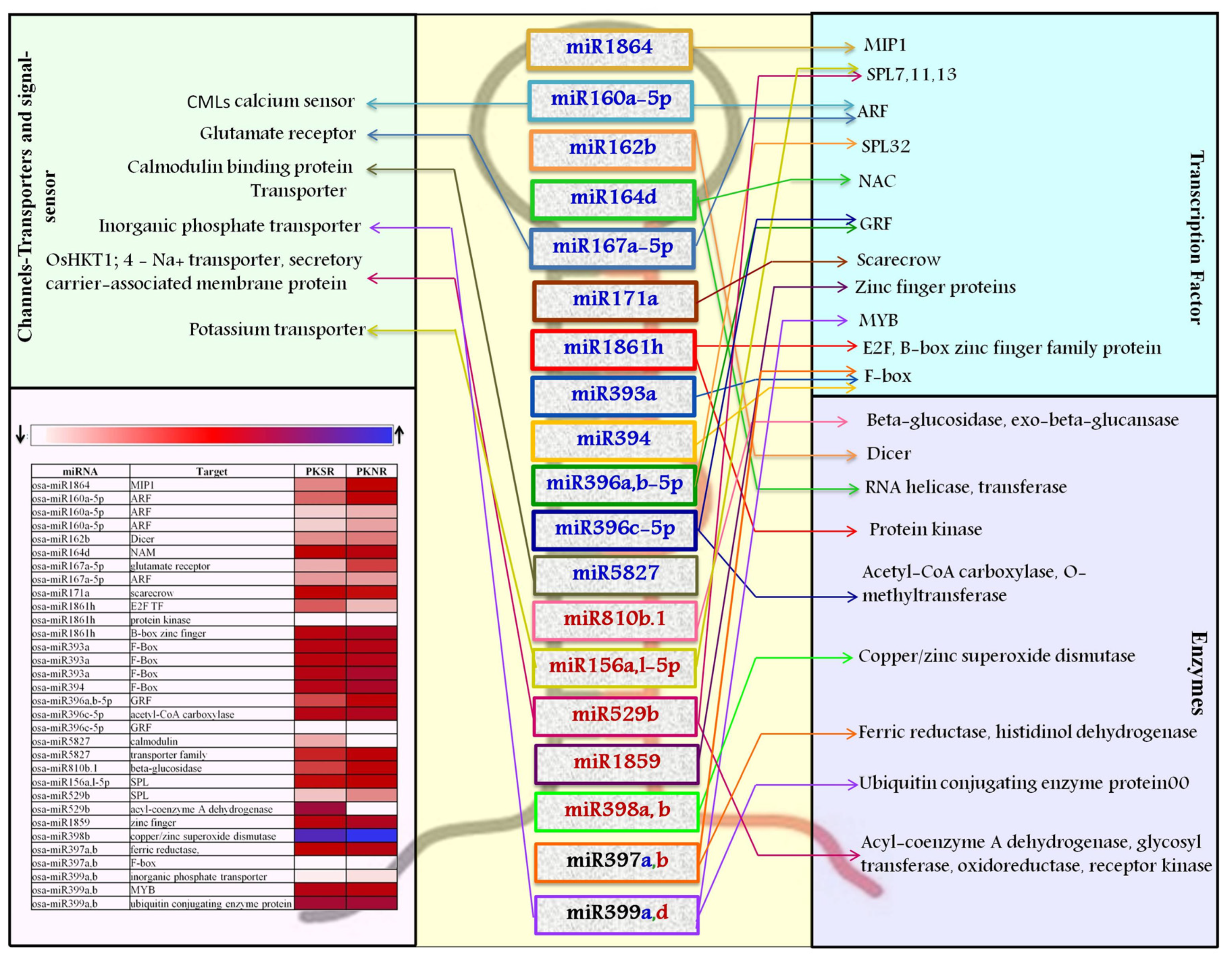
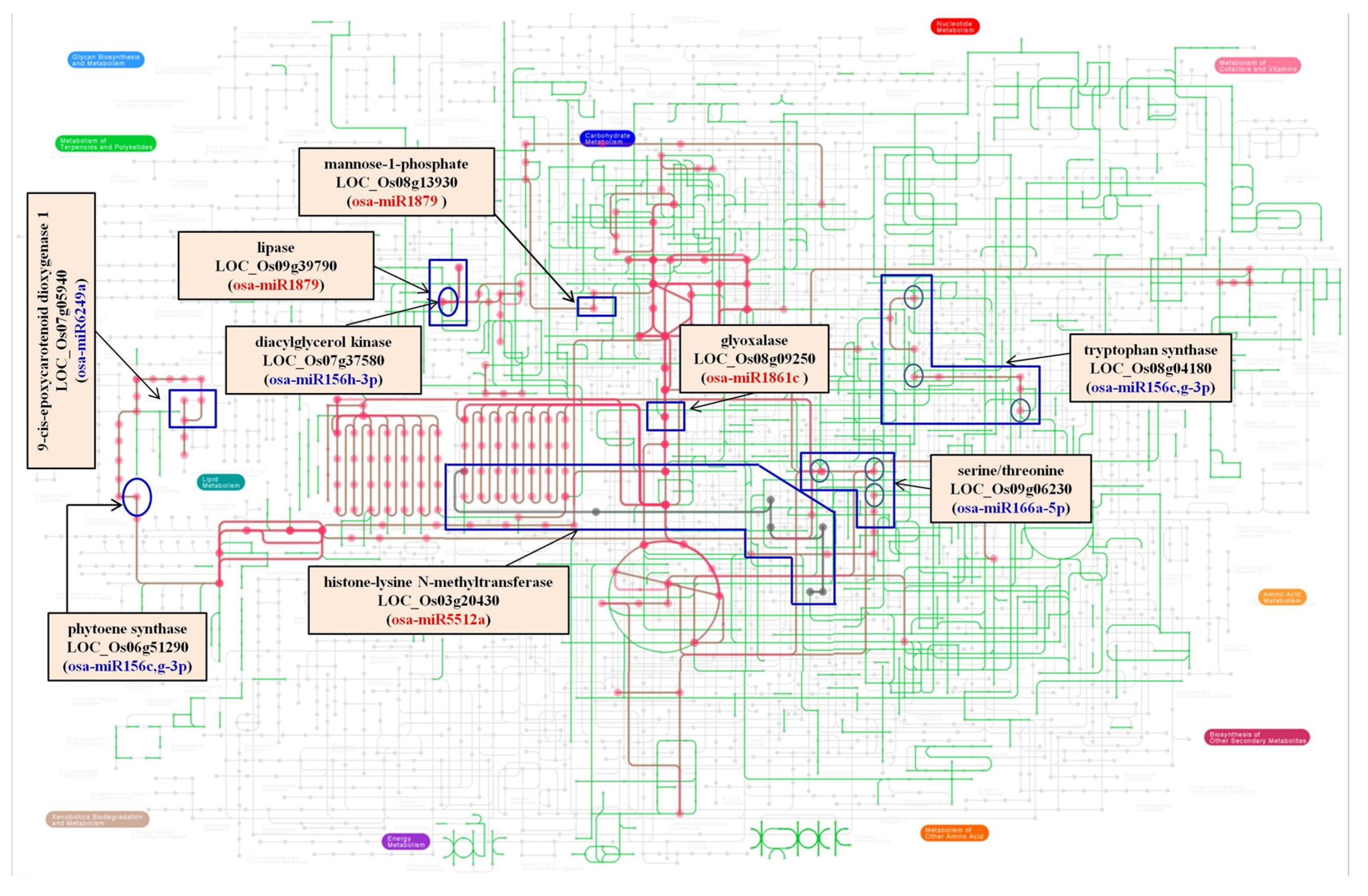
3.3. Target of miRs and Their Regulation in the Presence of Salt
3.4. Mapping of Integrating miR Expression Quantitative Trait Loci (miR-eQTL)
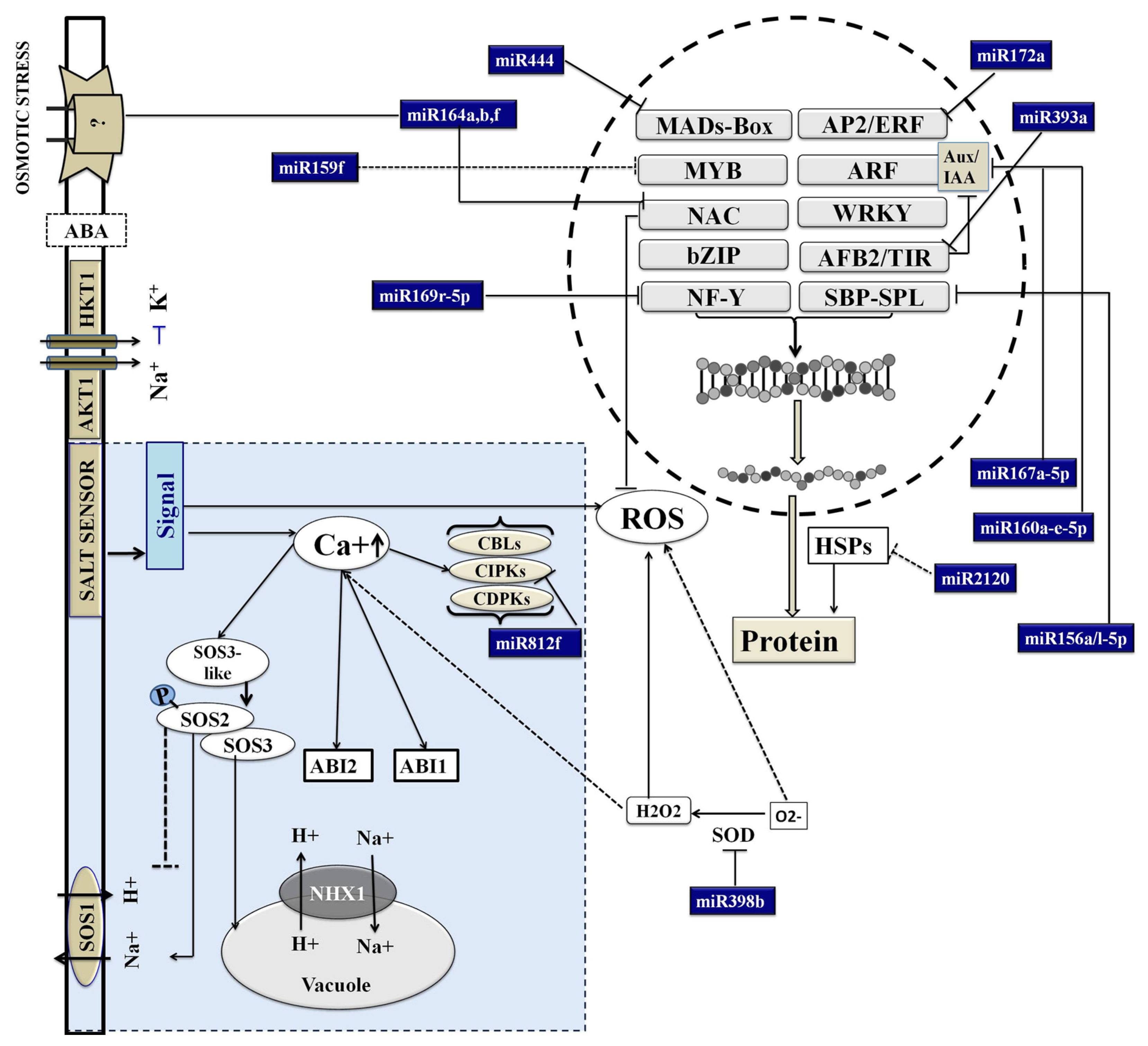
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genom. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Diédhiou, C.; Golldack, D. Salt-dependent regulation of chloride channel transcripts in rice. Plant Sci. 2006, 170, 793–800. [Google Scholar] [CrossRef]
- Kumar, R.; Kumar, S.; Sanan-Mishra, N. miRNAs: The game changer in producing salinity stress-tolerant crops. In Salinity Responses and Tolerance in Plants; Springer International Publishing: Basel, Switzerland, 2018; Volume 2, pp. 143–188. [Google Scholar]
- Joseph, E.A.; Mohanan, K. A study on the effect of salinity stress on the growth and yield of some native rice cultivars of Kerala state of India. Agric. For. Fish. 2013, 2, 141–150. [Google Scholar]
- Roychoudhury, A.; Chakraborty, M. Biochemical and molecular basis of varietal difference in plant salt tolerance. Annu. Res. Rev. Biol. 2013, 3, 422–454. [Google Scholar]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahmood, A.; Latif, T.; Khan, M.A. Effect of salinity on growth, yield and yield components in basmati rice germplasm. Pak. J. Bot. 2009, 41, 3035–3045. [Google Scholar]
- Hakim, M.; Juraimi, A.; Hanafi, M.; Ismail, M.; Rafii, M.; Islam, M.; Selamat, A. The effect of salinity on growth, ion accumulation and yield of rice varieties. J. Anim. Plant Sci. 2014, 24, 874–885. [Google Scholar]
- Babé, A.; Lavigne, T.; Séverin, J.-P.; Nagel, K.A.; Walter, A.; Chaumont, F.; Batoko, H.; Beeckman, T.; Draye, X. Repression of early lateral root initiation events by transient water deficit in barley and maize. Philos. Trans. R. Soc. Lond. Biol. Sci. 2012, 367, 1534–1541. [Google Scholar] [CrossRef] [Green Version]
- Singla-Pareek, S.L.; Yadav, S.K.; Pareek, A.; Reddy, M.K.; Sopory, S.K. Enhancing salt tolerance in a crop plant by overexpression of glyoxalase II. Transgenic Res. 2008, 17, 171–180. [Google Scholar] [CrossRef]
