Thrombolytic Potential of Novel Thiol-Dependent Fibrinolytic Protease from Bacillus cereus RSA1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Screening of Bacterial Strain Producing Fibrinolytic Protease
2.2. Fibrinolytic Activity of the Screened Strains
2.3. Identification of the Microbial Strain
2.4. Phylogenetic Analysis and Strain Identification
2.5. Fibrinolytic Protease Production
2.6. Activity Assay of the Fibrinolytic Protease Produced
2.7. Optimization of Media
2.8. Elucidation of Significant Variables
2.9. Central Composition Design
2.10. Purification of Fibrinolytic Protease
2.11. Diagnosis of Amino Acid Composition of Fibrinolytic Protease Using MALDI Mass Spectrometry
2.12. Characterization of the Purified Fibrinolytic Protease
2.12.1. Optimization of Temperature and pH for Maximum Fibrinolytic Protease Activity
2.12.2. Stability of Fibrinolytic Protease at Different Temperature and pH Range
2.12.3. Kinetic Studies
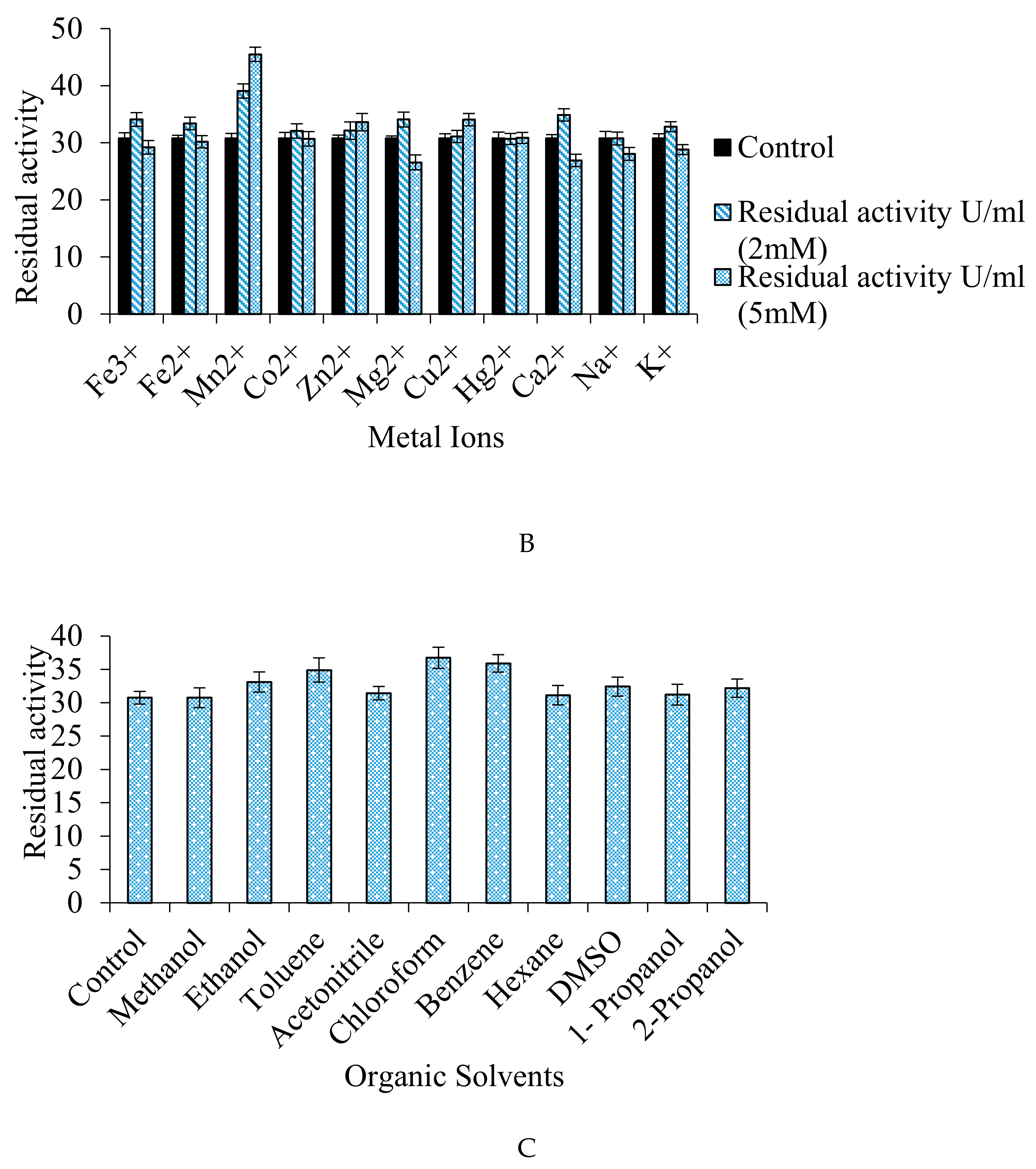
2.12.4. Effect of Different Metal Ions, Inhibitors/Activators, and Organic Solvents on the Fibrinolytic Protease Activity
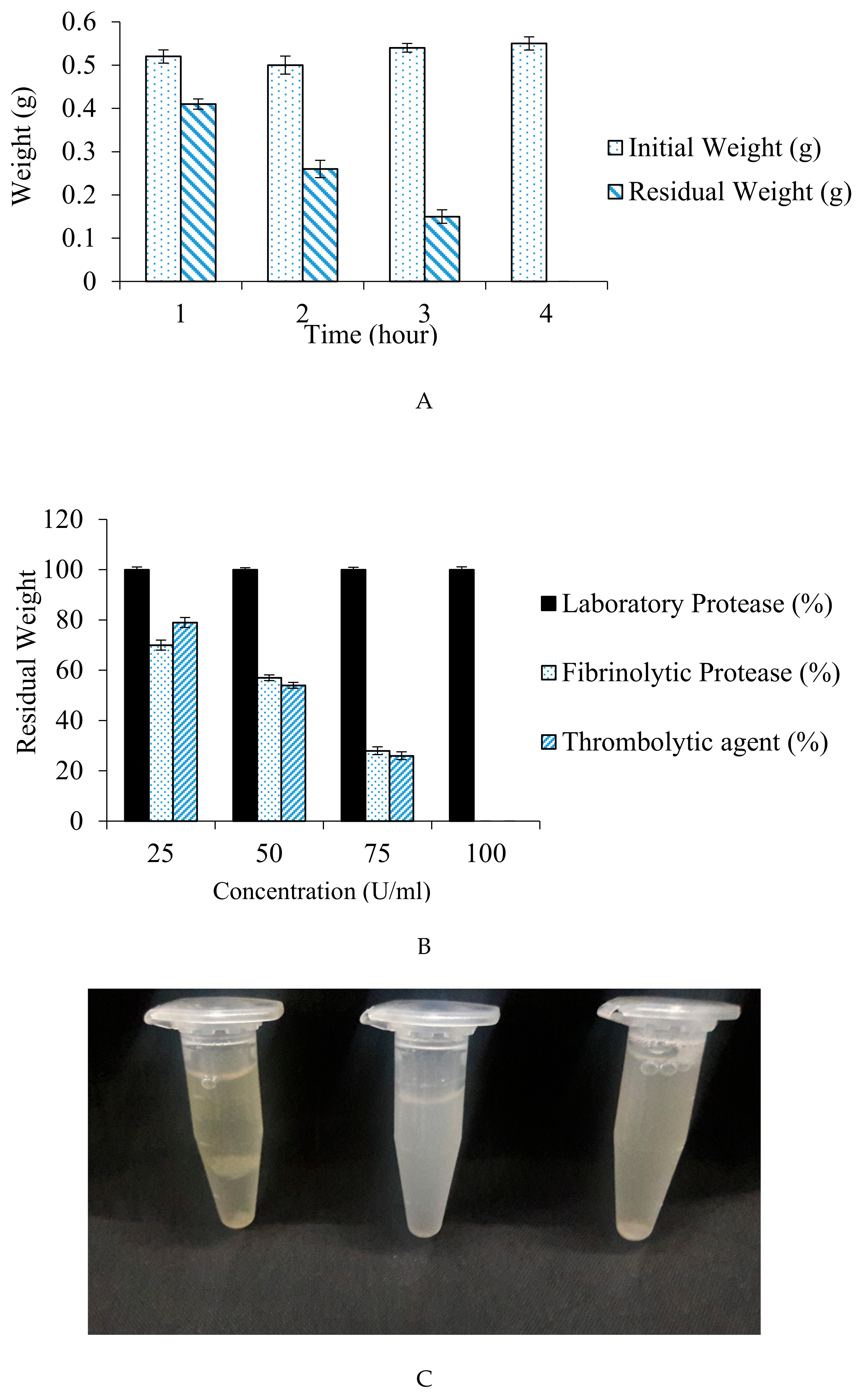
2.13. In Vitro Thrombolytic Activity of the Fibrinolytic Protease Using Blood Clot as Substrate
2.13.1. Blood Clot Preparation
2.13.2. Clot Lysis
3. Results and Discussion
3.1. Isolation and Screening of Bacterial Strain Producing Fibrinolytic Protease
