Exploration of Blood Metabolite Signatures of Colorectal Cancer and Polyposis through Integrated Statistical and Network Analysis
Abstract
:1. Introduction
2. Material and Methods
2.1. Dataset Description
2.2. Data Pre-Processing
2.2.1. Data Overview and Normalization
2.3. Univariate Analysis
2.4. Multivariate Analysis
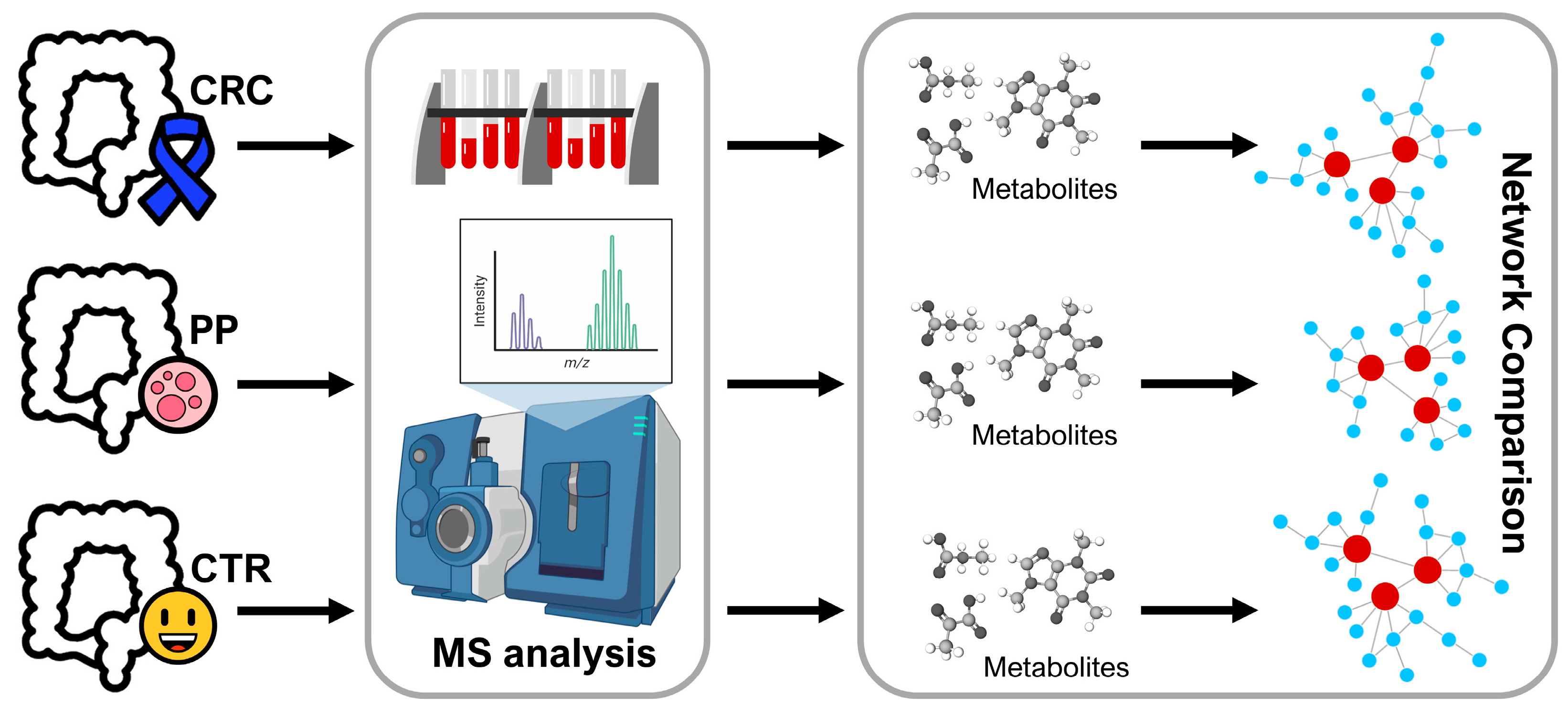
2.5. Network Analysis
2.5.1. Reconstruction of Metabolite-Metabolite Association Network
2.5.2. Pathway Enrichment Analysis
2.5.3. Measures for Network Topology
2.6. Software
3. Results
3.1. Exploratory Analysis of Serum Metabolomic Profiles of CRC, Polyposis, and Healthy Patients
3.2. Classification of CRC, PP, and CTR
3.3. Analysis of Metabolite-Metabolite Association Networks Specific to CRC, PP, and CTR
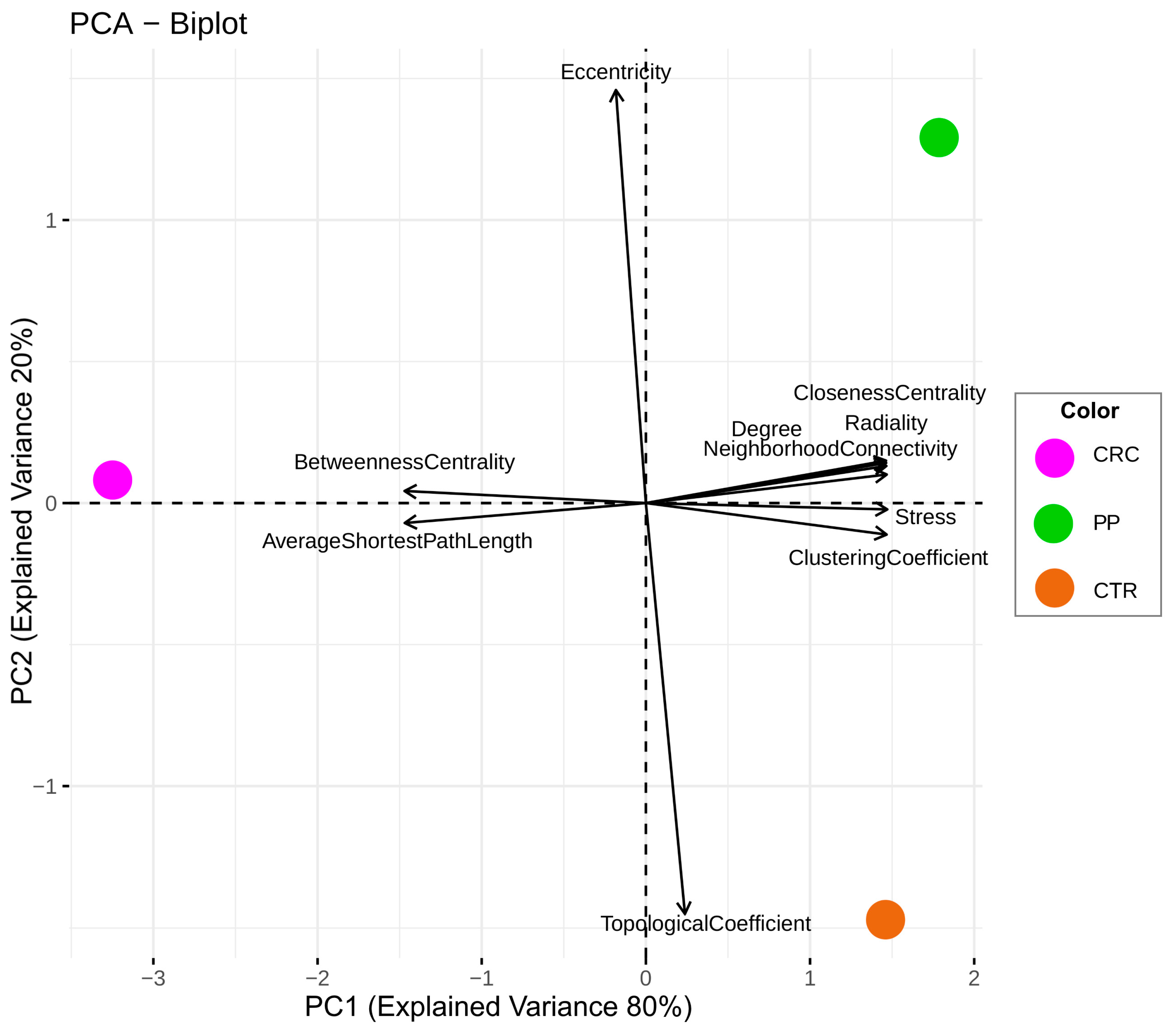
3.4. Comparison of the Topological Properties of the Metabolite-Metabolite Association Network
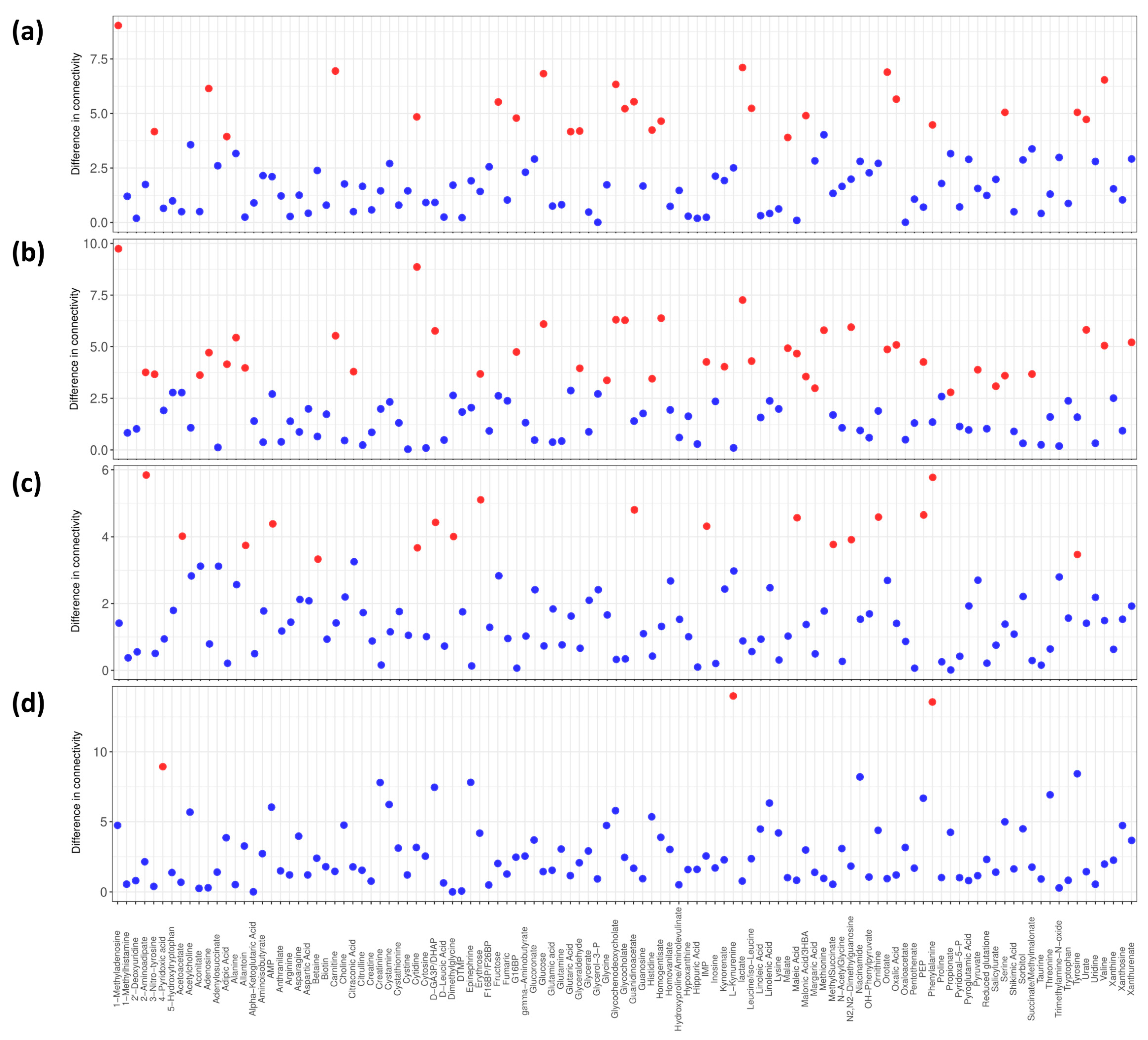
3.5. Differential Connectivity Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Rajani, C.; Jia, W. The Distinctive Serum Metabolomes of Gastric, Esophageal and Colorectal Cancers. Cancers 2021, 13, 720. [Google Scholar] [CrossRef] [PubMed]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet Lond. Engl. 2019, 394, 1467–1480. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.; Desantis, C.; Jemal, A. Colorectal cancer statistics, 2014. CA. Cancer J. Clin. 2014, 64, 104–117. [Google Scholar] [CrossRef]
- Nannini, G.; Meoni, G.; Amedei, A.; Tenori, L. Metabolomics profile in gastrointestinal cancers: Update and future perspectives. World J. Gastroenterol. 2020, 26, 2514–2532. [Google Scholar] [CrossRef]
- Issa, I.A.; Noureddine, M. Colorectal cancer screening: An updated review of the available options. World J. Gastroenterol. 2017, 23, 5086–5096. [Google Scholar] [CrossRef]
- Manne, U.; Shanmugam, C.; Katkoori, V.R.; Bumpers, H.L.; Grizzle, W.E. Development and progression of colorectal neoplasia. Cancer Biomark. 2011, 9, 235–265. [Google Scholar] [CrossRef] [Green Version]
- Aceto, G.M.; Catalano, T.; Curia, M.C. Molecular Aspects of Colorectal Adenomas: The Interplay among Microenvironment, Oxidative Stress, and Predisposition. BioMed Res. Int. 2020, 2020, 1726309. [Google Scholar] [CrossRef] [Green Version]