- Bertorello, A.M.; Zhu, J.-K. SIK1/SOS2 networks: Decoding sodium signals via calcium-responsive protein kinase pathways. Pflügers Arch. Eur. J. Physiol. 2009, 458, 613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.-K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant. Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Sakamoto, H.; Matsuda, O.; Iba, K. ITN1, a novel gene encoding an ankyrin-repeat protein that affects the ABA-mediated production of reactive oxygen species and is involved in salt-stress tolerance in Arabidopsis thaliana. Plant. J. 2008, 56, 411–422. [Google Scholar] [CrossRef] [PubMed]
- El-Shabrawi, H.; Kumar, B.; Kaul, T.; Reddy, M.K.; Singla-Pareek, S.L.; Sopory, S.K. Redox homeostasis, antioxidant defense, and methylglyoxal detoxification as markers for salt tolerance in Pokkali rice. Protoplasma 2010, 245, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Julkowska, M.M.; Hoefsloot, H.C.; Mol, S.; Feron, R.; de Boer, G.-J.; Haring, M.A.; Testerink, C. Capturing Arabidopsis root architecture dynamics with ROOT-FIT reveals diversity in responses to salinity. Plant. Physiol. 2014, 166, 1387–1402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, K.; Moe-Lange, J.; Hennet, L.; Feldman, L.J. Salt stress affects the redox status of Arabidopsis root meristems. Front. Plant. Sci. 2016, 7, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuenyong, W.; Chinpongpanich, A.; Comai, L.; Chadchawan, S.; Buaboocha, T. Downstream components of the calmodulin signaling pathway in the rice salt stress response revealed by transcriptome profiling and target identification. Bmc Plant. Biol. 2018, 18, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Dorlodot, S.; Forster, B.; Pagès, L.; Price, A.; Tuberosa, R.; Draye, X. Root system architecture: Opportunities and constraints for genetic improvement of crops. Trends Plant. Sci. 2007, 12, 474–481. [Google Scholar] [CrossRef]