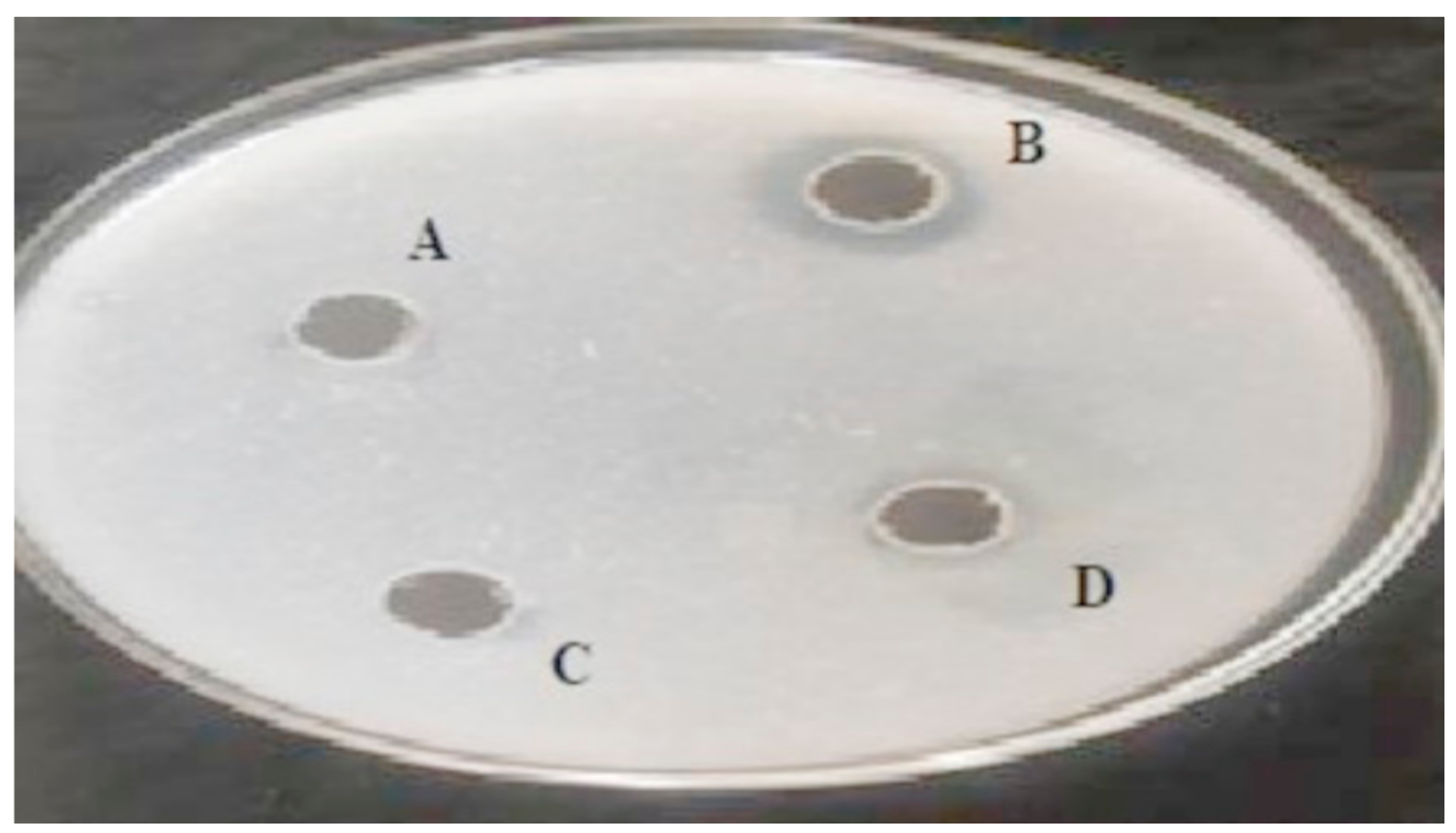
3.2. Fibrinolytic Activity of the Screened Bacterial Strains Using Fibrin Plate
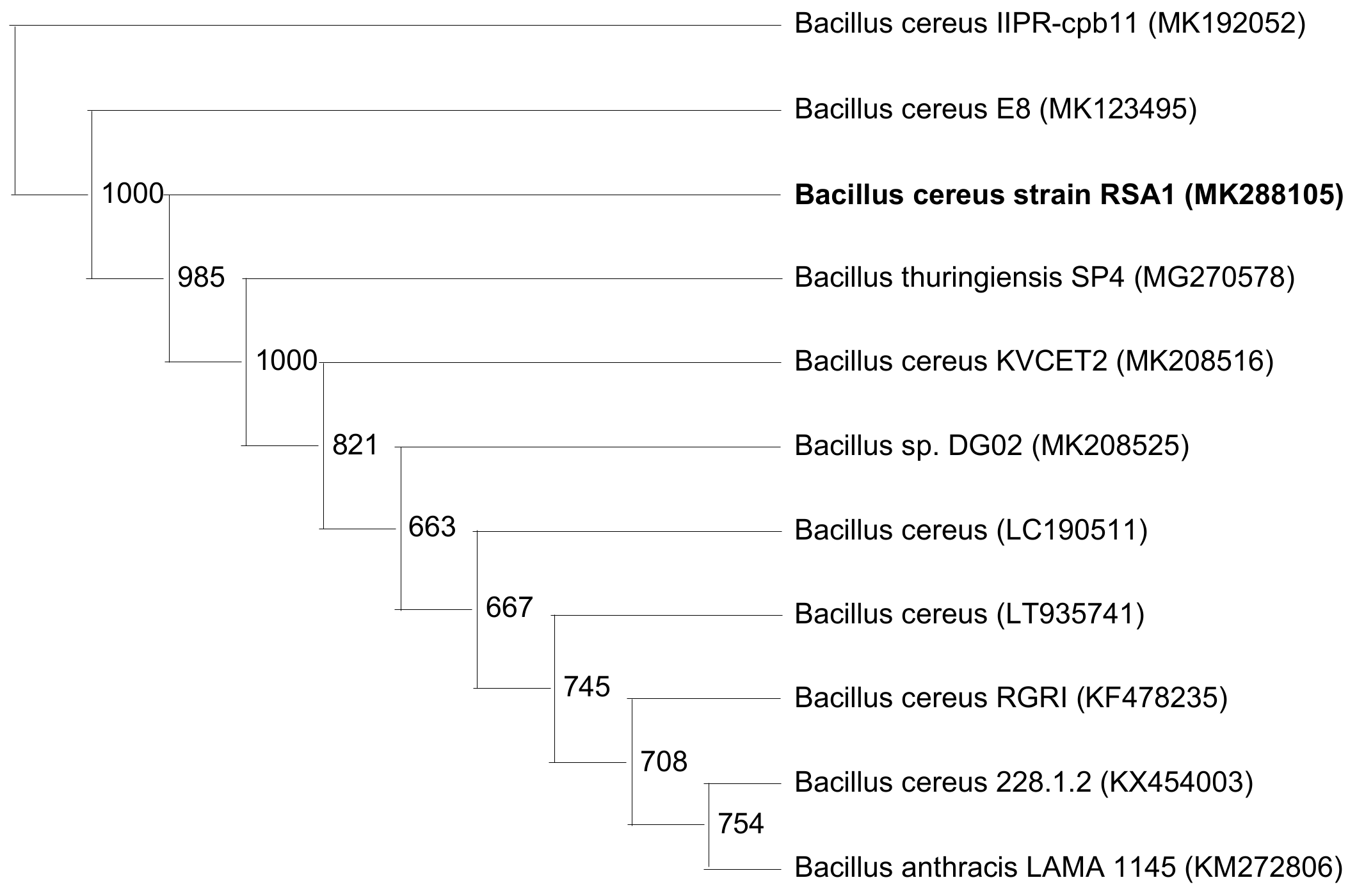
3.3. Identification and Phylogenetic Analysis of the Strain
3.4. Fibrinolytic Activity of Protease and Statistical Optimization of Media
3.4.1. Elucidation of Significant Variables
3.4.2. Optimization of Variables by Response Surface Methodology
3.4.3. Validation of the Design Model
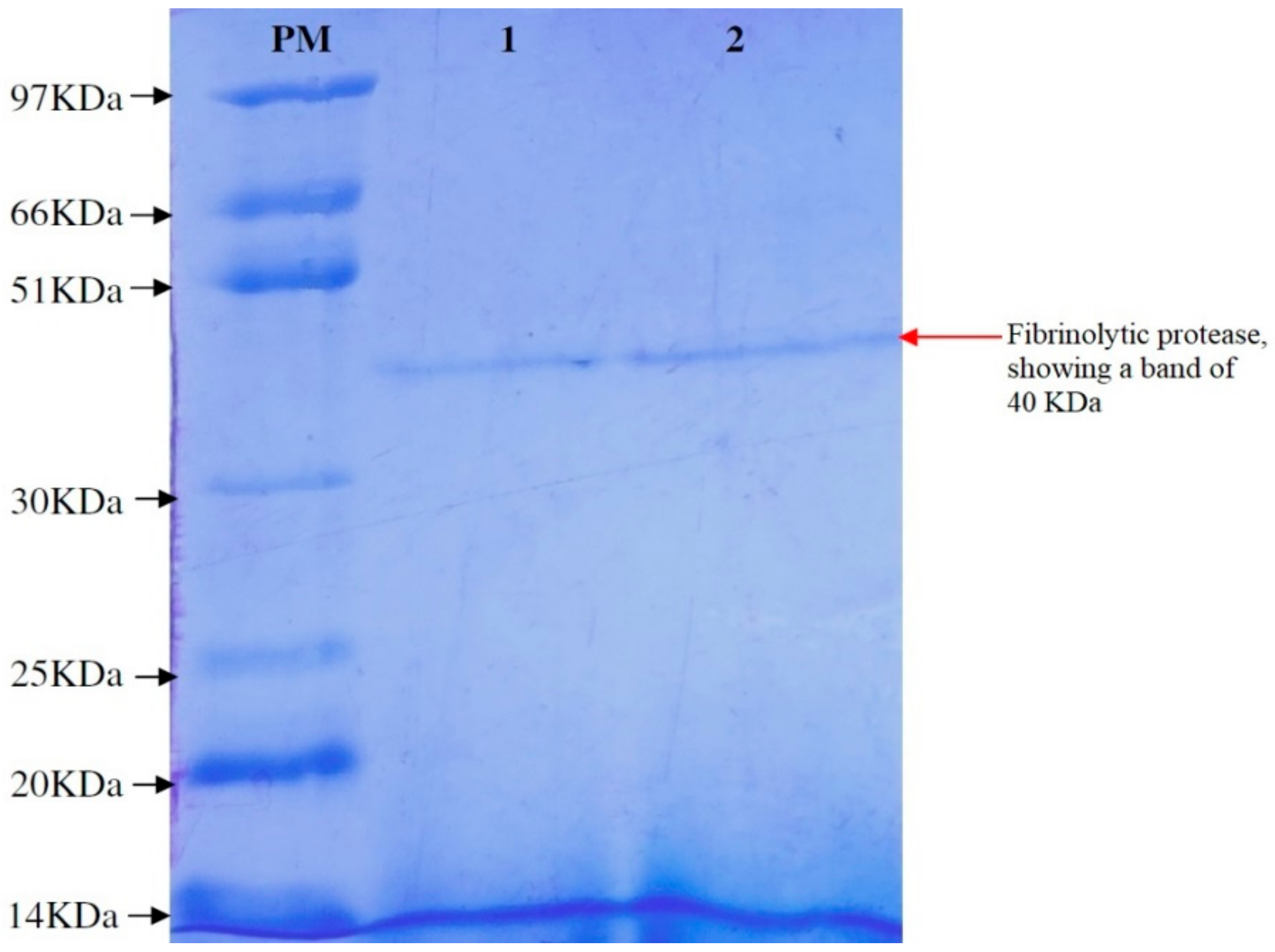
3.5. Purification of Fibrinolytic Protease
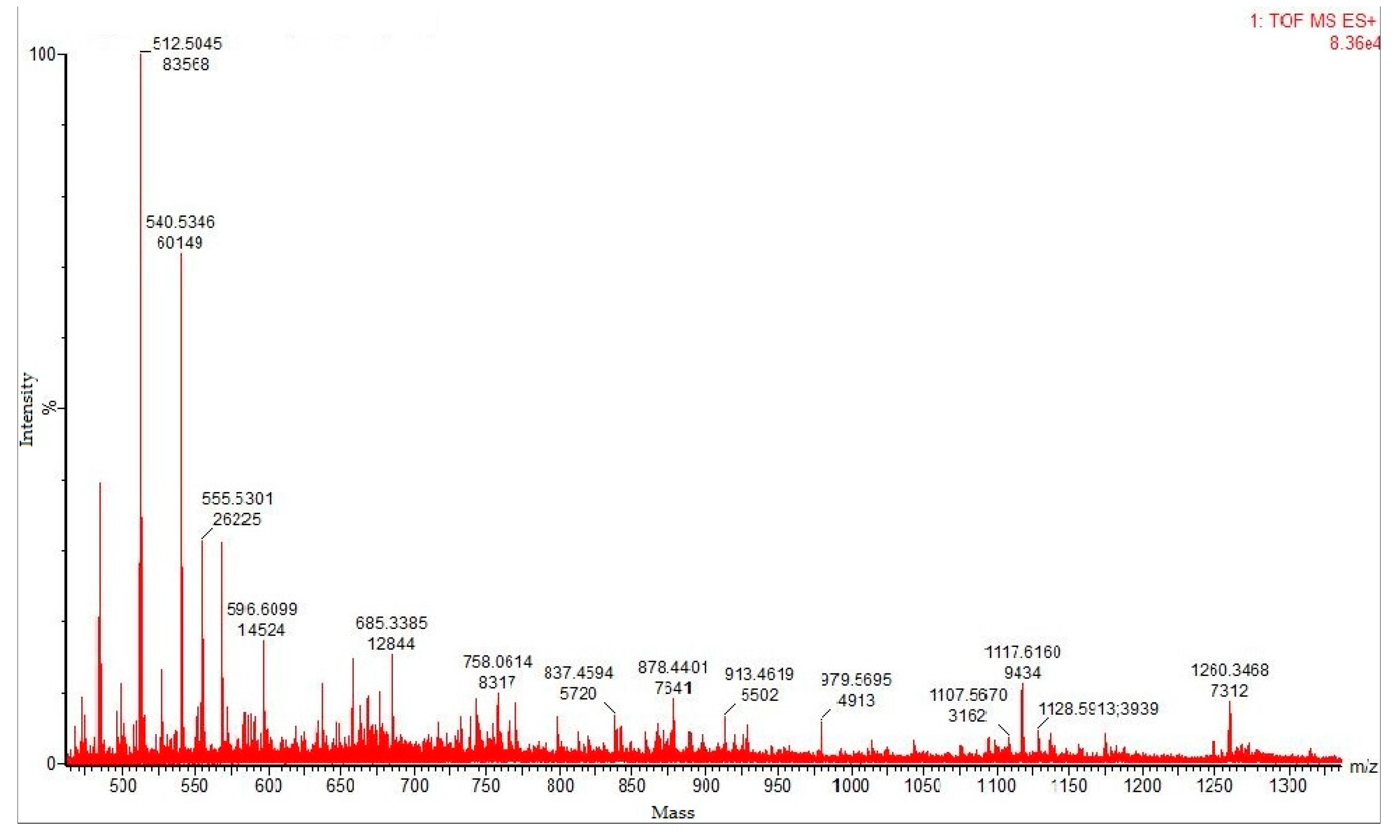
3.6. MALDI–TOF MS Analysis
3.7. Characterization of Purified Fibrinolytic Protease