- Haggar, F.A.; Preen, D.B.; Pereira, G.; Holman, C.D.J.; Einarsdottir, K. Cancer incidence and mortality trends in Australian adolescents and young adults, 1982-2007. BMC Cancer 2012, 12, 151. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Wang, S.; Lee, J.J.-K.; Lee, S.; Lee, E.; Shinbrot, E.; Wheeler, D.A.; Kucherlapati, R.; Park, P.J. An enhanced genetic model of colorectal cancer progression history. Genome Biol. 2019, 20, 168. [Google Scholar] [CrossRef] [Green Version]
- Vignoli, A.; Muraro, E.; Miolo, G.; Tenori, L.; Turano, P.; Di Gregorio, E.; Steffan, A.; Luchinat, C.; Corona, G. Effect of Estrogen Receptor Status on Circulatory Immune and Metabolomics Profiles of HER2-Positive Breast Cancer Patients Enrolled for Neoadjuvant Targeted Chemotherapy. Cancers 2020, 12, 314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Backshall, A.; Sharma, R.; Clarke, S.J.; Keun, H.C. Pharmacometabonomic profiling as a predictor of toxicity in patients with inoperable colorectal cancer treated with capecitabine. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2011, 17, 3019–3028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vignoli, A.; Risi, E.; McCartney, A.; Migliaccio, I.; Moretti, E.; Malorni, L.; Luchinat, C.; Biganzoli, L.; Tenori, L. Precision Oncology via NMR-Based Metabolomics: A Review on Breast Cancer. Int. J. Mol. Sci. 2021, 22, 4687. [Google Scholar] [CrossRef] [PubMed]
- Di Donato, S.; Vignoli, A.; Biagioni, C.; Malorni, L.; Mori, E.; Tenori, L.; Calamai, V.; Parnofiello, A.; Di Pierro, G.; Migliaccio, I.; et al. A Serum Metabolomics Classifier Derived from Elderly Patients with Metastatic Colorectal Cancer Predicts Relapse in the Adjuvant Setting. Cancers 2021, 13, 2762. [Google Scholar] [CrossRef]
- Rosato, A.; Tenori, L.; Cascante, M.; De Atauri Carulla, P.R.; Martins dos Santos, V.A.P.; Saccenti, E. From correlation to causation: Analysis of metabolomics data using systems biology approaches. Metabolomics 2018, 14, 37. [Google Scholar] [CrossRef] [Green Version]
- Saccenti, E.; Suarez-Diez, M.; Luchinat, C.; Santucci, C.; Tenori, L. Probabilistic Networks of Blood Metabolites in Healthy Subjects As Indicators of Latent Cardiovascular Risk. J. Proteome Res. 2015, 14, 1101–1111. [Google Scholar] [CrossRef]
