Ageing Investigation Using Two-Time-Point Metabolomics Data from KORA and CARLA Studies
Abstract
:1. Introduction
2. Results
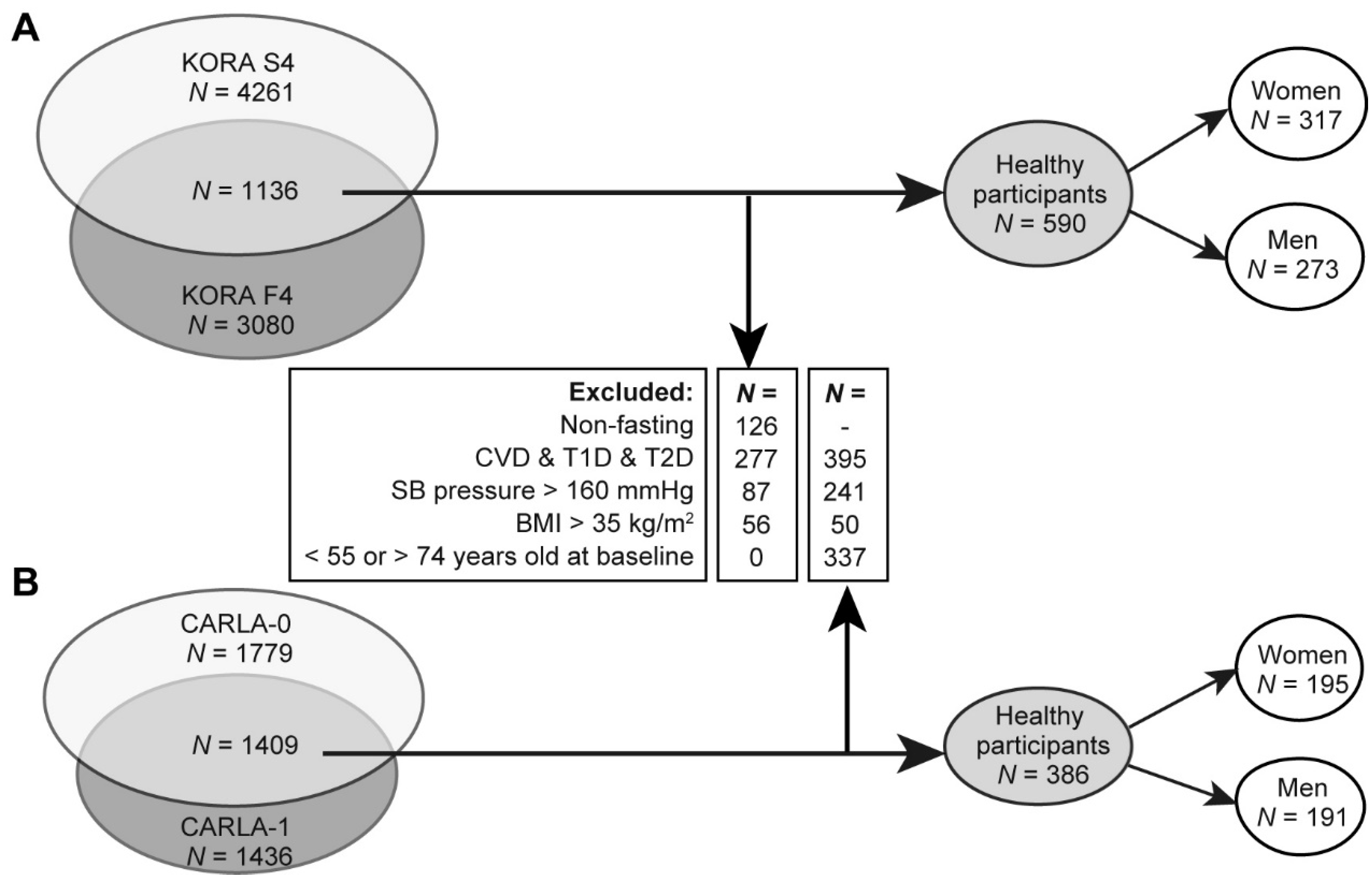
2.1. Characteristics for the Discovery and Replication Population
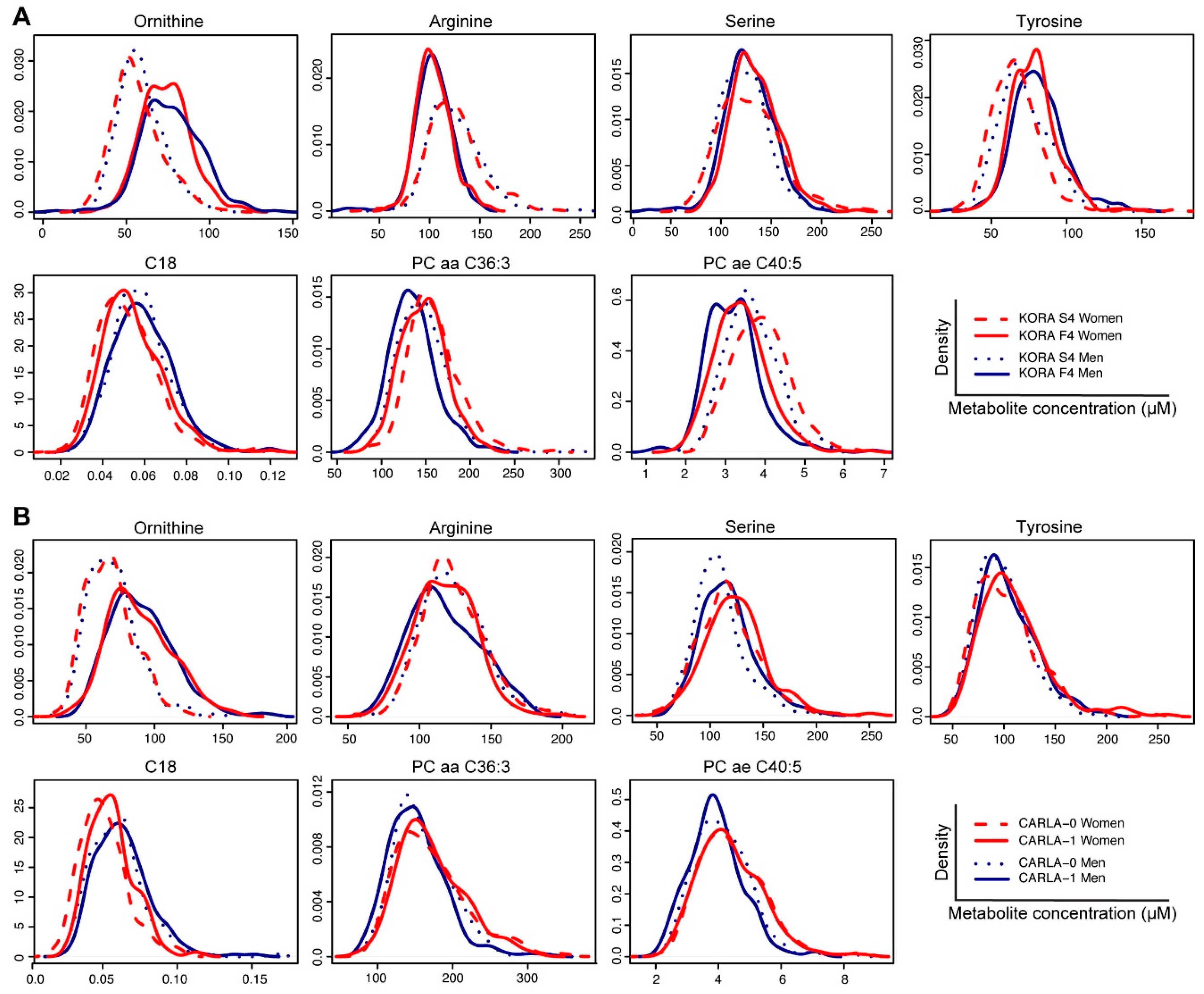
2.2. Identification of Ageing-Associated Metabolite
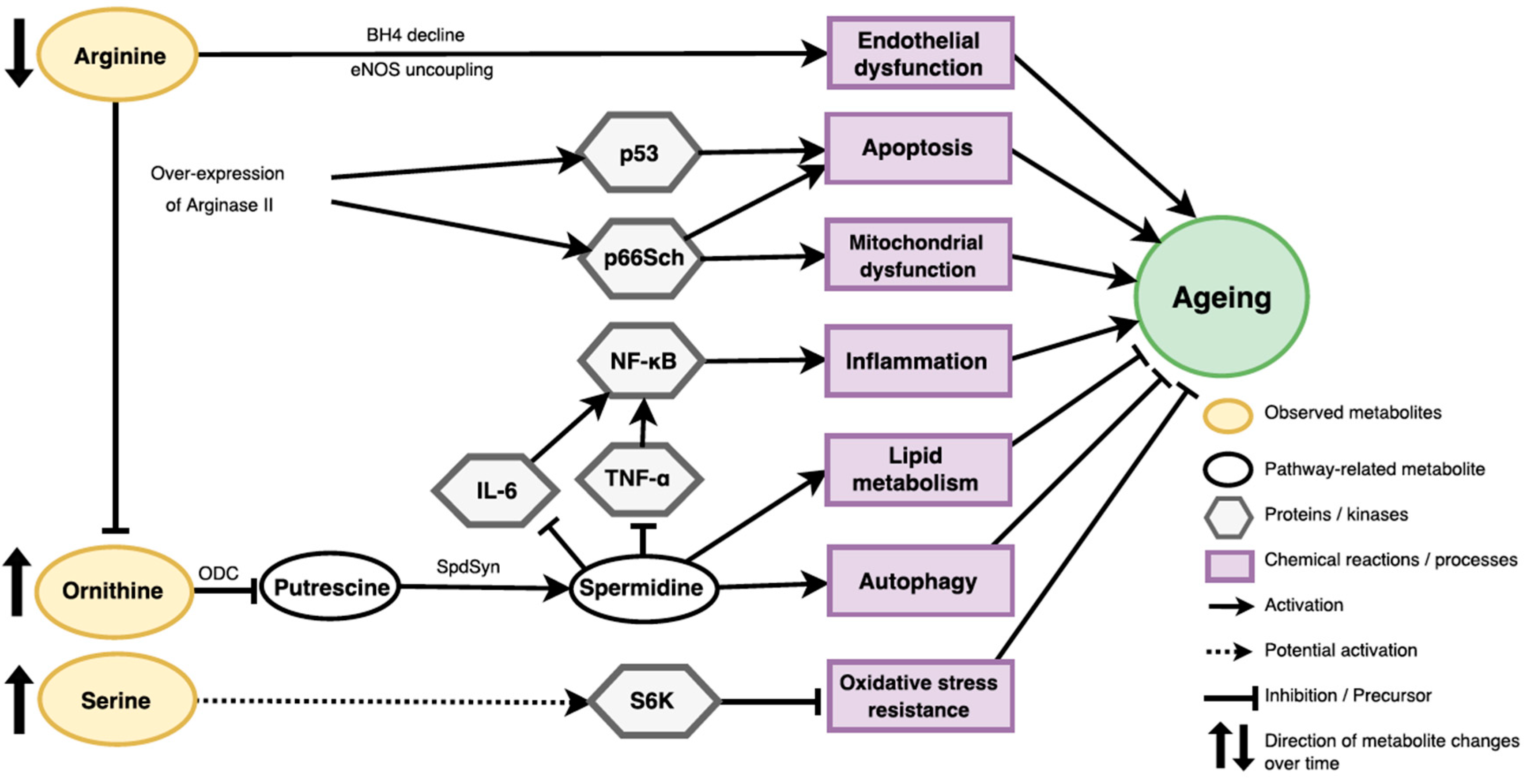
2.3. Ageing Related Pathways
3. Discussion
3.1. Potential Roles of Identified Metabolites in Ageing
3.2. Strengths and Limitations
4. Materials and Methods
4.1. Ethics Statement
4.2. CARLA Studies
4.3. Sample Collection
4.4. Metabolite Quantification
4.5. Pre-Processing Of Metabolite Data
4.6. Inclusion and Exclusion Criteria of Study Samples
4.7. Statistical Analysis
4.8. Pathway Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Financial Disclosure Statement
References
- Rodríguez-Rodero, S.; Fernández-Morera, J.L.; Menéndez-Torre, E.; Calvanese, V.; Fernández, A.F.; Fraga, M.F. Aging genetics and aging. Aging Dis. 2011, 2, 186–195. [Google Scholar] [PubMed]
- Bürkle, A.; Moreno-Villanueva, M.; Bernhard, J.; Blasco, M.; Zondag, G.; Hoeijmakers, J.H.J.; Toussaint, O.; Grubeck-Loebenstein, B.; Mocchegiani, E.; Collino, S.; et al. MARK-AGE biomarkers of ageing. Mech. Ageing Dev. 2015, 151, 2–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, J.M.; White, M. Biological ageing: A fundamental, biological link between socio-economic status and health? Eur. J. Public Health 2004, 14, 331–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niccoli, T.; Partridge, L. Ageing as a risk factor for disease. Curr. Biol. 2012, 22, R741–R752. [Google Scholar] [CrossRef] [PubMed]
- Hulbert, A.J.; Pamplona, R.; Buffenstein, R.; Buttemer, W.A. Life and death: Metabolic rate, membrane composition, and life span of animals. Physiol. Rev. 2007, 87, 1175–1213. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.M.; Glass, D.; Spector, T.D. Omics technologies and the study of human ageing. Nat. Rev. Genet. 