- Kinoshita, N.; Wang, H.; Kasahara, H.; Liu, J.; MacPherson, C.; Machida, Y.; Kamiya, Y.; Hannah, M.A.; Chua, N.-H. IAA-Ala Resistant3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant. Cell 2012, 24, 3590–3602. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, L.; Bussell, J.D.; Păcurar, D.I.; Schwambach, J.; Păcurar, M.; Bellini, C. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant. Cell 2009, 21, 3119–3132. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.S.; Gudimella, R.; Wong, G.R.; Tammi, M.T.; Khalid, N.; Harikrishna, J.A. Transcripts and microRNAs responding to salt stress in Musa acuminata Colla (AAA Group) cv. Berangan roots. PLoS ONE 2015, 10, e0127526. [Google Scholar] [CrossRef]
- Couzigou, J.M.; Combier, J.P. Plant microRNAs: Key regulators of root architecture and biotic interactions. New Phytol. 2016, 212, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Ma, X.; Chen, D.; Wu, P.; Chen, M. MicroRNA-mediated signaling involved in plant root development. Biochem. Biophys. Res. Commun. 2010, 393, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Shao, C.; Wang, H.; Jin, Y.; Meng, Y. Construction of small RNA-mediated gene regulatory networks in the roots of rice (Oryza sativa). BMC Genom. 2013, 14, 510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marín-González, E.; Suárez-López, P. “And yet it moves”: Cell-to-cell and long-distance signaling by plant microRNAs. Plant. Sci. 2012, 196, 18–30. [Google Scholar] [CrossRef]
- Ding, D.; Zhang, L.; Wang, H.; Liu, Z.; Zhang, Z.; Zheng, Y. Differential expression of miRNAs in response to salt stress in maize roots. Ann. Bot. 2009, 103, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Mallory, A.C.; Bartel, D.P.; Bartel, B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant. Cell 2005, 17, 1360–1375. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-W.; Wang, L.-J.; Mao, Y.-B.; Cai, W.-J.; Xue, H.-W.; Chen, X.-Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant. Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef] [Green Version]
- Yoon, E.K.; Yang, J.H.; Lim, J.; Kim, S.H.; Kim, S.-K.; Lee, W.S. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res. 2009, 38, 1382–1391. [Google Scholar] [CrossRef] [Green Version]
- Meng, Y.; Chen, D.; Ma, X.; Mao, C.; Cao, J.; Wu, P.; Chen, M. Mechanisms of microRNA-mediated auxin signaling inferred from the rice mutant osaxr. Plant. Signal. Behav. 2010, 5, 252–254. [Google Scholar] [CrossRef] [Green Version]
