Metabolomics and Cheminformatics Analysis of Antifungal Function of Plant Metabolites
Abstract
:1. Introduction
2. Results and Discussion
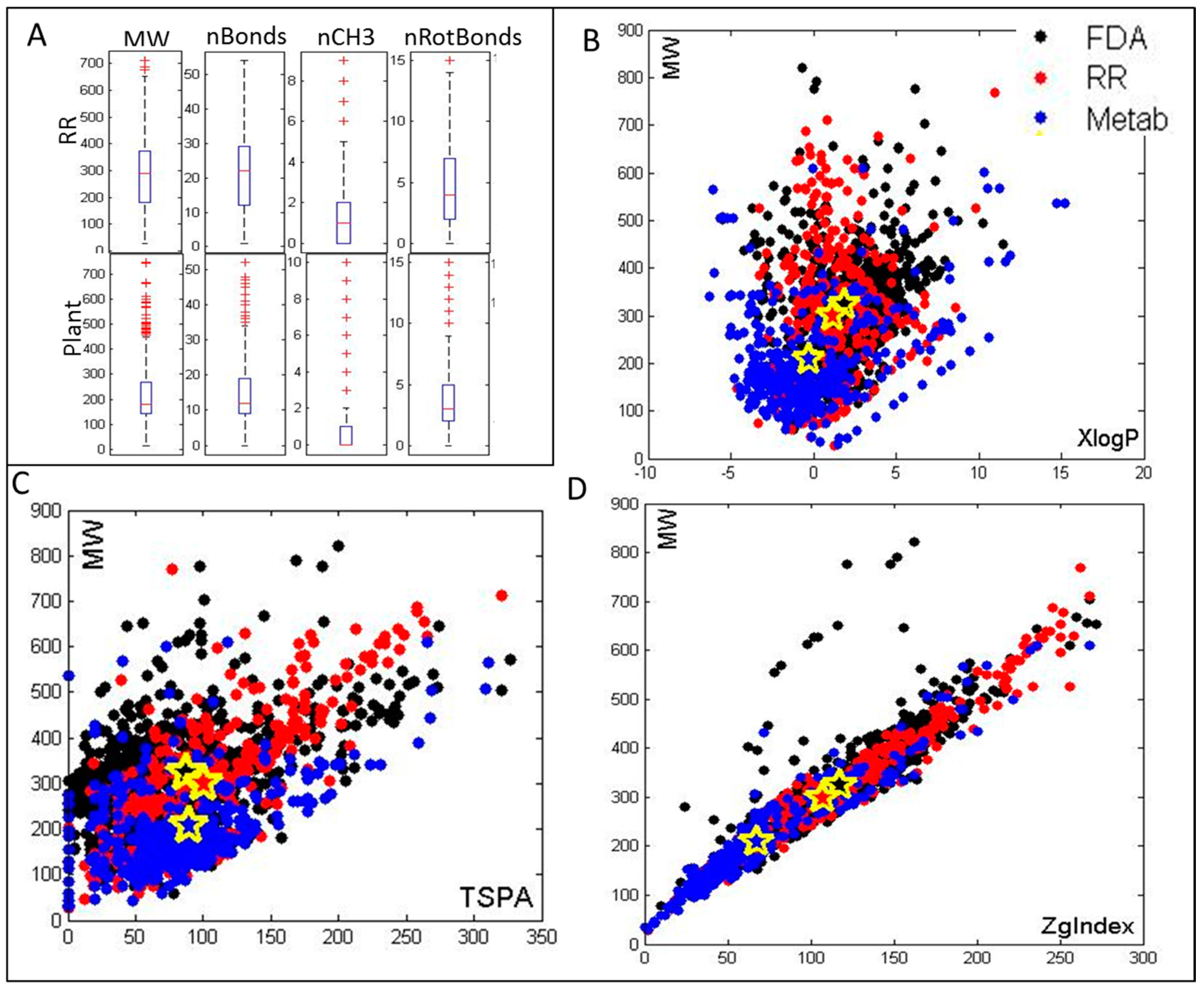
2.1. Selection of Metabolites and Cheminformatics Analysis of Their Molecular and Drug-Like Properties
2.2. Metabolite Activity
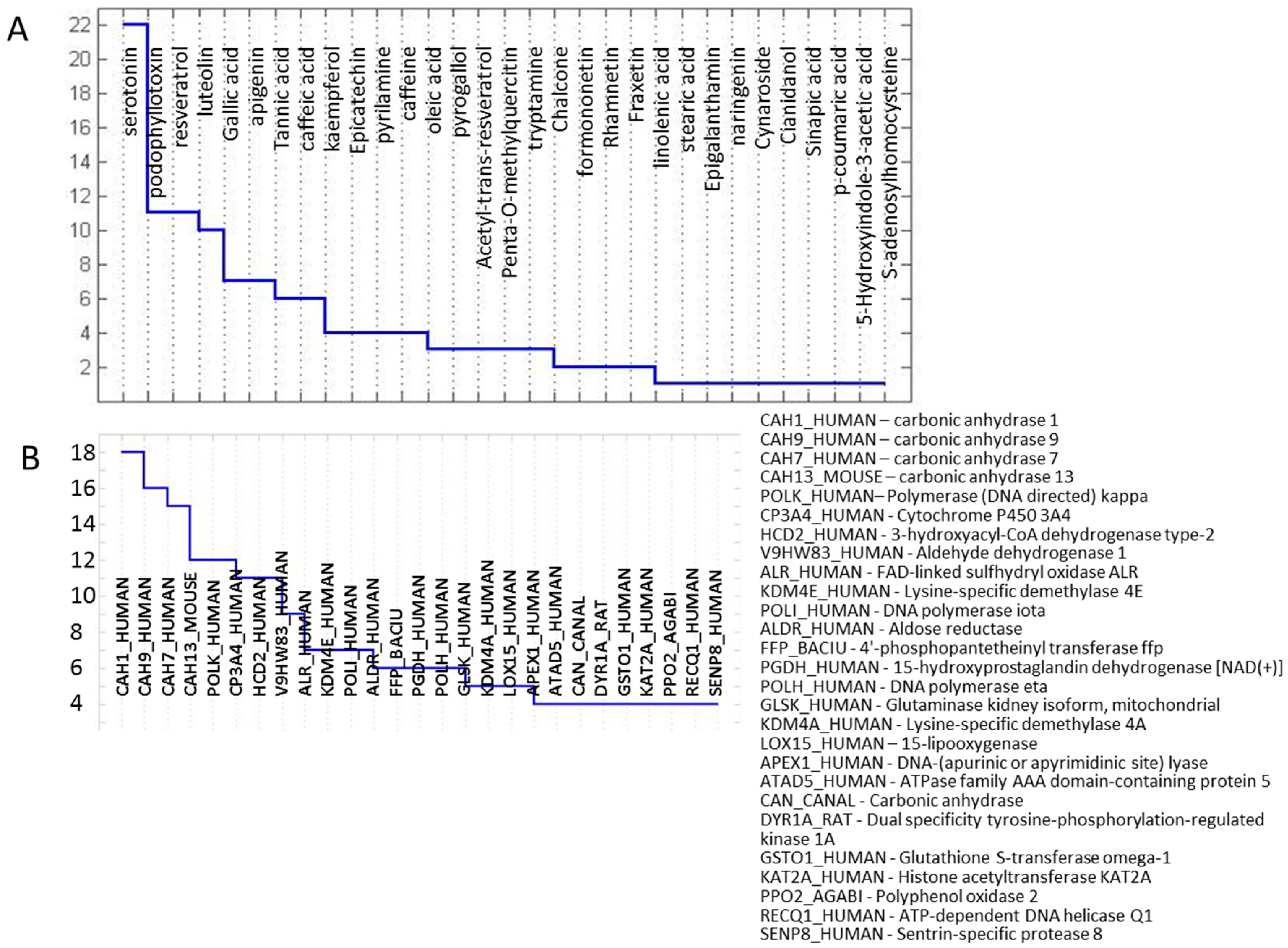
2.3. Protein Targets
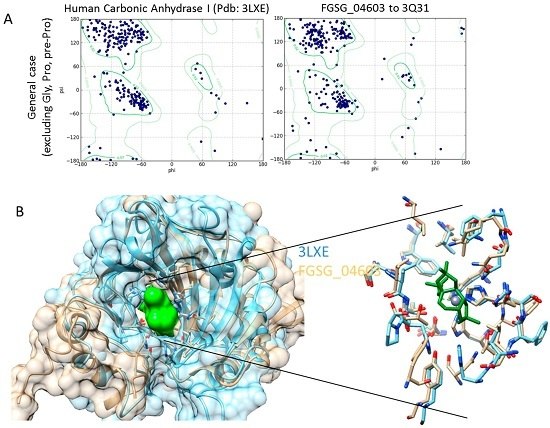
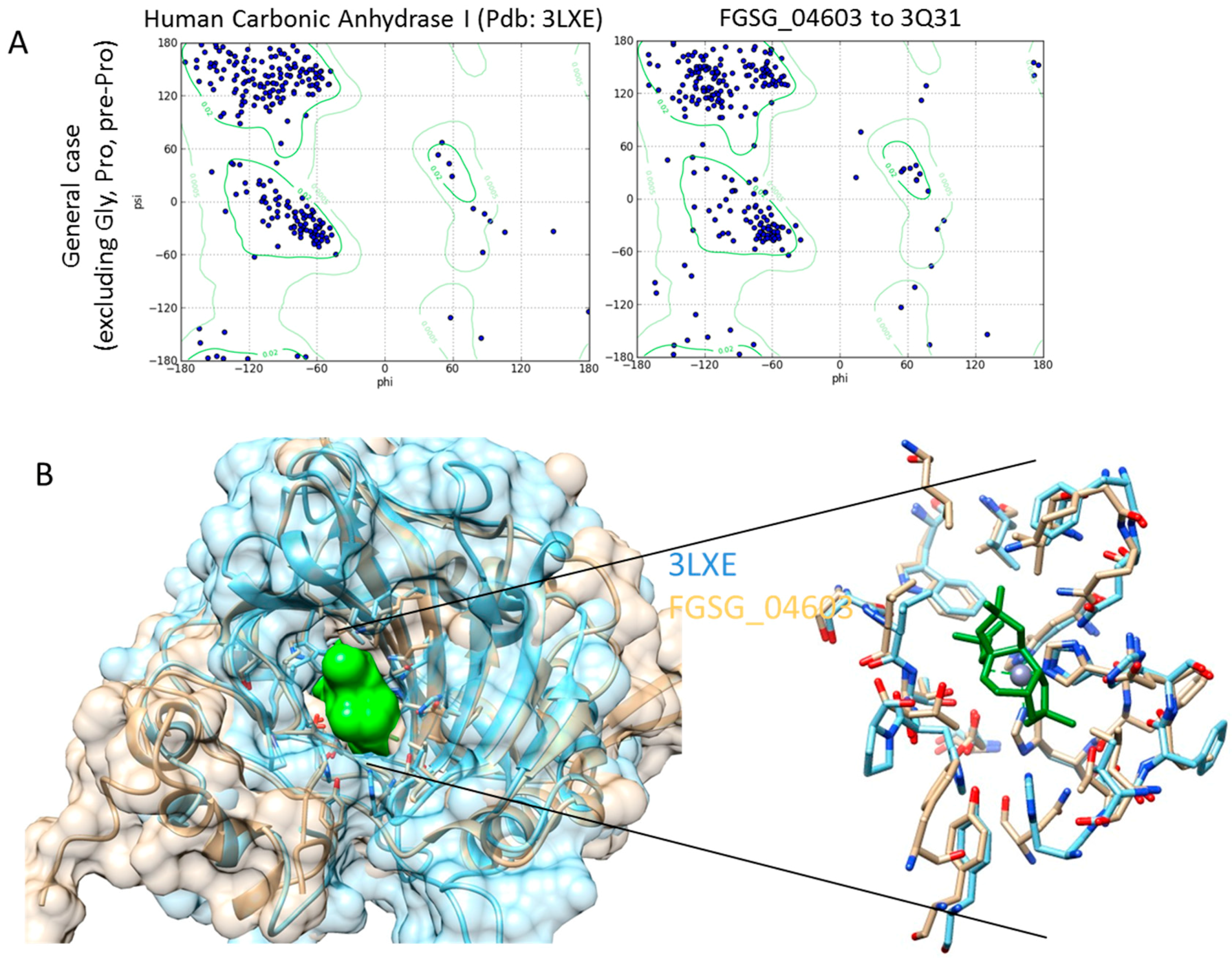
Homology Modelling and Docking Analysis—Activity of Metabolites against Fungal Targets
2.4. Experimental Analysis
3. Materials and Methods
3.1. Metabolite Characterization
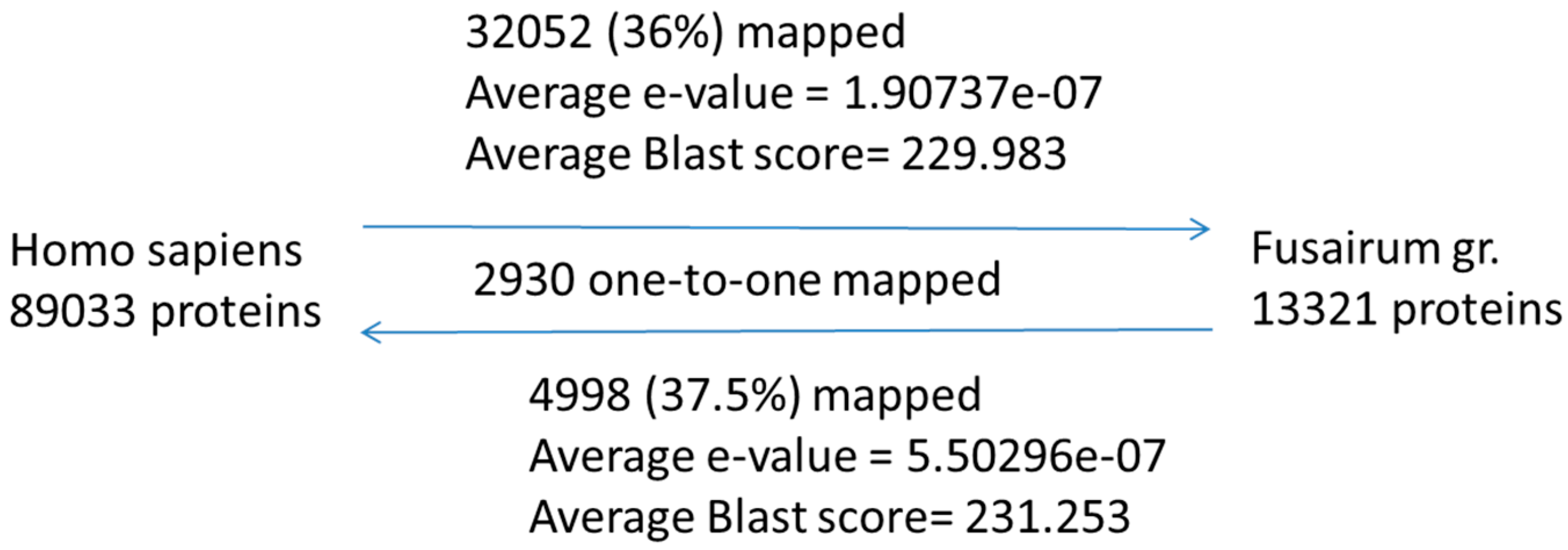
3.2. Protein Target Analysis
3.3. Sequence Comparison
3.4. Homology Modelling
3.5. Docking Analysis
3.6. Fusarium graminearum Dose-Response Growth Assays
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bai, G.; Shaner, G. Management and resistance in wheat and barley to fusarium head blight. Annu. Rev. Phytopathol. 2004, 42, 135–161. [Google Scholar] [CrossRef] [PubMed]
- Steffenson, B.J. Fusarium Head Blight of Barley: Impact, Epidemics, Management, and Strategies for Identifying and Utilizing Genetic Resistance; APS Press: St. Paul, MN, USA, 2003; pp. 241–295. [Google Scholar]
- Leonard, K.J.; Bushnell, W.R. Fusarium Head Blight of Wheat and Barley; APS Press: St. Paul, MN, USA, 2003; p. 512. [Google Scholar]
- Buerstmayr, H.; Ban, T.; Anderson, J.A. QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: A review. Plant Breed. 2009, 128. [Google Scholar] [CrossRef]