2013, 14, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, P.; Thyagarajan, B.; Sun, F.; Schupf, N.; Newman, A.B.; Montano, M.; Perls, T.T. Biomarker signatures of aging. Aging Cell 2017, 16, 329–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sprott, R.L. Biomarkers of aging and disease: Introduction and definitions. Exp. Gerontol. 2010, 45, 2–4. [Google Scholar] [CrossRef] [PubMed]
- Deelen, J.; Beekman, M.; Capri, M.; Franceschi, C.; Slagboom, P.E. Identifying the genomic determinants of aging and longevity in human population studies: Progress and challenges. BioEssays News Rev. Mol. Cell. Dev. Biol. 2013, 35, 386–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mäkinen, V.-P.; Ala-Korpela, M. Metabolomics of aging requires large-scale longitudinal studies with replication. Proc. Natl. Acad. Sci. USA 2016, 113, E3470. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhai, G.; Singmann, P.; He, Y.; Xu, T.; Prehn, C.; Römisch-Margl, W.; Lattka, E.; Gieger, C.; Soranzo, N.; et al. Human serum metabolic profiles are age dependent. Aging Cell 2012, 11, 960–967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, F.; Virtue, A.; Wang, H.; Yang, X.-F. Metabolomic analyses for atherosclerosis, diabetes, and obesity. Biomark. Res. 2013, 1, 17. [Google Scholar] [CrossRef] [PubMed]
- Sharman, A.; Zhumadilov, Z. The Scientific Basis for Healthy Aging and Antiaging Process; Mary Ann Liebert, Inc.: New York, NY, USA, 2011. [Google Scholar]
- Kristal, B.S.; Shurubor, Y.I. Metabolomics: Opening another window into aging. Sci. Aging Knowl. Environ. Sage Ke 2005, 2005, pe19. [Google Scholar] [CrossRef] [PubMed]
- Jové, M.; Maté, I.; Naudí, A.; Mota-Martorell, N.; Portero-Otín, M.; de La Fuente, M.; Pamplona, R. Human aging is a metabolome-related matter of gender. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2016, 71, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted metabolomics. Curr. Protoc. Mol. Biol. 2012, 98, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Wang-Sattler, R.; Yu, Z.; Herder, C.; Messias, A.C.; Floegel, A.; He, Y.; Heim, K.; Campillos, M.; Holzapfel, C.; Thorand, B.; et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol. Syst. Biol. 