3.7.1. Optimization of Temperature and pH
3.7.2. Stability of Fibrinolytic Protease at Different Temperatures and pH Ranges
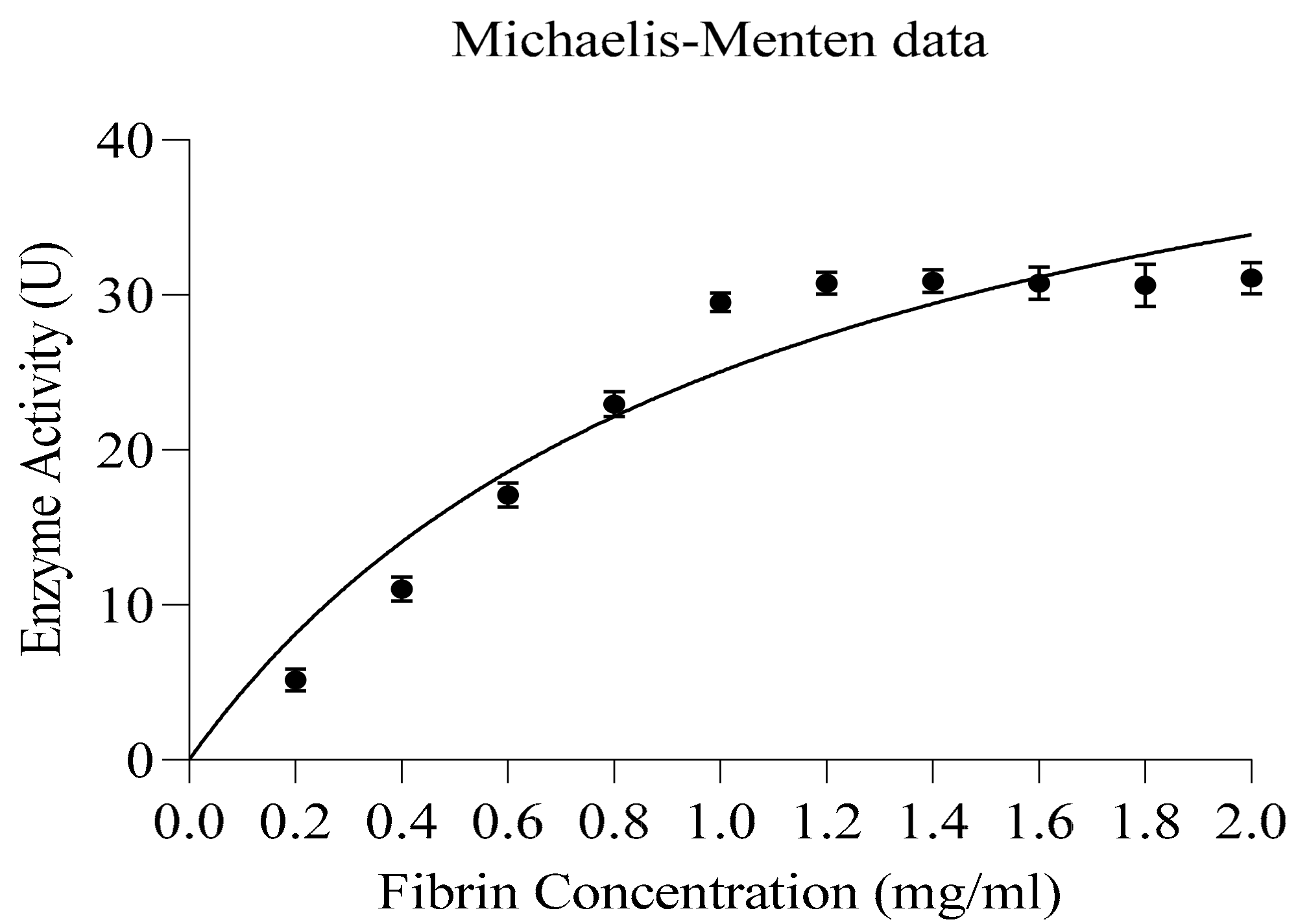
3.7.3. Kinetic Studies
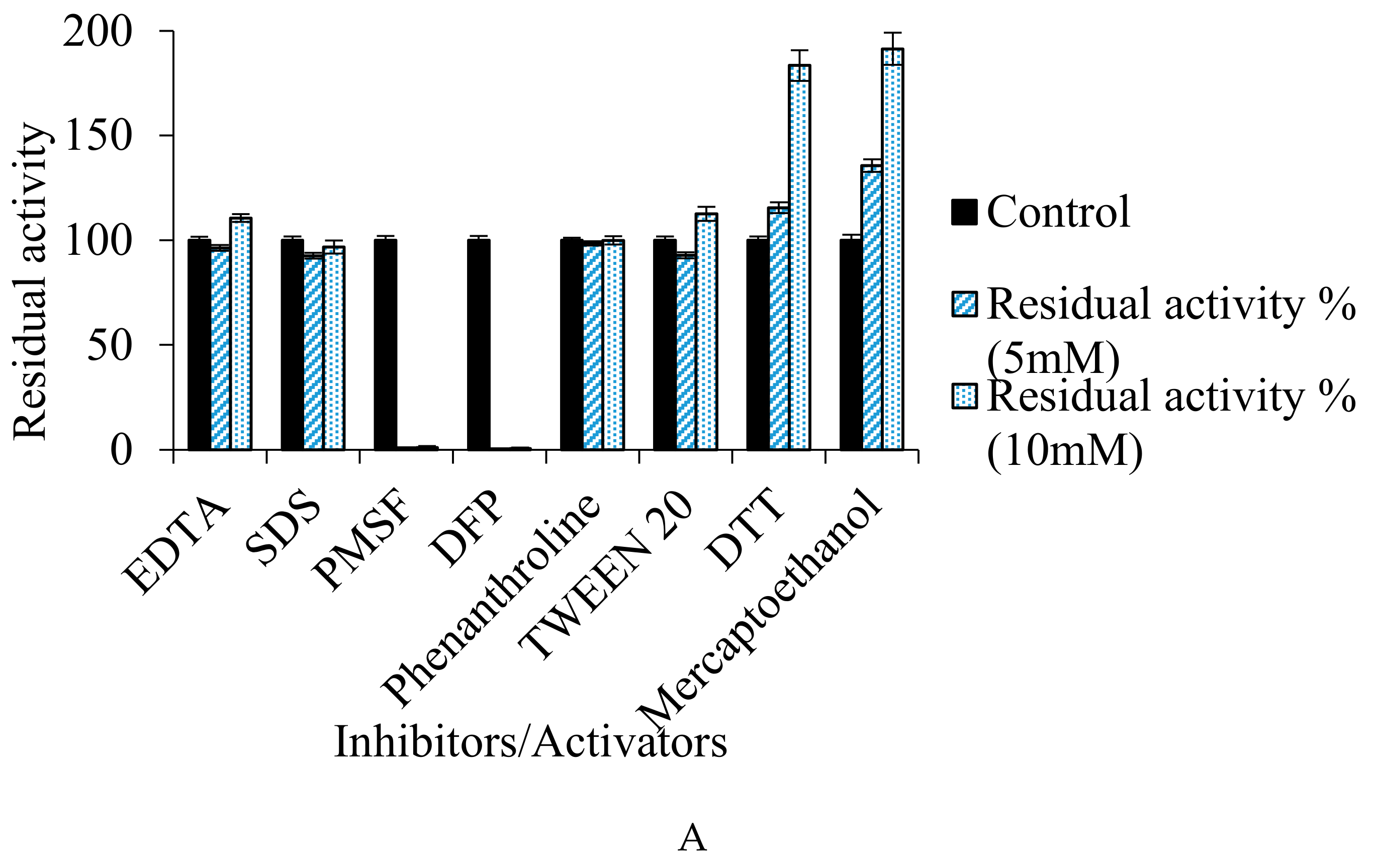
3.7.4. Effect of Inhibitors/Activators, Metal Ions, and Organic Solvents on Enzyme Activity
3.8. Fibrinolysis Assay for Blood Clot Dissolution
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Heart Federation. The World’s Most Common Cause of Death, Cardiovascular Diseases (CVDs), Global Facts and Figures. Available online: https://www.world-heart-federation.org/wp-content/uploads/2017/05/WCC2016_CVDs_infographic.pdf (accessed on 18 December 2019).