- Gunnaiah, R.; Kushalappa, A.C.; Duggavathi, R.; Fox, S.; Somers, D.J. Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Irving, H.R. Developing a model of plant hormone interactions. Plant Signal. Behav. 2011, 6, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defense responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.-F.; Johnston, A.; Balcerzak, M.; Rocheleau, H.; Harris, L.J.; Long, X.-Y.; Ouellet, T. Effect of salicylic acid on Fusarium graminearum, the major causal agent of fusarium head blight in wheat. Fungal Biol. 2012, 116, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Makandar, R.; Nalam, V.; Chaturvedi, R.; Jeannotte, R.; Sparks, A.A.; Shah, J. Involvement of salicylate and jasmonate signaling pathways in Arabidopsis interaction with Fusarium graminearum. Mol. Plant Microbe Interact. 2010, 23, 861–870. [Google Scholar] [CrossRef] [PubMed]
- Loake, G.; Grant, M. Salicylic acid in plant defence—The players and protagonists. Curr. Opin. Plant Biol. 2007, 10, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Reymond, P.; Farmer, E.E. Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1998, 1, 404–411. [Google Scholar] [CrossRef]
- Singh, B.; Usha, K. Salicylic acid induced physiological and biochemical changes in wheat seedlings under water stress. Plant Growth Regul. 2003, 39, 137–141. [Google Scholar] [CrossRef]
- Koornneef, A.; Pieterse, C.M.J. Cross talk in defense signaling. Plant Physiol. 2008, 146, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Laluk, K.; Mengiste, T. Necrotroph Attacks on Plants: Wanton Destruction or Covert Extortion? American Society of Plant Biologists: Rockville, MD, USA, 2010. [Google Scholar]