- Mittal., D.; Mukherjee, S.K.; Vasudevan, M.; Sanan-Mishra, N. Identification of tissue-preferential expression patterns of rice miRNAs. J. Cell. Biochem. 2013, 114, 2071–2081. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010, 32, 11.17.11–11.17.14. [Google Scholar] [CrossRef] [PubMed]
- Goswami, K.; Tripathi, A.; Sanan-Mishra, N. Comparative miRomics of salt-tolerant and salt-sensitive rice. J. Integr. Bioinform. 2017, 14, 1613–4516. [Google Scholar] [CrossRef] [PubMed]
- Addo-Quaye, C.; Miller, W.; Axtell, M.J. CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 2008, 25, 130–131. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2. 0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Tripathi, A.; Goswami, K.; Tiwari, M.; Mukherjee, S.K.; Sanan-Mishra, N. Identification and comparative analysis of microRNAs from tomato varieties showing contrasting response to ToLCV infections. Physiol. Mol. Biol. Plants 2018, 24, 185–202. [Google Scholar] [CrossRef]
- Voorrips, R. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.-H.; Tian, X.; Li, Y.-J.; Wu, C.-A.; Zheng, C.-C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef] [Green Version]
- Sunkar, R.; Li, Y.-F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant. Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef]
- Sun, Z.; Wang, Y.; Mou, F.; Tian, Y.; Chen, L.; Zhang, S.; Jiang, Q.; Li, X. Genome-wide small RNA analysis of soybean reveals auxin-responsive microRNAs that are differentially expressed in response to salt stress in root apex. Front. Plant. Sci. 2016, 6, 1273. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Duan, H.; Li, J.; Deng, X.W.; Yin, W.; Xia, X. Global identification of miRNAs and targets in Populus euphratica under salt stress. Plant. Mol. Biol. 2013, 81, 525–539. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Kundu, A.; Pal, A. Identification and validation of conserved microRNAs along with their differential expression in roots of Vigna unguiculata grown under salt stress. Plant. Celltissue Organ. Cult. 2011, 105, 233–242. [Google Scholar] [CrossRef]