2012, 8, 615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, A.; Sun, H.; Xu, H.; Qiu, S.; Wang, X. Cell metabolomics. Omics J. Integr. Biol. 2013, 17, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Slupsky, C.M.; Rankin, K.N.; Wagner, J.; Fu, H.; Chang, D.; Weljie, A.M.; Saude, E.J.; Lix, B.; Adamko, D.J.; Shah, S.; et al. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal. Chem. 2007, 79, 6995–7004. [Google Scholar] [CrossRef] [PubMed]
- Lawton, K.A.; Berger, A.; Mitchell, M.; Milgram, K.E.; Evans, A.M.; Guo, L.; Hanson, R.W.; Kalhan, S.C.; Ryals, J.A.; Milburn, M.V. Analysis of the adult human plasma metabolome. Pharmacogenomics 2008, 9, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Psihogios, N.G.; Gazi, I.F.; Elisaf, M.S.; Seferiadis, K.I.; Bairaktari, E.T. Gender-related and age-related urinalysis of healthy subjects by NMR-based metabonomics. Nmr Biomed. 2008, 21, 195–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Adamo, P.; Ulivi, S.; Beneduci, A.; Pontoni, G.; Capasso, G.; Lanzara, C.; Andrighetto, G.; Hladnik, U.; Nunes, V.; Palacin, M.; et al. Metabonomics and population studies: Age-related amino acids excretion and inferring networks through the study of urine samples in two Italian isolated populations. Amino Acids 2010, 38, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Menni, C.; Kastenmüller, G.; Petersen, A.K.; Bell, J.T.; Psatha, M.; Tsai, P.-C.; Gieger, C.; Schulz, H.; Erte, I.; John, S.; et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int. J. Epidemiol. 2013, 42, 1111–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.H.; Park, S.; Kim, H.-S.; Jung, B.H. Metabolomic approaches to the normal aging process. Metabolomics 2014, 10, 1268–1292. [Google Scholar] [CrossRef]