- Thaler, J.S.; Humphrey, P.T.; Whiteman, N.K. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 2012, 17, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-T.; Wu, K. Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behav. 2010, 5, 1318–1320. [Google Scholar] [CrossRef] [PubMed]
- Mutka, A.M.; Fawley, S.; Tsao, T.; Kunkel, B.N. Auxin promotes susceptibility to Pseudomonas syringae via a mechanism independent of suppression of salicylic acid-mediated defenses. Plant J. Cell Mol. Biol. 2013, 74, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Petti, C.; Reiber, K.; Ali, S.S.; Berney, M.; Doohan, F.M. Auxin as a player in the biocontrol of Fusarium head blight disease of barley and its potential as a disease control agent. BMC Plant Biol. 2012, 12. [Google Scholar] [CrossRef] [PubMed]
- Pierre-Jerome, E.; Moss, B.L.; Nemhauser, J.L. Tuning the auxin transcriptional response. J. Exp. Bot. 2013, 64, 2557–2563. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P.; Sponsel, V. A Century of Gibberellin Research. J. Plant Growth Regul. 2015, 34, 740–760. [Google Scholar] [CrossRef] [PubMed]
- Lattanzio, V.; Lattanzio, V.M.T.; Cardinali, A.; Amendola, V. Role of phenolics in the resistance mechanisms of plants against fungal pathogens and insects. Phytochemistry 2006, 37, 23–67. [Google Scholar]
- Kumaraswamy, K.G.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S. Mass spectrometry based metabolomics to identify potential biomarkers for resistance in barley against fusarium head blight (Fusarium graminearum). J. Chem. Ecol. 2011, 37, 846–856. [Google Scholar] [CrossRef] [PubMed]