- Patade, V.Y.; Suprasanna, P. Short-term salt and PEG stresses regulate expression of MicroRNA, miR159 in sugarcane leaves. J. Crop. Sci. Biotechnol. 2010, 13, 177–182. [Google Scholar] [CrossRef]
- Yu, N.; Niu, Q.W.; Ng, K.H.; Chua, N.H. The role of miR156/SPL s modules in Arabidopsis lateral root development. Plant. J. 2015, 83, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Roy, S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant. Signal. Behav. 2016, 11, e1117723. [Google Scholar] [CrossRef] [Green Version]
- Khraiwesh, B.; Zhu, J.-K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Wang, H. The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol. Plant. 2015, 8, 677–688. [Google Scholar] [CrossRef] [Green Version]
- Gao, P.; Bai, X.; Yang, L.; Lv, D.; Pan, X.; Li, Y.; Cai, H.; Ji, W.; Chen, Q.; Zhu, Y. osa-MIR393: A salinity-and alkaline stress-related microRNA gene. Mol. Biol. Rep. 2011, 38, 237–242. [Google Scholar] [CrossRef]
- Sunkar, R. MicroRNAs with macro-effects on plant stress responses. In Proceedings of Seminars in Cell and Developmental Biology 2010, 21, 805–811. [Google Scholar] [CrossRef]
- Zhao, B.; Ge, L.; Liang, R.; Li, W.; Ruan, K.; Lin, H.; Jin, Y. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. Bmc Mol. Biol. 2009, 10, 29. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.-S.; Chen, M.; Li, L.-C.; Ma, Y.-Z. Functions of the ERF transcription factor family in plants. Botany 2008, 86, 969–977. [Google Scholar] [CrossRef]
- Fang, Y.; You, J.; Xie, K.; Xie, W.; Xiong, L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genom. 2008, 280, 547–563. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Ren, W.; Zhi, D.; Wang, L.; Xia, G. Arabidopsis DREB1A/CBF3 bestowed transgenic tall fescue increased tolerance to drought stress. Plant. Cell Rep. 2007, 26, 1521–1528. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Chacon, O.; Singla-Pareek, S.L.; Sopory, S.K.; Sanan-Mishra, N. Mapping the microRNA expression profiles in glyoxalase over-expressing salinity tolerant rice. Curr. Genom. 2018, 19, 21–35. [Google Scholar]
- Bhalerao, R.P.; Eklöf, J.; Ljung, K.; Marchant, A.; Bennett, M.; Sandberg, G. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant. J. 2002, 29, 325–332. [Google Scholar] [CrossRef]