- Vignoli, A.; Tenori, L.; Luchinat, C.; Saccenti, E. Differential Network Analysis Reveals Molecular Determinants Associated with Blood Pressure and Heart Rate in Healthy Subjects. J. Proteome Res. 2021, 20, 1040–1051. [Google Scholar] [CrossRef]
- Vignoli, A.; Tenori, L.; Giusti, B.; Valente, S.; Carrabba, N.; Baizi, D.; Barchielli, A.; Marchionni, N.; Gensini, G.F.; Marcucci, R.; et al. Differential Network Analysis Reveals Metabolic Determinants Associated with Mortality in Acute Myocardial Infarction Patients and Suggests Potential Mechanisms Underlying Different Clinical Scores Used To Predict Death. J. Proteome Res. 2020, 19, 949–961. [Google Scholar] [CrossRef]
- Balder, Y.; Vignoli, A.; Tenori, L.; Luchinat, C.; Saccenti, E. Exploration of Blood Lipoprotein and Lipid Fraction Profiles in Healthy Subjects through Integrated Univariate, Multivariate, and Network Analysis Reveals Association of Lipase Activity and Cholesterol Esterification with Sex and Age. Metabolites 2021, 11, 326. [Google Scholar] [CrossRef]
- Zhu, J.; Djukovic, D.; Deng, L.; Gu, H.; Himmati, F.; Chiorean, E.G.; Raftery, D. Colorectal Cancer Detection Using Targeted Serum Metabolic Profiling. J. Proteome Res. 2014, 13, 4120–4130. [Google Scholar] [CrossRef]
- Ludwig, O. Blom, Gunnar: Statistical estimates and transformed beta-variables. Wiley/New York, Almquist und Wiksell/Stockholm 1958; 176 S., Kr. 20,—. Biom. Z. 1961, 3, 285. [Google Scholar] [CrossRef]
- van der Vaart, A.W. Asymptotic Statistics; Cambridge University Press: Cambridge UK, 2000. [Google Scholar]
- Bliss, C.I. The Method of Probits. Science 1934, 79, 38–39. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Student, B. The Probable Error of a Mean. Biometrika 1908, 6, 1–25. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Hotelling, H. Analysis of a complex of statistical variables into principal components. J. Educ. Psychol. 1933, 24, 417–441. [Google Scholar] [CrossRef]