- Bollina, V.; Kushalappa, A.C.; Choo, T.M.; Dion, Y.; Rioux, S. Identification of metabolites related to mechanisms of resistance in barley against Fusarium graminearum, based on mass spectrometry. Plant Mol. Biol. 2011, 77, 355–370. [Google Scholar] [CrossRef] [PubMed]
- Hamzehzarghani, H.; Kushalappa, C.; Dion, Y.; Rioux, S.; Comeau, A.; Yaylayan, V.; Marshall, W.D.; Mather, D.E. Metabolic profiling and factor analysis to discriminate quantitative resistance in wheat cultivars against fusarium head blight. Physiol. Mol. Plant Pathol. 2005, 66, 119–133. [Google Scholar] [CrossRef]
- Paranidharan, V.; Abu-Nada, Y. Resistance-related metabolites in wheat against Fusarium graminearum and the virulence factor deoxynivalenol (DON). Botany 2008, 1179, 1168–1179. [Google Scholar] [CrossRef]
- Browne, R.A.; Brindle, K. 1H NMR-based metabolite profiling as a potential selection tool for breeding passive resistance against Fusarium head blight (FHB) in wheat. Mol. Plant Pathol. 2007, 8, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Cruz, A.F.; Hamel, C.; Yang, C.; Matsubara, T.; Gan, Y.; Singh, A.K.; Kuwada, K.; Ishii, T. Phytochemicals to suppress Fusarium head blight in wheat-chickpea rotation. Phytochemistry 2012, 78, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Boutigny, A.-L.; Atanasova-Pénichon, V.; Benet, M.; Barreau, C.; Richard-Forget, F. Natural phenolic acids from wheat bran inhibit Fusarium culmorum trichothecene biosynthesis in vitro by repressing Tri gene expression. Eur. J. Plant Pathol. 2010, 127, 275–286. [Google Scholar] [CrossRef]
- Boutigny, A.-L.; Barreau, C.; Atanasova-Penichon, V.; Verdal-Bonnin, M.-N.; Pinson-Gadais, L.; Richard-Forget, F. Ferulic acid, an efficient inhibitor of type B trichothecene biosynthesis and Tri gene expression in Fusarium liquid cultures. Mycol. Res. 2009, 113, 746–753. [Google Scholar] [CrossRef] [PubMed]