- Zolla, G.; Heimer, Y.M.; Barak, S. Mild salinity stimulates a stress-induced morphogenic response in Arabidopsis thaliana roots. J. Exp. Bot. 2009, 61, 211–224. [Google Scholar] [CrossRef] [Green Version]
- Hernandez, Y.; Sanan-Mishra, N. miRNA mediated regulation of NAC Transcription factors in plant development. Plant. Gene 2017, 11B, 190–198. [Google Scholar] [CrossRef]
- Chen, Z.; Hu, L.; Han, N.; Hu, J.; Yang, Y.; Xiang, T.; Zhang, X.; Wang, L. Overexpression of a miR393-resistant form of transport inhibitor response protein 1 (mTIR1) enhances salt tolerance by increased osmoregulation and Na+ exclusion in Arabidopsis thaliana. Plant. Cell Physiol. 2014, 56, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Knauer, S.; Holt, A.L.; Rubio-Somoza, I.; Tucker, E.J.; Hinze, A.; Pisch, M.; Javelle, M.; Timmermans, M.C.; Tucker, M.R.; Laux, T. A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell 2013, 24, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Song, J.B.; Gao, S.; Sun, D.; Li, H.; Shu, X.X.; Yang, Z.M. miR394 and LCR are involved in Arabidopsis salt and drought stress responses in an abscisic acid-dependent manner. BMC Plant. Biol. 2013, 13, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Leng, X.; Sun, X.; Mu, Q.; Wang, B.; Li, X.; Wang, C.; Fang, J. Discovery of conservation and diversification of miR171 genes by phylogenetic analysis based on global genomes. Plant. Genome 2015, 8. [Google Scholar] [CrossRef] [Green Version]
- Henriksson, E.; Olsson, A.S.; Johannesson, H.; Johansson, H.; Hanson, J.; Engström, P.; Söderman, E. Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant. Physiol. 2005, 139, 509–518. [Google Scholar]
- Canales, J.; Contreras-López, O.; Álvarez, J.M.; Gutiérrez, R.A. Nitrate induction of root hair density is mediated by TGA 1/TGA 4 and CPC transcription factors in Arabidopsis thaliana. Plant. J. 2017, 92, 305–316. [Google Scholar] [CrossRef] [Green Version]
- Kurihara, Y.; Watanabe, Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 2004, 101, 12753–12758. [Google Scholar] [CrossRef] [Green Version]