- Benjamin, E.J.; Virani, S.S.; Callaway, C.W.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Chiuve, S.E.; Cushman, M.; Delling, F.N.; Deo, R.; et al. Heart disease and stroke statistics-2018 update: A report from the American Heart Association. Circulation 2018, 137, 67. [Google Scholar] [CrossRef] [PubMed]
- Riddel, J.P., Jr.; Aouizerat, B.E.; Miaskowski, C.; Lillicrap, D.P. Theories of blood coagulation. J. Pediatr. Oncol. Nurs. 2007, 24, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.K.; Kim, J.H.; Gal, S.W.; Kim, J.E.; Park, S.S.; Chung, K.T.; Kim, Y.H.; Kim, B.W.; Joo, W.H. Molecular cloning and characterization of the gene encoding a fibrinolytic enzyme from Bacillus subtilis strain A1. World J. Microbiol. Biotechnol. 2004, 20, 711–717. [Google Scholar] [CrossRef]
- Mukherjee, A.K.; Rai, S.K.; Thakur, R.; Chattopadhyay, P.; Kar, S.K. Bafibrinase: A non-toxic, non-hemorrhagic, direct-acting fibrinolytic serine protease from Bacillus sp. strain AS-S20-I exhibits in vivo anticoagulant activity and thrombolytic potency. Biochimie 2012, 94, 1300–1308. [Google Scholar] [CrossRef]
- Mahajan, P.M.; Nayak, S.; Lele, S.S. Fibrinolytic enzyme from newly isolated marine Bacillus subtilis ICTF-1: Media optimization, purification and characterization. J. Biosci. Bioeng. 2012, 113, 307–314. [Google Scholar] [CrossRef]
- Gurewich, V. Thrombolysis: A critical first-line therapy with an unfulfilled potential. Am. J. Med. 2016, 129, 573–575. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.H.; Sapkota, K.; Park, S.E.; Kim, S.; Kim, S.J. Thrombolytic, anticoagulant and antiplatelet activities of codiase, a bifunctional fibrinolytic enzyme from Codium fragile. Biochimie 2013, 95, 1266–1277. [Google Scholar] [CrossRef]
- Kang, S.R.; Choi, J.H.; Kim, D.W.; Park, S.E.; Sapkota, K.; Kim, S.; Kim, S.J. A bifunctional protease from green alga Ulva pertusa with anticoagulant properties: Partial purification and characterization. J. Appl. Phycol. 2016, 28, 599–607. [Google Scholar] [CrossRef]
- Fenton, J.W.; Ofosu, F.A.; Brezniak, D.V.; Hassouna, H.I. Understanding thrombin and hemostasis. Hematol. Oncol. Clin. N. Am. 1993, 7, 1107–1119. [Google Scholar] [CrossRef]
- Turner, N.A.; Moake, J. Assembly and activation of alternative complement components on endothelial cell-anchored ultra-large von Willebrand factor links complement and hemostasis-thrombosis. PLoS ONE 2013, 8, 59372. [Google Scholar] [CrossRef] [Green Version]
- Monroe, D.M.; Hofman, M.; Roberts, H.R. Platelets and thrombin generation. Arterioscler. Thromb. Vasc. Biol. 2002, 22, 1381–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabriela, C.M.; Hajjar, K.A. Molecular mechanisms of fibrinolysis. Br. J. Haematol. 2005, 129, 307–321. [Google Scholar]
- Liu, X.L.; Zheng, X.Q.; Qian, P.Z.; Kopparapu, N.K.; Deng, Y.P.; Nonaka, M.; Harada, N. Purification and characterization of a novel fibrinolytic enzyme from culture supernatant of Pleurotus ostreatus. J. Microbiol. Biotechnol. 2014, 24, 245–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajaj, B.K.; Singh, S.; Khullar, M.; Singh, K.; Bhardwaj, S. Optimization of fibrinolytic protease production from Bacillus subtilis I-2 using agro-residues. Arch. Biol. Technol. 2014, 57, 653–662. [Google Scholar] [CrossRef] [Green Version]
- Simkhada, J.R.; Mander, P.; Cho, S.S.; Yoo, J.C. A novel fibrinolytic protease from Streptomyces sp. CS684. Process. Biochem. 2010, 45, 88–93. [Google Scholar] [CrossRef]
- Kotb, E. The biotechnological potential of fibrinolytic enzymes in the dissolution of endogenous blood thrombi. Biotechnol. Progr. 2014, 30, 656–672. [Google Scholar] [CrossRef]
- Nazari, J.; Davison, R.; Kaplan, K.; Fintel, D. Adverse reactions to thrombolytic agents implications for coronary reperfusion following myocardial infarction. Med. Toxicol. Adverse. Drug Exp. 1987, 2, 274–286. [Google Scholar]
- Bonnard, T.; Law, L.S.; Tennant, Z.; Hagemeyer, C.E. Development and validation of a high throughput whole blood thrombolysis plate assay. Sci. Rep. 2017, 7, 2346. [Google Scholar] [CrossRef]
- Bhargavi, P.L.; Prakasham, R.S. A fibrinolytic, alkaline and thermostable metalloprotease from the newly isolated Serratia sp. RSPB11. Int. J. Biol. Macromol. 2013, 61, 479–486. [Google Scholar] [CrossRef]
- Lal, V. Fibrinolytic drug therapy in the management of intravascular thrombosis, especially acute myocardial infarction—A review. Asian J. Pharm. Clin. Res. 2017, 2, 555593. [Google Scholar]
- Bayoudh, A.; Gharsallah, N.; Chamkha, M.; Dhouib, A.; Ammar, S.; Nasri, M. Purification and characterization of an alkaline protease from Pseudomonas aeruginosa MN1. J. Ind. Microbiol. Biotechnol. 2000, 24, 291–295. [Google Scholar] [CrossRef]
- Patel, R.; Dodia, M.; Singh, S.P. Extracellular alkaline protease from a newly isolated haloalkaliphilic Bacillus sp. production and optimization. Process. Biochem. 2005, 40, 3569–3575. [Google Scholar] [CrossRef]
- Demina, N.S.; Lysenko, S.V. Micro-organisms synthesizing enzymes with thrombolytic action. Nauchnye. Doki. Vyss. Shkoly Biol. Nauki. 1991, 9, 136–153. [Google Scholar]
- Kotb, E. Activity assessment of microbial fibrinolytic enzymes. Appl. Microbiol. Biotechnol. 2013, 97, 6647–6665. [Google Scholar] [CrossRef]
- Kotb, E. Fibrinolytic bacterial enzymes with thrombolytic activity. In Fibrinolytic Bacterial Enzymes with Thrombolytic Activity; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–74. [Google Scholar]
- Fayek, K.I.; EI-Sayed, A.T. Some properties of two purified fibrinolytic enzymes from Bacillus subtilis and B. polymyxa. J. Basic Microbiol. 1980, 20, 383–387. [Google Scholar] [CrossRef]
- Malke, H.; Ferretti, J.J. Streptokinase cloning expression and excretion by Escherichia coli. Proc. Natl. Acad. Sci. USA 1984, 81, 3557–3561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Chen, H.; Liang, T.; Lin, Y. A novel nattokinase produced by Pseudomonas sp. TKU015 using shrimp shells as substrate. Process. Biochem. 2009, 44, 70–76. [Google Scholar] [CrossRef]