- Knudson, L. Tannic acid fermentation. I. J. Biol. Chem. 1913, 14, 159–184. [Google Scholar]
- Henis, Y.; Tagari, H.; Volcani, R. Effect of water extracts of carob pods, tannic acid, and their derivatives on the morphology and growth of microorganisms. Appl. Microbiol. Biotechnol. 1964, 12, 204–209. [Google Scholar]
- McKeehen, J.D.; Busch, R.H.; Fulcher, R.G. Evaluation of wheat (Triticum aestivum L.) phenolic acids during grain development and their contribution to Fusarium resistance. J. Agric. Food Chem. 1999, 47, 1476–1482. [Google Scholar] [CrossRef] [PubMed]
- Gunnaiah, R.; Kushalappa, A.C. Metabolomics deciphers the host resistance mechanisms in wheat cultivar Sumai-3, against trichothecene producing and non-producing isolates of Fusarium graminearum. Plant Physiol. Biochem. 2014, 83, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Forrer, H.-R.; Musa, T.; Schwab, F.; Jenny, E.; Bucheli, T.D.; Wettstein, F.E.; Vogelgsang, S. Fusarium head blight control and prevention of mycotoxin contamination in wheat with botanicals and tannic acid. Toxins 2014, 6, 830–849. [Google Scholar] [CrossRef] [PubMed]
- Afendi, F.M.; Okada, T.; Yamazaki, M.; Hirai-Morita, A.; Nakamura, Y.; Nakamura, K.; Ikeda, S.; Takahashi, H.; Altaf-Ul-Amin, M.; Darusman, L.K.; et al. KNApSAcK Family Databases: Integrated Metabolite-Plant Species Databases for Multifaceted Plant Research. Plant Cell Physiol. 2012, 53, e1. [Google Scholar] [CrossRef] [PubMed]