- Sedlmeier, A.; Kluttig, A.; Giegling, I.; Prehn, C.; Adamski, J.; Kastenmüller, G.; Lacruz, M.E. The human metabolic profile reflects macro- and micronutrient intake distinctly according to fasting time. Sci. Rep. 2018, 8, 12262. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.K.; Sloane, R.; Bain, J.R.; Stevens, R.D.; Newgard, C.B.; Pieper, C.F.; Kraus, V.B. Daily variation of serum acylcarnitines and amino acids. Metabolomics 2012, 8, 556–565. [Google Scholar] [CrossRef] [PubMed]
- Krug, S.; Kastenmüller, G.; Stückler, F.; Rist, M.J.; Skurk, T.; Sailer, M.; Raffler, J.; Römisch-Margl, W.; Adamski, J.; Prehn, C.; et al. The dynamic range of the human metabolome revealed by challenges. Faseb J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2012, 26, 2607–2619. [Google Scholar] [CrossRef] [PubMed]
- Mittelstrass, K.; Ried, J.S.; Yu, Z.; Krumsiek, J.; Gieger, C.; Prehn, C.; Roemisch-Margl, W.; Polonikov, A.; Peters, A.; Theis, F.J.; et al. Discovery of sexual dimorphisms in metabolic and genetic biomarkers. PLoS Genet. 2011, 7, e1002215. [Google Scholar] [CrossRef] [PubMed]
- Trabado, S.; Al-Salameh, A.; Croixmarie, V.; Masson, P.; Corruble, E.; Fève, B.; Colle, R.; Ripoll, L.; Walther, B.; Boursier-Neyret, C.; et al. The human plasma-metabolome: Reference values in 800 French healthy volunteers; impact of cholesterol, gender and age. PLoS ONE 2017, 12, e0173615. [Google Scholar] [CrossRef] [PubMed]
- Maeba, R.; Maeda, T.; Kinoshita, M.; Takao, K.; Takenaka, H.; Kusano, J.; Yoshimura, N.; Takeoka, Y.; Yasuda, D.; Okazaki, T.; et al. Plasmalogens in human serum positively correlate with high-density lipoprotein and decrease with aging. J. Atheroscler. Thromb. 2007, 14, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Mauvais-Jarvis, F. Sex differences in metabolic homeostasis, diabetes, and obesity. Biol. Sex Differ. 2015, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Auro, K.; Joensuu, A.; Fischer, K.; Kettunen, J.; Salo, P.; Mattsson, H.; Niironen, M.; Kaprio, J.; Eriksson, J.G.; Lehtimäki, T.; et al. A metabolic view on menopause and ageing. Nat. Commun. 2014, 5, 4708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, C.; Hou, Y.; Zhang, H.; Yang, K.; Wang, J.; Guo, B.; Zhang, F.; Li, H.; Zhou, X.; Li, Y.; et al. Plasma metabolic profiles in women are menopause dependent. PLoS ONE 2015, 10, e0141743. [Google Scholar] [CrossRef] [PubMed]