- Kaur, C.; Singla-Pareek, S.L.; Sopory, S.K. Glyoxalase and methylglyoxal as biomarkers for plant stress tolerance. Crit. Rev. Plant. Sci. 2014, 33, 429–456. [Google Scholar] [CrossRef]
- Virdi, A.S.; Singh, S.; Singh, P. Abiotic stress responses in plants: Roles of calmodulin-regulated proteins. Front. Plant. Sci. 2015, 6, 809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arshad, M.; Feyissa, B.A.; Amyot, L.; Aung, B.; Hannoufa, A. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant. Sci. 2017, 258, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Yue, E.; Liu, Z.; Li, C.; Li, Y.; Liu, Q.; Xu, J.-H. Overexpression Mir529a Confers Enhanced Resistance Oxidative Stress Rice (Oryza Sativa L.). Plant. Cell Rep. 2017, 36, 1171–1182. [Google Scholar]
- Prashanth, S.; Sadhasivam, V.; Parida, A. Over expression of cytosolic copper/zinc superoxide dismutase from a mangrove plant Avicennia marina in indica rice var Pusa Basmati-1 confers abiotic stress tolerance. Transgenic Res. 2008, 17, 281–291. [Google Scholar] [CrossRef]
- Wang, X.; Hou, C.; Liu, J.; He, W.; Nan, W.; Gong, H.; Bi, Y. Hydrogen peroxide is involved in the regulation of rice (Oryza sativa L.) tolerance to salt stress. Acta Physiol. Plant. 2013, 35, 891–900. [Google Scholar] [CrossRef]
- Rodriguez, R.E.; Ercoli, M.F.; Debernardi, J.M.; Breakfield, N.W.; Mecchia, M.A.; Sabatini, M.; Cools, T.; De Veylder, L.; Benfey, P.N.; Palatnik, J.F. MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. Plant. Cell 2015, 27, 3354–3366. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Shimada, T.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. KATAMARI1/MURUS3 is a novel Golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant. Cell 2005, 17, 1764–1776. [Google Scholar] [CrossRef] [Green Version]
- Bazin, J.; Khan, G.A.; Combier, J.P.; Bustos-Sanmamed, P.; Debernardi, J.M.; Rodriguez, R.; Sorin, C.; Palatnik, J.; Hartmann, C.; Crespi, M. miR396 affects mycorrhization and root meristem activity in the legume M edicago truncatula. Plant. J. 2013, 74, 920–934. [Google Scholar] [CrossRef] [PubMed]