- Wall, M.E.; Rechtsteiner, A.; Rocha, L.M. Singular Value Decomposition and Principal Component Analysis. In A Practical Approach to Microarray Data Analysis; Berrar, D.P., Dubitzky, W., Granzow, M., Eds.; Springer US: Boston, MA, USA, 2003; pp. 91–109. ISBN 978-0-306-47815-4. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Ali, J.; Khan, R.; Ahmad, N.; Maqsood, I. Random Forests and Decision Trees. Int. J. Comput. Sci. IssuesIJCSI 2012, 9, 272. [Google Scholar]
- Cheng, S.; Shah, S.H.; Corwin, E.J.; Fiehn, O.; Fitzgerald, R.L.; Gerszten, R.E.; Illig, T.; Rhee, E.P.; Srinivas, P.R.; Wang, T.J.; et al. Potential Impact and Study Considerations of Metabolomics in Cardiovascular Health and Disease A Scientific Statement From the American Heart Association. Circ. Cardiovasc. Genet. 2017, 10, e000032. [Google Scholar] [CrossRef] [Green Version]
- Amaratunga, D.; Cabrera, J.; Lee, Y.-S. Enriched random forests. Bioinformatics 2008, 24, 2010–2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Cesare, F.; Tenori, L.; Luchinat, C.; Saccenti, E. Association of Plasma Metabolites and Lipoproteins with Rh and ABO Blood Systems in Healthy Subjects. J. Proteome Res. 2022, 21, 2655–2663. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Diez, M.; Saccenti, E. Effects of Sample Size and Dimensionality on the Performance of Four Algorithms for Inference of Association Networks in Metabonomics. J. Proteome Res. 2015, 14, 5119–5130. [Google Scholar] [CrossRef]
- Akhand, M.A.H.; Nandi, R.N.; Amran, S.M.; Murase, K. Context likelihood of relatedness with maximal information coefficient for Gene Regulatory Network inference. In Proceedings of the 2015 18th International Conference on Computer and Information Technology (ICCIT), Dhaka, Bangladesh, 21–23 December 2015; pp. 312–316. [Google Scholar]
- Jahagirdar, S.; Suarez-Diez, M.; Saccenti, E. Simulation and Reconstruction of Metabolite-Metabolite Association Networks Using a Metabolic Dynamic Model and Correlation Based Algorithms. J. Proteome Res. 2019, 18, 1099–1113. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Wieder, C.; Frainay, C.; Poupin, N.; Rodríguez-Mier, P.; Vinson, F.; Cooke, J.; Lai, R.P.; Bundy, J.G.; Jourdan, F.; Ebbels, T. Pathway analysis in metabolomics: Recommendations for the use of over-representation analysis. PLoS Comput. Biol. 2021, 17, e1009105. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Assenov, Y.; Domingues, F.S.; Albrecht, M. Topological analysis and interactive visualization of biological networks and protein structures. Nat. Protoc. 2012, 7, 670–685. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- McCaw, Z.R.; Lane, J.M.; Saxena, R.; Redline, S.; Lin, X. Operating characteristics of the rank-based inverse normal transformation for quantitative trait analysis in genome-wide association studies. Biometrics 2020, 76, 1262–1272. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Levine, J.S.; Ahnen, D.J. Clinical practice. Adenomatous polyps of the colon. N. Engl. J. Med. 2006, 355, 2551–2557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, L.; Guo, F.; Cheng, K.-K.; Zhu, J.; Gu, H.; Raftery, D.; Dong, J. Identifying Significant Metabolic Pathways Using Multi-Block Partial Least-Squares Analysis. J. Proteome Res. 2020, 19, 1965–1974. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Xu, Z.; Zhu, X.; Zhang, J. New insights into molecules and pathways of cancer metabolism and therapeutic implications. Cancer Commun. 2021, 41, 16–36. [Google Scholar] [CrossRef]
- La Vecchia, S.; Sebastián, C. Metabolic pathways regulating colorectal cancer initiation and progression. Semin. Cell Dev. Biol. 2020, 98, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Gowda, G.A.N.; Zhu, J.; Deng, L.; Gu, H.; Chiorean, E.G.; Zaid, M.A.; Harrison, M.; Zhang, D.; Zhang, M.; et al. Altered metabolite levels and correlations in patients with colorectal cancer and polyps detected using seemingly unrelated regression analysis. Metabolomics 2017, 13, 125. [Google Scholar] [CrossRef]
- Neurauter, G.; Grahmann, A.V.; Klieber, M.; Zeimet, A.; Ledochowski, M.; Sperner-Unterweger, B.; Fuchs, D. Serum phenylalanine concentrations in patients with ovarian carcinoma correlate with concentrations of immune activation markers and of isoprostane-8. Cancer Lett. 2008, 272, 141–147. [Google Scholar] [CrossRef]
- Ploder, M.; Neurauter, G.; Spittler, A.; Schroecksnadel, K.; Roth, E.; Fuchs, D. Serum phenylalanine in patients post trauma and with sepsis correlate to neopterin concentrations. Amino Acids 2008, 35, 303–307. [Google Scholar] [CrossRef]