- Minois, N. Molecular basis of the ‘anti-aging’ effect of spermidine and other natural polyamines-a mini-review. Gerontology 2014, 60, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Santhanam, L.; Christianson, D.W.; Nyhan, D.; Berkowitz, D.E. Arginase and vascular aging. J. Appl. Physiol. 2008, 105, 1632–1642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Ming, X.-F. Arginase: The emerging therapeutic target for vascular oxidative stress and inflammation. Front. Immunol. 2013, 4, 149. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Liu, X.J. Deficiency of ovarian ornithine decarboxylase contributes to aging-related egg aneuploidy in mice. Aging Cell 2013, 12, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Aichler, M.; Borgmann, D.; Krumsiek, J.; Buck, A.; MacDonald, P.E.; Fox, J.E.M.; Lyon, J.; Light, P.E.; Keipert, S.; Jastroch, M.; et al. N-acyl taurines and acylcarnitines cause an imbalance in insulin synthesis and secretion provoking β cell dysfunction in type 2 diabetes. Cell Metab. 2017, 25, 1334–1347.e4. [Google Scholar] [CrossRef] [PubMed]
- Mirisola, M.G.; Taormina, G.; Fabrizio, P.; Wei, M.; Hu, J.; Longo, V.D. Serine-and threonine/valine-dependent activation of PDK and Tor orthologs converge on Sch9 to promote aging. PLoS Genet. 2014, 10, e1004113. [Google Scholar] [CrossRef] [PubMed]
- Gamella-Pozuelo, L.; Grande, M.T.; Clemente-Lorenzo, M.; Murillo-Gómez, C.; de Pablo, F.; López-Novoa, J.M.; Hernández-Sánchez, C. Tyrosine hydroxylase haploinsufficiency prevents age-associated arterial pressure elevation and increases half-life in mice. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Hertel, J.; Friedrich, N.; Wittfeld, K.; Pietzner, M.; Budde, K.; van der Auwera, S.; Lohmann, T.; Teumer, A.; Völzke, H.; Nauck, M.; et al. Measuring biological age via metabonomics: The metabolic age score. J. Proteome Res. 2016, 15, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Rotter, M.; Brandmaier, S.; Covic, M.; Burek, K.; Hertel, J.; Troll, M.; Bader, E.; Adam, J.; Prehn, C.; Rathkolb, B.; et al. Night shift work affects urine metabolite profiles of nurses with early chronotype. Metabolites 2018, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Meisinger, C.; Stöckl, D.; Rückert, I.M.; Döring, A.; Thorand, B.; Heier, M.; Huth, C.; Belcredi, P.; Kowall, B.; Rathmann, W. Serum potassium is associated with prediabetes and newly diagnosed diabetes in hypertensive adults from the general population: The KORA F4-study. Diabetologia 2013, 56, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Brandmaier, S.; Messias, A.C.; Herder, C.; Draisma, H.H.M.; Demirkan, A.; Yu, Z.; Ried, J.S.; Haller, T.; Heier, M.; et al. Effects of metformin on metabolite profiles and LDL cholesterol in patients with type 2 diabetes. Diabetes Care 2015, 38, 1858–1867. [Google Scholar] [CrossRef] [PubMed]
- Greiser, K.H.; Kluttig, A.; Schumann, B.; Kors, J.A.; Swenne, C.A.; Kuss, O.; Werdan, K.; Haerting, J. Cardiovascular disease, risk factors and heart rate variability in the elderly general population: Design and objectives of the CARdiovascular disease, living and ageing in halle (CARLA) study. BMC Cardiovasc. Disord. 2005, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Lacruz, M.E.; Kluttig, A.; Tiller, D.; Medenwald, D.; Giegling, I.; Rujescu, D.; Prehn, C.; Adamski, J.; Frantz, S.; Greiser, K.H.; et al. Cardiovascular risk factors associated with blood metabolite concentrations and their alterations during a 4-year period in a population-based cohort. Circ. Cardiovasc. Genet. 2016, 9, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Römisch-Margl, W.; Prehn, C.; Bogumil, R.; Röhring, C.; Suhre, K.; Adamski, J. Procedure for tissue sample preparation and metabolite extraction for high-throughput targeted metabolomics. Metabolomics 2012, 8, 133–142. [Google Scholar] [CrossRef]
- Zukunft, S.; Sorgenfrei, M.; Prehn, C.; Möller, G.; Adamski, J. Targeted metabolomics of dried blood spot extracts. Chromatographia 2013, 76, 1295–1305. [Google Scholar] [CrossRef]