- Kant, S.; Peng, M.; Rothstein, S.J. Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. Plo Genet. 2011, 7, e1002021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawa, D.; Julkowska, M.M.; Sommerfeld, H.M.; ter Horst, A.; Haring, M.A.; Testerink, C. Phosphate-dependent root system architecture responses to salt stress. Plant. Physiol. 2016, 172, 690–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, M.; Takahashi, H.; Tamura, K.; Huang, J.; Yu, L.-H.; Kawai-Yamada, M.; Tezuka, T.; Uchimiya, H. Enhanced dihydroflavonol-4-reductase activity and NAD homeostasis leading to cell death tolerance in transgenic rice. Proc. Natl. Acad. Sci. USA 2005, 102, 7020–7025. [Google Scholar] [CrossRef] [Green Version]
- Akram, N.A.; Shafiq, F.; Ashraf, M. Ascorbic acid-a potential oxidant scavenger and its role in plant development and abiotic stress tolerance. Front. Plant. Sci. 2017, 8, 613. [Google Scholar] [CrossRef]
- Zsigmond, L.; Szepesi, Á.; Tari, I.; Rigó, G.; Király, A.; Szabados, L. Overexpression of the mitochondrial PPR40 gene improves salt tolerance in Arabidopsis. Plant. Sci. 2012, 182, 87–93. [Google Scholar] [CrossRef]
- Li, Y.-F.; Zheng, Y.; Vemireddy, L.R.; Panda, S.K.; Jose, S.; Ranjan, A.; Panda, P.; Govindan, G.; Cui, J.; Wei, K.; et al. Comparative transcriptome and translatome analysis in contrasting rice genotypes reveals differential mRNA translation in salt-tolerant Pokkali under salt stress. BMC Genomics 2018, 19, 935. [Google Scholar] [CrossRef] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goswami, K.; Mittal, D.; Gautam, B.; Sopory, S.K.; Sanan-Mishra, N. Mapping the Salt Stress-Induced Changes in the Root miRNome in Pokkali Rice. Biomolecules 2020, 10, 498. https://doi.org/10.3390/biom10040498
Goswami K, Mittal D, Gautam B, Sopory SK, Sanan-Mishra N. Mapping the Salt Stress-Induced Changes in the Root miRNome in Pokkali Rice. Biomolecules. 2020; 10(4):498. https://doi.org/10.3390/biom10040498
Chicago/Turabian StyleGoswami, Kavita, Deepti Mittal, Budhayash Gautam, Sudhir K. Sopory, and Neeti Sanan-Mishra. 2020. "Mapping the Salt Stress-Induced Changes in the Root miRNome in Pokkali Rice" Biomolecules 10, no. 4: 498. https://doi.org/10.3390/biom10040498