- Qiu, Y.; Cai, G.; Su, M.; Chen, T.; Zheng, X.; Xu, Y.; Ni, Y.; Zhao, A.; Xu, L.X.; Cai, S.; et al. Serum metabolite profiling of human colorectal cancer using GC-TOFMS and UPLC-QTOFMS. J. Proteome Res. 2009, 8, 4844–4850. [Google Scholar] [CrossRef]
- Tan, B.; Qiu, Y.; Zou, X.; Chen, T.; Xie, G.; Cheng, Y.; Dong, T.; Zhao, L.; Feng, B.; Hu, X.; et al. Metabonomics Identifies Serum Metabolite Markers of Colorectal Cancer. J. Proteome Res. 2013, 12, 3000–3009. [Google Scholar] [CrossRef] [Green Version]
- Deng, L.; Ma, L.; Cheng, K.-K.; Xu, X.; Raftery, D.; Dong, J. Sparse PLS-Based Method for Overlapping Metabolite Set Enrichment Analysis. J. Proteome Res. 2021, 20, 3204–3213. [Google Scholar] [CrossRef]
- Yamada, H.; Kumagai, H. Synthesis of L-Tyrosine-Related Amino Acids by β-Tyrosinase11Systematic name: Tyrosine phenol-lyase (deaminating) (EC 4.1.99.2). In Advances in Applied Microbiology; Perlman, D., Ed.; Academic Press: Cambridge, MA, USA, 1975; Volume 19, pp. 249–288. [Google Scholar]
- Leichtle, A.B.; Nuoffer, J.-M.; Ceglarek, U.; Kase, J.; Conrad, T.; Witzigmann, H.; Thiery, J.; Fiedler, G.M. Serum amino acid profiles and their alterations in colorectal cancer. Metabolomics 2012, 8, 643–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiggins, T.; Kumar, S.; Markar, S.R.; Antonowicz, S.; Hanna, G.B. Tyrosine, Phenylalanine, and Tryptophan in Gastroesophageal Malignancy: A Systematic Review. Cancer Epidemiol. Prev. Biomark. 2015, 24, 32–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Contorno, S.; Darienzo, R.E.; Tannenbaum, R. Evaluation of aromatic amino acids as potential biomarkers in breast cancer by Raman spectroscopy analysis. Sci. Rep. 2021, 11, 1698. [Google Scholar] [CrossRef] [PubMed]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of ‘small-world’ networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Blumer, A.; Lee, K. An algorithm for modularity analysis of directed and weighted biological networks based on edge-betweenness centrality. Bioinformatics 2006, 22, 3106–3108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freeman, L.C. Centrality in social networks conceptual clarification. Soc. Netw. 1978, 1, 215–239. [Google Scholar] [CrossRef] [Green Version]
- Barabási, A.-L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef]