- Pasquet, J.-C.; Chaouch, S.; Macadré, C.; Balzergue, S.; Huguet, S.; Martin-Magniette, M.-L.; Bellvert, F.; Deguercy, X.; Thareau, V.; Heintz, D.; et al. Differential gene expression and metabolomic analyses of Brachypodium distachyon infected by deoxynivalenol producing and non-producing strains of Fusarium graminearum. BMC Genom. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Chamarthi, S.K.; Kumar, K.; Gunnaiah, R.; Kushalappa, A.C.; Dion, Y.; Choo, T.M. Identification of fusarium head blight resistance related metabolites specific to doubled-haploid lines in barley. Eur. J. Plant Pathol. 2014, 138, 67–78. [Google Scholar] [CrossRef]
- PubChem. Available online: http://pubchem.ncbi.nlm.nih.gov (accessed on 29 September 2016).
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2009, 37, D603–D610. [Google Scholar] [CrossRef] [PubMed]
- Hastings, J.; Owen, G.; Dekker, A.; Ennis, M.; Kale, N.; Muthukrishnan, V.; Turner, S.; Swainston, N.; Mendes, P.; Steinbeck, C. ChEBI in 2016: Improved services and an expanding collection of metabolites. Nucleic Acids Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmüller, E.; Dörmann, P.; Weckwerth, W.; Gibon, Y.; Stitt, M.; et al. GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Cruz, I.; Cheetham, J.J.; Arnason, J.T.; Yack, J.E.; Smith, M.L. Alkamides from Echinacea distrupt the fungal cell wall-membrane complex. Phytomedicine 2014, 2, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Bolstad, E.S.; Coleman, R.G. ZINC: A free tool to discover chemistry for biology. J. Chem. Inf. Model. 2012, 52, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- The R Project for Statistical Computing. Available online: http://www.R-project.org/ (accessed on 30 September 2016).