- Nakamura, E.; Miyao, K. Sex differences in human biological aging. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2008, 63, 936–944. [Google Scholar] [CrossRef]
- Ostan, R.; Monti, D.; Gueresi, P.; Bussolotto, M.; Franceschi, C.; Baggio, G. Gender, aging and longevity in humans: An update of an intriguing/neglected scenario paving the way to a gender-specific medicine. Clin. Sci. (Lond.) 2016, 130, 1711–1725. [Google Scholar] [CrossRef] [PubMed]
| Women | Men | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Women in Discovery KORA (N = 317) | Women in Replication CARLA (N = 195) | Men in Discovery KORA (N = 273) | Men in Replication CARLA (N = 191) | |||||||||
| Variables | KORA S4 (Baseline) | KORA F4 (Follow-up) | p-Value a | CARLA-0 (Baseline) | CARLA-1 (Follow-up) | p-Value a | KORA S4 (Baseline) | KORA F4 (Follow-up) | p-Value a | CARLA-0 (Baseline) | CARLA-1 (Follow-up) | p-Value a |
| Chronological age, years (mean (range)) | 62.71 (55–74) | 69.71 (62–81) | - | 63.50 (55–74) | 67.51 (59–78) | - | 62.69 (55–74) | 69.69 (62–81) | - | 63.40 (55–74) | 67.41 (59–79) | - |
| (mean (SD)) | 26.88 (3.41) | 27.21 (3.53) | 7.0 × 10−4 | 26.87 (3.34) | 27.24 (3.43) | 9.6 × 10−5 | 27.36 (2.82) | 27.49 (3.00) | 0.115 | 27.31 (3.08) | 27.51 (3.26) | 0.01 |
| Physically active (%) b | 52.7 | 58.7 | 0.0402 | 42.6 | 53.9 | 0.003 | 47.0 | 55.3 | 0.017 | 33.5 | 42.9 | 0.009 |
| Non-smokers (%) | 89.9 | 93.4 | 0.0055 | 84.6 | 86.7 | 0.1573 | 82.7 | 89.7 | 1.0 × 10−4 | 83.2 | 86.4 | 0.01 |
| Low alcohol intake (%) c | 86.8 | 89.0 | 0.3711 | 94.4 | 94.4 | 0.99 | 77.4 | 82.8 | 0.037 | 88.0 | 92.2 | 0.05 |
| SB pressure, mmHg (mean (SD)) | 125.54 (15.94) | 121.38 (16.37) | 3.2 × 10−5 | 132.34 (14.76) | 129.99 (13.38) | 0.03 | 131.79 (14.53) | 127.58 (14.93) | 2.3 × 10−5 | 138.99 (12.65) | 133.14 (13.93) | 1.4 × 10−11 |
| Metabolites | Women in Discovery KORA (N = 317) | Women in Replication CARLA (N = 195) | ||||
| ß (95% CI) | p-Value | pFDR | ß (95% CI) | p-Value | pFDR | |