- Ravasz, E.; Somera, A.L.; Mongru, D.A.; Oltvai, Z.N.; Barabási, A.-L. Hierarchical Organization of Modularity in Metabolic Networks. Science 2002, 297, 1551–1555. [Google Scholar] [CrossRef] [Green Version]
- Maslov, S.; Sneppen, K. Specificity and stability in topology of protein networks. Science 2002, 296, 910–913. [Google Scholar] [CrossRef] [Green Version]
- Brandes, U. A faster algorithm for betweenness centrality. J. Math. Sociol. 2001, 25, 163–177. [Google Scholar] [CrossRef]
- Stelzl, U.; Worm, U.; Lalowski, M.; Haenig, C.; Brembeck, F.H.; Goehler, H.; Stroedicke, M.; Zenkner, M.; Schoenherr, A.; Koeppen, S.; et al. A Human Protein-Protein Interaction Network: A Resource for Annotating the Proteome. Cell 2005, 122, 957–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Mean Accuracy % (95% CI, p-Value) | Mean Specificity % (95% CI, p-Value) | Mean Sensitivity % (95% CI, p-Value) | AUC (95% CI, p-Value) | |
|---|---|---|---|---|
| CRC vs. CTR | 78.2 (77.8–78.5, 0.001) | 79.0 (78.6–79.3, 0.001) | 77.1 (76.6–77.7, 0.001) | 0.875 (0.873–0.877, 0.001) |
| CRC vs. PP | 79.5 (79.2–79.9, 0.001) | 77.7 (77.1–78.3, 0.002) | 81.2 (70.8–81.7, 0.003) | 0.871 (0.869–0.872, 0.001) |
| PP vs. CTR | 62.2 (61.8–62.6, 0.004) | 63.1 (62.3–63.8, 0.001) | 61.5 (60.9–63.8, 0.012) | 0.661 (0.658–0.665, 0.004) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Cesare, F.; Vignoli, A.; Luchinat, C.; Tenori, L.; Saccenti, E. Exploration of Blood Metabolite Signatures of Colorectal Cancer and Polyposis through Integrated Statistical and Network Analysis. Metabolites 2023, 13, 296. https://doi.org/10.3390/metabo13020296
Di Cesare F, Vignoli A, Luchinat C, Tenori L, Saccenti E. Exploration of Blood Metabolite Signatures of Colorectal Cancer and Polyposis through Integrated Statistical and Network Analysis. Metabolites. 2023; 13(2):296. https://doi.org/10.3390/metabo13020296
Chicago/Turabian StyleDi Cesare, Francesca, Alessia Vignoli, Claudio Luchinat, Leonardo Tenori, and Edoardo Saccenti. 2023. "Exploration of Blood Metabolite Signatures of Colorectal Cancer and Polyposis through Integrated Statistical and Network Analysis" Metabolites 13, no. 2: 296. https://doi.org/10.3390/metabo13020296