- Vijayaraghavan, P.; Vincent, P.; Gnana, S. Statistical optimization of fibrinolytic enzyme production using agroresidues by Bacillus cereus IND1 and its thrombolytic activity in vitro. BioMed Res. Int. 2014, 2014, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uesugi, Y.; Usuki, H.; Iwabuchi, M.; Hatanaka, T. Highly potent fibrinolytic serine protease from Streptomyces. Enzyme Microb. Technol. 2011, 48, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Vijayaraghavan, P.; Vincent, S.G.P.; Arasu, M.V. Purification, characterization of a novel fibrinolytic enzyme from Paenibacillus sp. IND8, and its in vitro thrombolytic activity. South J. Biol. Sci. 2016, 2, 434–444. [Google Scholar] [CrossRef]
- Taneja, K.; Bajaj, B.K.; Kumar, S.; Dilbaghi, N. Production, purification and characterization of fibrinolytic enzyme from Serratia sp. KG-2-1 using optimized media. 3 Biotech. 2017, 7, 184. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Chatterjee, S.; Keziah, M.S.; Devi, S.C. Fibrinolytic Protease from Marine Streptomyces rubiginosus VITPSS1. Cardiovasc. Hematol. Agents Med. Chem. 2018, 16, 44–45. [Google Scholar] [CrossRef] [PubMed]
- EI-Aassar, S.A.; EI-Badry, H.M.; Abdel-Fattah, A.F. The biosynthesis of proteases with fibrinolytic activity in immobilized cultures of Penicillium chrysogenum H9. Appl. Microbiol. Biotechnol. 1990, 33, 26–30. [Google Scholar]
- Wu, B.; Wu, L.; Chen, D.; Yang, Z.; Luo, M. Purification and characterization of a novel fibrinolytic protease from Fusarium sp. CPCC 480097. J. Ind. Microbiol. Biotechnol. 2009, 36, 451–459. [Google Scholar] [CrossRef]
- Ueda, M.; Kubo, T.; Miyatake, K.; Nakamura, T. Purification and characterization of fibrinolytic alkaline protease from Fusarium sp. BLB. Appl. Microbiol. Biotechnol. 2007, 74, 331–338. [Google Scholar] [CrossRef]
- Batomunkueva, B.P.; Egorov, N.S. Isolation, purification and resolution of the extracellular proteinase complex of Aspergillus ochraceus 513 with fibrinolytic and anticoagulant activities. Microbiology 2001, 70, 519–522. [Google Scholar] [CrossRef]
- Shirasaka, N.; Naitou, M.; Okamura, K.; Fukuta, Y.; Terashita, T.; Kusuda, M. Purification and characterization of a fibrinolytic protease from Aspergillus oryzae KSK-3. Mycoscience 2012, 53, 354–364. [Google Scholar] [CrossRef]
- Xiao-Lan, L.; Lian-xiang, D.; Fu-ping, L.; Xi-qun, Z.; Jing, X. Purification and characterization of a novel fibrinolytic enzyme from Rhizopus chinensis 12. Appl. Microbiol. Biotechnol. 2005, 67, 209–214. [Google Scholar] [CrossRef]
- Liu, X.; Kopparapu, N.K.; Li, Y.; Deng, Y.; Zheng, X. Biochemical characterization of a novel fibrinolytic enzyme from Cordyceps militaris. Int. J. Biol. Macromol. 2016, 94, 793–801. [Google Scholar] [CrossRef]
- Nehete, P.N.; Shah, V.D.; Kothari, R.M. Profiles of alkaline protease production as a function of composition of the slant, age, transfer and isolate number and physiological state of culture. Biotechnol. Lett. 1985, 7, 413–418. [Google Scholar] [CrossRef]
- Narasimhan, M.K.; Chandrasekaran, M.; Rajesh, M. Fibrinolytic enzyme production by newly isolated Bacillus cereus SRM-001 with enhanced in-vitro blood clot lysis potential 2015. J. Gen. Appl. Microbiol. 2015, 61, 157–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxena, R.; Singh, R. Statistical optimization of conditions for protease production from Bacillus sp. Acta Biol. Szeged 2010, 54, 135–141. [Google Scholar]
- Plackett, R.L.; Burman, J.P. The design of optimum multifactorial experiments. Biometrika 1946, 33, 305–325. [Google Scholar] [CrossRef]
- Ghanem, N.B.; Yusef, H.H.; Mahrouse, H.K. Production of Aspergillus terreus xylanase in solid-state cultures: Application of the Plackett-Burman experimental design to evaluate nutritional requirements. Bioresour. Technol. 2000, 73, 113–121. [Google Scholar] [CrossRef]
- Astrup, T.; Mullertz, S. The fibrin plate method for estimating fibrinolytic activity. Arch. Biochem. Biophys. 1952, 40, 346–351. [Google Scholar] [CrossRef]
- Moore, E.; Arnscheidt, A.; Krüger, A.; Strömpl, C.; Mau, M. Simplified protocols for the preparation of genomic DNA from bacterial cultures. In Molecular Microbial Ecology Manual; Kluwer Academic Publishers: Dodrecht, The Netherlands, 1999; pp. 1–15. [Google Scholar]
- Saxena, R.; Singh, R. Contemporaneous production of amylase and protease through CCD response surface methodology by newly isolated Bacillus megaterium Strain B69. Enzyme Res. 2014, 2014, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method—A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Wang, Y.H.; Feng, J.T.; Zhang, Q.; Zhang, X. Optimization of fermentation condition for antibiotic production by Xenorhabdus nematophila with response surface methodology. J. Appl. Microbiol. 2007, 104, 735–744. [Google Scholar] [CrossRef]
- Saxena, R.; Singh, R. Metal ion and pH Stable Protease Production Using Agro-industrial Waste. J. Ecobiotechnol. 2010, 2, 1–5. [Google Scholar]
- Nadeem, M.; Qazi, J.I.; Syed, Q.; Gulsher, M. Purification and characterization of an alkaline protease from Bacillus licheniformis UV-9 for detergent formulations. Songklanakarin J. Sci. Technol. 2013, 35, 187–195. [Google Scholar]
- Saxena, R.; Singh, R. MALDI-TOF MS and CD Spectral Analysis for Identification and Structure Prediction of a Purified, Novel, Organic Solvent Stable, Fibrinolytic Metalloprotease from Bacillus cereus B80. BioMed Res. Int. 2015, 2015, 527015. [Google Scholar] [CrossRef] [PubMed]
- Afifah, D.N.; Rustanti, N.; Anjani, G.; Syah, D.; Suhartono, M.T. Proteomics study of extracellular fibrinolytic proteases from Bacillus licheniformis RO3 and Bacillus pumilus 2.g isolated from Indonesian fermented food. IOP Conf. Ser. Earth Environ. Sci. 2017, 55, 012025. [Google Scholar] [CrossRef]
- Paik, H.D.; Lee, S.K.; Heo, S.; Kim, S.Y.; Lee, H.H.; Kwon, T.J. Purification and characterization of the fibrinolytic enzyme produced by Bacillus subtilis KCK-7 from Chungkookjang. J. Microbiol. Biotechn. 2004, 14, 829–835. [Google Scholar]
- Peng, Y.; Huang, Q.; Zhang, R.H.; Zhang, Y.Z. Purification and characterization of a fibrinolytic enzyme produced by Bacillus amyloliquefaciens DC-4 screened from douchi, a traditional Chinese soybean food. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2003, 134, 45–52. [Google Scholar] [CrossRef]