| Ornithine | 0.14 (0.13, 0.16) | 4.6 × 10−82 | 1.1 × 10−80 | 0.24 (0.21, 0.27) | 6.9 × 10−48 | 4.0 × 10−46 |
| Arginine | −0.10 (−0.12, −0.09) | 3.2 × 10−30 | 1.5 × 10−29 | −0.06 (−0.10, −0.03) | 1.1 × 10−3 | 1.0 × 10−2 |
| Serine | 0.04 (0.02, 0.06) | 3.0× 10−4 | 5.1× 10−4 | 0.01 (0.01, 0.02) | 1.1 × 10−3 | 1.1 × 10−2 |
| Tyrosine | 0.11 (0.09, 0.13) | 1.6 × 10−36 | 9.2 × 10−36 | 0.05 (0.01, 0.08) | 6.8 × 10−3 | 4.9 × 10−2 |
| C18 | 0.03 (0.01, 0.05) | 3.7 × 10−4 | 6.2 × 10−4 | 0.03 (0.02, 0.04) | 6.1 × 10−8 | 1.2 × 10−6 |
| Metabolites | Men in Discovery KORA (N = 273) | Men in Replication CARLA (N = 191) | ||||
| ß (95% CI) | p-value | pFDR | ß (95% CI) | p-value | pFDR | |
| Ornithine | 0.14 (0.12, 0.16) | 2.7 × 10−56 | 4.2 × 10−55 | 0.22 (0.19, 0.25) | 2.8 × 10−41 | 1.6 × 10−39 |
| Arginine | −0.12 (−0.14, −0.09) | 5.1 × 10−29 | 2.4 × 10−28 | −0.07 (−0.10, −0.03) | 7.0 × 10−5 | 1.4 × 10−3 |
| PC aa C36:3 | −0.05 (−0.07, −0.03) | 2.2 × 10−8 | 4.9 × 10−8 | −0.05 (−0.08, −0.02) | 2.3 × 10−3 | 2.1 × 10−2 |
| PC ae C40:5 | −0.09 (−0.11, −0.08) | 1.2 × 10−31 | 5.6 × 10−31 | −0.06 (−0.09, −0.03) | 5.4 × 10−4 | 6.2 × 10−3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chak, C.M.; Lacruz, M.E.; Adam, J.; Brandmaier, S.; Covic, M.; Huang, J.; Meisinger, C.; Tiller, D.; Prehn, C.; Adamski, J.; et al. Ageing Investigation Using Two-Time-Point Metabolomics Data from KORA and CARLA Studies. Metabolites 2019, 9, 44. https://doi.org/10.3390/metabo9030044
Chak CM, Lacruz ME, Adam J, Brandmaier S, Covic M, Huang J, Meisinger C, Tiller D, Prehn C, Adamski J, et al. Ageing Investigation Using Two-Time-Point Metabolomics Data from KORA and CARLA Studies. Metabolites. 2019; 9(3):44. https://doi.org/10.3390/metabo9030044
Chicago/Turabian StyleChak, Choiwai Maggie, Maria Elena Lacruz, Jonathan Adam, Stefan Brandmaier, Marcela Covic, Jialing Huang, Christa Meisinger, Daniel Tiller, Cornelia Prehn, Jerzy Adamski, and et al. 2019. "Ageing Investigation Using Two-Time-Point Metabolomics Data from KORA and CARLA Studies" Metabolites 9, no. 3: 44. https://doi.org/10.3390/metabo9030044