- Ertl, R.; Rohde, B.; Selzer, P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 2000, 43, 3714–3717. [Google Scholar] [CrossRef] [PubMed]
- Palm, K.; Stenberg, P.; Luthman, K.; Artursson, P. Polar molecular surface properties predict the intestinal absorption of drugs in humans. Pharm. Res. 1997, 14, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.; Gattass, C.R. Topological polar surface area defines substrate transport by multidrug resistance associated protein 1 (MRP1/ABCC1). J. Med. Chem. 2009, 52, 1214–1218. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Jin, X.; Jia, X.; Wang, H.; Cao, A.; Zhao, W.; Pei, H.; Xue, Z.; He, L.; Chen, Q.; et al. Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genom. 2013, 14, 197. [Google Scholar] [CrossRef] [PubMed]
- Gutman, I.; Das, K.C. The first Zagreb index 30 years after. MATCDY 2004, 50, 83–92. [Google Scholar]
- Van Rhee, A.M. Method for Producing Chemical Libraries Enhanced with Biologically Active Molecules. U.S. Patent 20030120430 A1, 26 June 2003. [Google Scholar]
- Available online: ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/pan_proteomes/ (accessed on 29 September 2016).
- Kang, J.; Park, J.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC Transporters. Arabidopsis Book Am. Soc. Plant Biol. 2011, 9, e0153. [Google Scholar] [CrossRef] [PubMed]
- Pelagio-Flores, R.; Ortíz-Castro, R.; Méndez-Bravo, A.; Macías-Rodríguez, L.; López-Bucio, J. Serotonin, a tryptophan-derived signal conserved in plants and animals, regulates root system architecture probably acting as a natural auxin inhibitor in Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 490–508. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, A.; Hashimoto, Y.; Tanaka, C.; Dubouzet, J.G.; Nakao, T.; Matsuda, F.; Nishioka, T.; Miyagawa, H.; Wakasa, K. The tryptophan pathway is involved in the defense responses of rice against pathogenic infection via serotonin production. Plant J. 2008, 54, 481–495. [Google Scholar] [CrossRef] [PubMed]
- Broad Institute. Available online: http://www.broadinstitute.org/ (accessed on 29 September 2016).