- Huang, S.; Pan, S.; Chen, G.; Huang, S.; Zhang, Z.; Li, Y.; Liang, Z. Biochemical characteristics of a fibrinolytic enzyme purified from a marine bacterium, Bacillus subtilis HQS-3. Int. J. Biol. Macromol. 2013, 62, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Vijayaraghavan, P.; Raj, S.R.F.; Vincent, S.G.P. Purification and characterization of fibrinolytic enzyme from Pseudoalteromonas sp., IND11 and its in vitro activity on blood clot. J. Biol. Chem. 2015, 9, 11–20. [Google Scholar] [CrossRef] [Green Version]
- Lateef, A.; Adelere, I.A.; Kana, E.B.G. Bacillus safensis LAU 13: A new source of keratinase and its multi-functional biocatalytic applications. Biotechnol. Biotec. Equip. 2015, 29, 54–63. [Google Scholar] [CrossRef]
- Mitsuiki, S.; Ichikawa, M.; Oka, T.; Sakai, M.; Moriyama, Y.; Sameshima, Y.; Goto, M.; Furukawa, K. Molecular characterization of a keratinolytic enzyme from an alkaliphilic Nocardiopsis sp. TOA-1. Enzyme Microb. Technol. 2004, 34, 482–489. [Google Scholar] [CrossRef]
- Kim, H.C.; Choi, B.S.; Sapkota, K.; Kim, S.; Lee, H.J.; Yoo, J.C.; Kim, S.J. Purification and characterization of a novel, highly potent fibrinolytic enzyme from Paecilomyces tenuipes. Process. Biochem. 2011, 46, 1545–1553. [Google Scholar] [CrossRef]
- Liu, X.; Kopparapu, N.K.; Zheng, H.; Katrolia, P.; Deng, Y.; Zheng, X. Purification and characterization of a fibrinolytic enzyme from the food-grade fungus, Neurospora sitophila. J. Mol. Catal. B Enzym. 2016, 134, 98–104. [Google Scholar]
- Devaraj, Y.; Rajender, S.K.; Halami, P.M. Purification and characterization of fibrinolytic protease from Bacillus amyloliquefaciens MCC2606 and analysis of fibrin degradation product by MS/MS. Prep. Biochem. Biotechnol. 2018, 48, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Ju, X.; Cao, X.; Sun, Y.; Wang, Z.; Cao, C.; Liu, J.; Jiang, J. Purification and characterization of a fibrinolytic enzyme from Streptomyces sp. XZNUM 00004. World J. Microbiol. Biotechnol. 2012, 28, 2479–2486. [Google Scholar] [CrossRef] [PubMed]
- Rashad, M.M.; Mahmoud, A.E.; Al-Kashef, A.S.; Nooman, M.U. Purification and characterization of a novel fibrinolytic enzyme by Candida guilliermondii grown on sunflower oil cake. JASR 2012, 8, 635–645. [Google Scholar]
- Cha, W.S.; Park, S.S.; Kim, S.J.; Choi, D. Biochemical and enzymatic properties of a fibrinolytic enzyme from Pleurotus eryngii cultivated under solid-state conditions using corn cob. Bioresour. Technol. 2010, 101, 6475–6481. [Google Scholar] [CrossRef]
- Nascimento, T.P.; Sales, A.E.; Porto, T.S.; Costa, R.M.P.B.; Breydo, L.; Uversky, V.N.; Porto, A.L.F.; Converti, A. Purification, biochemical, and structural characterization of a novel fibrinolytic enzyme from Mucor subtilissimus UCP 1262. Bioproc. Biosyst. Eng. 2017, 40, 1209–1219. [Google Scholar] [CrossRef]
- Afifah, D.N.; Sulchan, M.; Syah, D. Purification and characterization of a fibrinolytic enzyme from Bacillus pumilus 2. g isolated from Gembus, an Indonesian fermented food. Prev. Nutr. Food Sci. 2014, 19, 213–219. [Google Scholar] [CrossRef] [Green Version]
- Esakkiraj, P.; Meleppat, B.; Lakra, A.K.; Ayyannaa, R.; Arul, V. Cloning, expression, characterization and application of protease produced by Bacillus cereus PMW8. RSC Adv. 2016, 6, 38611–38616. [Google Scholar] [CrossRef]
- Kasana, R.C.; Yadav, S.K. Isolation of a psychrotrophic Exiguobacterium sp. SKPB5 (MTCC 7803) and characterization of its alkaline protease. Curr. Microbiol. 2007, 54, 224–229. [Google Scholar] [CrossRef]
- Beg, Q.K.; Gupta, R. Purification and characterization of an oxidation-stable, thiol-dependent serine alkaline protease from Bacillus mojavensis. Enzyme Microb. Technol. 2003, 32, 294–304. [Google Scholar] [CrossRef]
- Joshi, S.; Satyanarayana, T. Characteristics and applications of a recombinant alkaline serine protease from a novel bacterium Bacillus lehensis. Bioresour. Technol. 2013, 131, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Sinha, R.; Khare, S.K. Characterization of detergent compatible protease of a halophilic Bacillus sp. EMB9: Differential role of metal ions in stability and activity. Bioresour. Technol. 2013, 145, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Sari, E.; Logoglu, E.; Oktemer, A. Purification and characterization of organic solvent stable serine alkaline protease from newly isolated Bacillus circulans M34. Biomed. Chromatogr. 2015, 29, 1356–1363. [Google Scholar] [CrossRef] [PubMed]
- Hassanein, W.A.; Kotb, E.; Awny, N.M.; El-Zawahry, Y.A. Fibrinolysis and anticoagulant potential of a metallo protease produced by Bacillus subtilis K42. J. Biosci. 2011, 36, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.L.; Chen, S.N. Magnesium Enhanced Fibrinolytic Activity of Protease from Schizophyllum commune. Taiwania 2010, 55, 117–122. [Google Scholar]
- Yogesh, D.; Halami, P.M. Fibrinolytic enzymes of Bacillus spp.: An overview. Int. Food Res. J. 2017, 24, 35–47. [Google Scholar]
- Lucy, J.; Raharjo, P.F.; Elvina, E.; Florencia, L.; Susanti, A.I.; Pinontoan, R. Clot Lysis Activity of Bacillus subtilis G8 Isolated from Japanese Fermented Natto Soybeans. Appl. Food Biotechnol. 2019, 6, 101–109. [Google Scholar]


| Run | Peptone (g) | Yeast Extract (g) | Casien (g) | Glucose (g) | Sucrose (g) | K2HPO4 (g) | MgSO4 (g) | Speed (rpm) | pH | Time (h) | Inoculum (%) | Response (U/mL) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5 | 5 | 2 | 0.5 | 5 | 4 | 1 | 100 | 7 | 24 | 5 | 16.5 |
| 2 | 7.5 | 3.5 | 1 | 2.75 | 2.5 | 2.5 | 0.75 | 140 | 8 | 48 | 3 | 17.9 |
| 3 | 5 | 2 | 0 | 0.5 | 0 | 1 | 0.5 | 100 | 7 | 24 | 1 | 6.5 |
| 4 | 10 | 2 | 0 | 0.5 | 5 | 1 | 1 | 180 | 7 | 72 | 5 | 13.2 |
| 5 | 10 | 2 | 2 | 5 | 5 | 1 | 0.5 | 100 | 9 | 24 | 5 | 24.7 |
| 6 | 10 | 5 | 0 | 5 | 5 | 4 | 0.5 | 100 | 7 | 72 | 1 | 23.0 |
| 7 | 5 | 2 | 0 | 5 | 0 | 4 | 1 | 100 | 9 | 72 | 5 | 17.6 |
| 8 | 5 | 5 | 0 | 5 | 5 | 1 | 1 | 180 | 9 | 24 | 1 | 14.6 |
| 9 | 5 | 5 | 2 | 5 | 0 | 1 | 0.5 | 180 | 7 | 72 | 5 | 16.9 |
| 10 | 10 | 5 | 0 | 0.5 | 0 | 4 | 0.5 | 180 | 9 | 24 | 5 | 17.8 |
| 11 | 5 | 2 | 2 | 0.5 | 5 | 4 | 0.5 | 180 | 9 | 72 | 1 | 10.6 |
| 12 | 10 | 2 | 2 | 5 | 0 | 4 | 1 | 180 | 7 | 24 | 1 | 26.1 |
| 13 | 10 | 5 | 2 | 0.5 | 0 | 1 | 1 | 100 | 9 | 72 | 1 | 28.6 |
| Variable | Component | Mi+ | Mi− | E(xi) | Absolute E(xi) | Ranking |
|---|---|---|---|---|---|---|
| A | Peptone | 133.4 | 82.7 | 8.45 | 8.45 | 1 |
| B | Yeast extract | 117.4 | 98.7 | 3.11 | 3.11 | 4 |
| C | Casein | 123.4 | 92.7 | 5.116 | 5.116 | 2 |
| D | Glucose | 122.9 | 93.2 | 4.95 | 4.95 | 3 |
| E | Sucrose | 102.6 | 113.5 | −1.8166 | 1.8166 | 9 |
| F | K2HPO4 | 111.6 | 104.5 | 1.1833 | 1.833 | 8 |
| G | MgSO4 | 116.6 | 99.5 | 2.85 | 2.85 | 6 |
| H | RPM | 116.9 | 99.2 | 2.95 | 2.95 | 5 |
| J | pH | 113.9 | 102.2 | 1.95 | 1.95 | 7 |
| K | Incubation time | 109.9 | 106.2 | 0.616 | 0.616 | 10 |
| L | Inoculum size | 106.7 | 109.4 | −0.45 | 0.45 | 11 |
| Source | Sum of Squares | Degree of Freedom | Mean Square | F Value | p-Value Prob > F | |
|---|---|---|---|---|---|---|
| Model | 473.10 | 11 | 43.01 | 12,455.28 | 0.0070 | Significant |
| Peptone | 213.72 | 1 | 213.72 | 61,893.00 | 0.0026 | |
| Yeast extract | 29.42 | 1 | 29.42 | 8519.68 | 0.0069 | |
| Casein | 79.19 | 1 | 79.19 | 22,932.00 | 0.0042 | |
| Glucose | 73.27 | 1 | 73.27 | 21,218.08 | 0.0044 | |
| Sucrose | 9.83 | 1 | 9.83 | 2847.52 | 0.0119 | |
| K2HPO4 | 4.31 | 1 | 4.31 | 1248.52 | 0.0180 | |
| MgSO4 | 24.16 | 1 | 24.16 | 6997.12 | 0.0076 | |
| RPM | 26.29 | 1 | 26.29 | 7613.32 | 0.0073 | |
| pH | 11.21 | 1 | 11.21 | 3245.32 | 0.0112 | |
| Incubation time | 1.12 | 1 | 1.12 | 325.00 | 0.0353 | |
| Inoculum size | 0.58 | 1 | 0.58 | 168.48 | 0.0489 | |
| Residual | 0.00345 | 1 | 0.00345 | |||
| Cor total | 473.10 | 12 |
| Variable | Name | Units | Type | Subtype | Minimum | Maximum | −1 Actual | +1 Actual | Mean | Std. Dev. |
|---|---|---|---|---|---|---|---|---|---|---|
| A | Peptone | G | Numeric | Continuous | 2.5 | 12.5 | 5 | 10 | 7.5 | 2.27429 |
| B | Casein | G | Numeric | Continuous | −1 | 3 | 0 | 2 | 1 | 0.909718 |
| C | Yeast extract | G | Numeric | Continuous | 0.5 | 6.5 | 2 | 5 | 3.5 | 1.36458 |
| D | Glucose | G | Numeric | Continuous | −1.75 | 7.25 | 0.5 | 5 | 2.75 | 2.04686 |
| Response Name | Units | Obs | Analysis | Minimum | Maximum | Mean | Std. Dev. | Ratio | Trans | Model |
|---|---|---|---|---|---|---|---|---|---|---|
| Fibrinolytic protease activity | U/mL | 30 | Polynomial | 15.1193 | 30.7523 | 22.4807 | 4.25754 | 2.03398 | None | 2FI |
| Source | Sum of Squares | Degree of Freedom | Mean Square | F Value | p-Value Prob > F | |
|---|---|---|---|---|---|---|
| Model | 317.12 | 10 | 31.71 | 2.89 | 0.0224 | Significant |
| A—Peptone | 1.40 | 1 | 1.40 | 0.13 | 0.7249 | |
| B—Casein | 5.25 | 1 | 5.25 | 0.48 | 0.4974 | |
| C—Yeast extract | 2.67 | 1 | 2.67 | 0.24 | 0.6277 | |
| D—Glucose | 116.03 | 1 | 116.03 | 10.57 | 0.0042 | |
| AB | 8.73 | 1 | 8.73 | 0.80 | 0.3837 | |
| AC | 9.17 | 1 | 9.17 | 0.84 | 0.3723 | |
| AD | 65.48 | 1 | 65.48 | 5.97 | 0.0245 | |
| BC | 38.69 | 1 | 38.69 | 3.52 | 0.0759 | |
| BD | 12.28 | 1 | 12.28 | 1.12 | 0.3034 | |
| CD | 57.43 | 1 | 57.43 | 5.23 | 0.0338 | |
| Residual | 208.55 | 19 | 10.98 | |||
| Lack of fit | 138.92 | 14 | 9.92 | 0.71 | 0.7178 | Not Significant |
| Pure error | 69.64 | 5 | 13.93 | |||
| Cor total | 525.67 | 29 |
| Purification Steps | Volume (mL) | Total Activity (U) | Total Protein (mg) | Specific Activity (U/mg) | Yield or Recovery (%) | Fold Purification |
|---|---|---|---|---|---|---|
| Crude | 100 | 190 | 15.4 | 12.337 ± 0.32 | 100 | 1 |
| EtOH-treated | 100 | 86 | 5.45 | 15.779 ± 0.45 | 35.39 | 1.28 |
| Sephadex-75 | 100 | 146 | 5.1 | 28.627 ± 0.33 | 33.11 | 2.32 |
| Start | End | Observed | Mr(expt) | Mr(calc) | ppm | M | Peptide |
|---|---|---|---|---|---|---|---|
| 1 | 5 | 649.3323 | 648.3250 | 648.3741 | −75.7 | 2 | -.MRSKK.L |
| 5 | 32 | 3073.6745 | 3072.6672 | 3072.6544 | 4.17 | 1 | K.KLWISLLFALTLIFTMAFSNMSAQAAGK.S |
| 6 | 32 | 2945.5436 | 2944.5363 | 2944.5594 | −7.85 | 0 | K.LWISLLFALTLIFTMAFSNMSAQAAGK.S |
| 33 | 44 | 1386.6593 | 1385.6520 | 1385.7554 | −74.6 | 2 | K.SSTEKKYIVGFK.Q |
| 38 | 44 | 854.5358 | 853.5285 | 853.5062 | 26.2 | 1 | K.KYIVGFK.Q |
| 39 | 54 | 1748.8624 | 1747.8551 | 1747.8637 | −4.90 | 1 | K.YIVGFKQTMSAMSSAK.K |
| 45 | 54 | 1041.4160 | 1040.4087 | 1040.4630 | −52.2 | 0 | K.QTMSAMSSAK.K |
| 45 | 55 | 1169.5683 | 1168.5610 | 1168.5580 | 2.59 | 1 | K.QTMSAMSSAKK.K |
| 55 | 62 | 946.5210 | 945.5137 | 945.5495 | −37.8 | 2 | K.KKDVISEK.G |
| 56 | 65 | 1060.5299 | 1059.5226 | 1059.5924 | −65.9 | 2 | K.KDVISEKGGK.V |
| 57 | 65 | 932.5373 | 931.5300 | 931.4975 | 35.0 | 1 | K.DVISEKGGK.V |
| 57 | 68 | 1287.7175 | 1286.7102 | 1286.7194 | −7.14 | 2 | K.DVISEKGGKVQK.Q |
| 63 | 71 | 1019.6070 | 1018.5997 | 1018.5924 | 7.23 | 2 | K.GGKVQKQFK.Y |
| 66 | 71 | 777.4271 | 776.4198 | 776.4545 | −44.6 | 1 | K.VQKQFK.Y |
| 66 | 83 | 2024.3001 | 2023.2928 | 2023.0738 | 108 | 2 | K.VQKQFKYVNAAAATLDEK.A |
| 69 | 86 | 1967.2133 | 1966.2060 | 1966.0523 | 78.2 | 2 | K.QFKYVNAAAATLDEKAVK.E |
| 72 | 83 | 1265.6468 | 1264.6395 | 1264.6299 | 7.61 | 0 | K.YVNAAAATLDEK.A |
| 72 | 86 | 1563.7549 | 1562.7476 | 1562.8304 | −53.0 | 1 | K.YVNAAAATLDEKAVK.E |
| 91 | 118 | 3145.4957 | 3144.4884 | 3144.5040 | −4.95 | 0 | K.DPSVAYVEEDHIAHEYAQSVPYGISQIK.A |
| 134 | 151 | 1893.9589 | 1892.9516 | 1892.9592 | −4.01 | 0 | K.VAVIDSGIDSSHPDLNVR.G |
| 248 | 276 | 2595.2804 | 2594.2731 | 2594.2824 | −3.57 | 0 | K.AVSSGIVVAAAAGNEGSSGSTSTVGYPAK.Y |
| 277 | 292 | 1663.7859 | 1662.7786 | 1662.8325 | −32.4 | 0 | K.YPSTIAVGAVNSSNQR.A |
| 344 | 353 | 1209.5433 | 1208.5360 | 1208.6051 | −57.1 | 0 | K.HPTWTNAQVR.D |
| 344 | 355 | 1480.7394 | 1479.7321 | 1479.7331 | −0.67 | 1 | K.HPTWTNAQVRDR.L |
| 354 | 371 | 2084.9103 | 2083.9030 | 2083.9851 | −39.4 | 1 | R.DRLESTATYLGNSFYYGK.G |
| 356 | 381 | 2779.7205 | 2778.7132 | 2778.3864 | 118 | 1 | R.LESTATYLGNSFYYGKGLINVQAAAQ.- |
| 372 | 381 | 984.5695 | 983.5622 | 983.5400 | 22.6 | 0 | K.GLINVQAAAQ.- |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, C.; Salem, G.E.M.; Sharma, N.; Gautam, P.; Singh, R. Thrombolytic Potential of Novel Thiol-Dependent Fibrinolytic Protease from Bacillus cereus RSA1. Biomolecules 2020, 10, 3. https://doi.org/10.3390/biom10010003
Sharma C, Salem GEM, Sharma N, Gautam P, Singh R. Thrombolytic Potential of Novel Thiol-Dependent Fibrinolytic Protease from Bacillus cereus RSA1. Biomolecules. 2020; 10(1):3. https://doi.org/10.3390/biom10010003
Chicago/Turabian StyleSharma, Chhavi, Gad Elsayed Mohamed Salem, Neha Sharma, Prerna Gautam, and Rajni Singh. 2020. "Thrombolytic Potential of Novel Thiol-Dependent Fibrinolytic Protease from Bacillus cereus RSA1" Biomolecules 10, no. 1: 3. https://doi.org/10.3390/biom10010003