- Iyer, R.; Barrese, A.A.; Parakh, S.; Parker, C.N.; Tripp, B.C. Inhibition profiling of human carbonic anhydrase II by high-throughput screening of structurally diverse, biologically active compounds. J. Biomol. Screen. 2006, 11, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Sarikaya, S.B.; Gülçin, I.; Supuran, C.T. Carbonic anhydrase inhibitors: Inhibition of human erythrocyte isozymes I and II with a series of phenolic acids. Chem. Biol. Drug Des. 2010, 75, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. Bacterial, fungal and protozoan carbonic anhydrases as drug targets. Expert Opin. Ther. Targets 2015, 19, 1689–1704. [Google Scholar] [CrossRef] [PubMed]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL Workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30 (Suppl. S1), 162–173. [Google Scholar] [CrossRef] [PubMed]
- Jacob, R.B.; Andersen, T.; McDougal, O.M. Accessible high-throughput virtual screening molecular docking software for students and educators. PLoS Comput. Biol. 2012, 8, e1002499. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Olson, A.J. Using AutoDock for Ligand-receptor docking. Curr. Protoc. Bioinform. 2008. [Google Scholar] [CrossRef]
- Cao, Y.; Charisi, A.; Cheng, L.C.; Jiang, T.; Girke, T. ChemmineR: A compound mining framework for R. Bioinformatics 2008, 24, 1733–1734. [Google Scholar] [CrossRef] [PubMed]
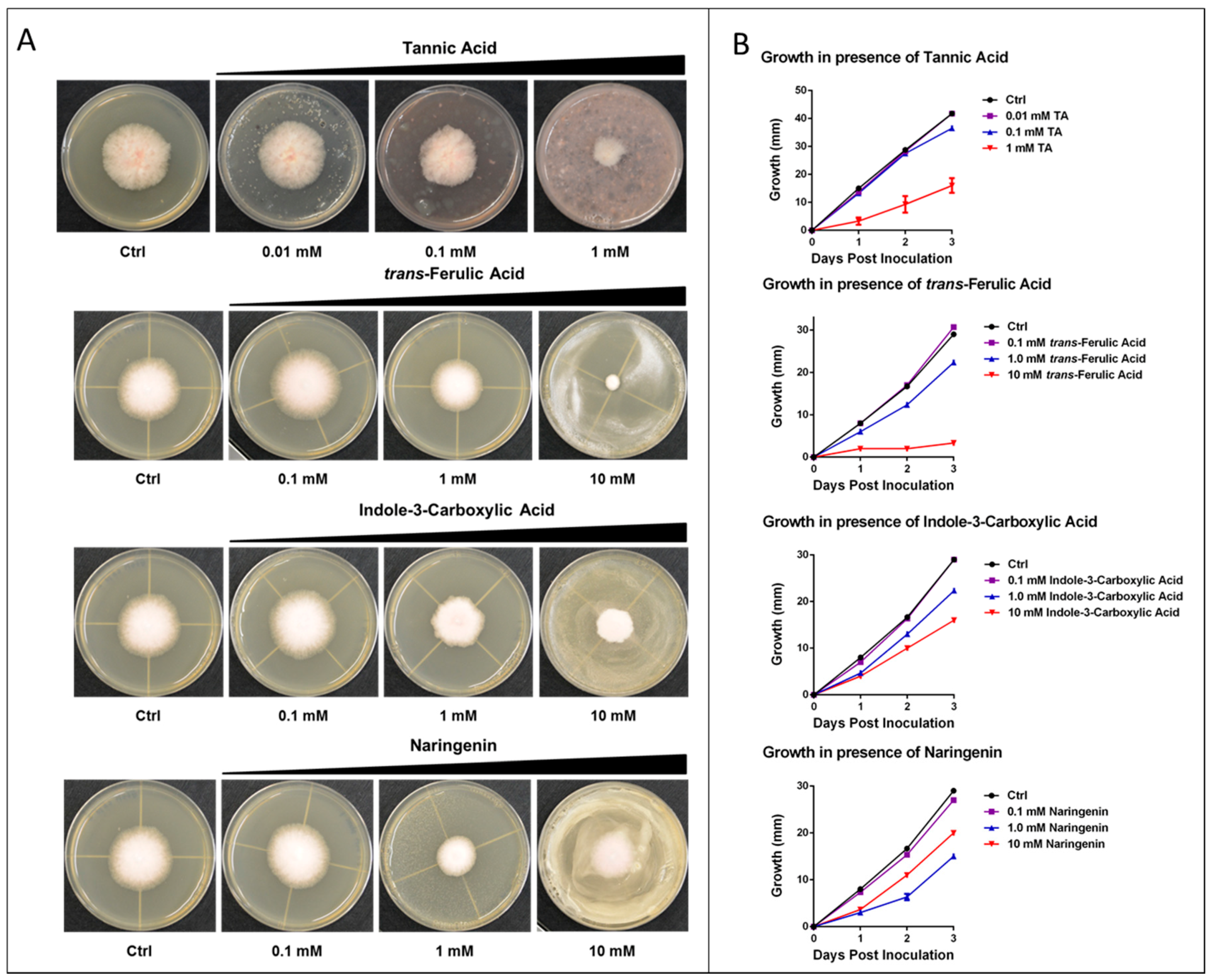
| Compound | IC50 (mM) |
|---|---|
| Tannic Acid | 0.58 |
| trans-Ferulic Acid | 3.1 |
| Naringenin | ~ 1 |
| indole-3-carboxylic acid | > 10 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuperlovic-Culf, M.; Rajagopalan, N.; Tulpan, D.; Loewen, M.C. Metabolomics and Cheminformatics Analysis of Antifungal Function of Plant Metabolites. Metabolites 2016, 6, 31. https://doi.org/10.3390/metabo6040031
Cuperlovic-Culf M, Rajagopalan N, Tulpan D, Loewen MC. Metabolomics and Cheminformatics Analysis of Antifungal Function of Plant Metabolites. Metabolites. 2016; 6(4):31. https://doi.org/10.3390/metabo6040031
Chicago/Turabian StyleCuperlovic-Culf, Miroslava, NandhaKishore Rajagopalan, Dan Tulpan, and Michele C. Loewen. 2016. "Metabolomics and Cheminformatics Analysis of Antifungal Function of Plant Metabolites" Metabolites 6, no. 4: 31. https://doi.org/10.3390/metabo6040031
APA StyleCuperlovic-Culf, M., Rajagopalan, N., Tulpan, D., & Loewen, M. C. (2016). Metabolomics and Cheminformatics Analysis of Antifungal Function of Plant Metabolites. Metabolites, 6(4), 31. https://doi.org/10.3390/metabo6040031