Modulation of MicroRNAs and Exosomal MicroRNAs after Dietary Interventions for Obesity and Insulin Resistance: A Narrative Review
Abstract
:1. Introduction
2. Biogenesis and Mechanism of Action of miRNAs
3. Circulating miRNAs: Novel Cell-Cell Communication
3.1. Extracellular Vesicles: Exosomes
3.2. miRNA-Binding Proteins and Lipoproteins
4. miRNAs in Obesity and Insulin Resistance
4.1. Obesity
4.2. Insulin Resistance
5. Regulation of miRNAs by Diet
5.1. Energy Restriction
5.2. High-Protein Diet
5.3. Ketogenic Diet
5.4. Diet with Supplementation
5.5. Other Related Interventions
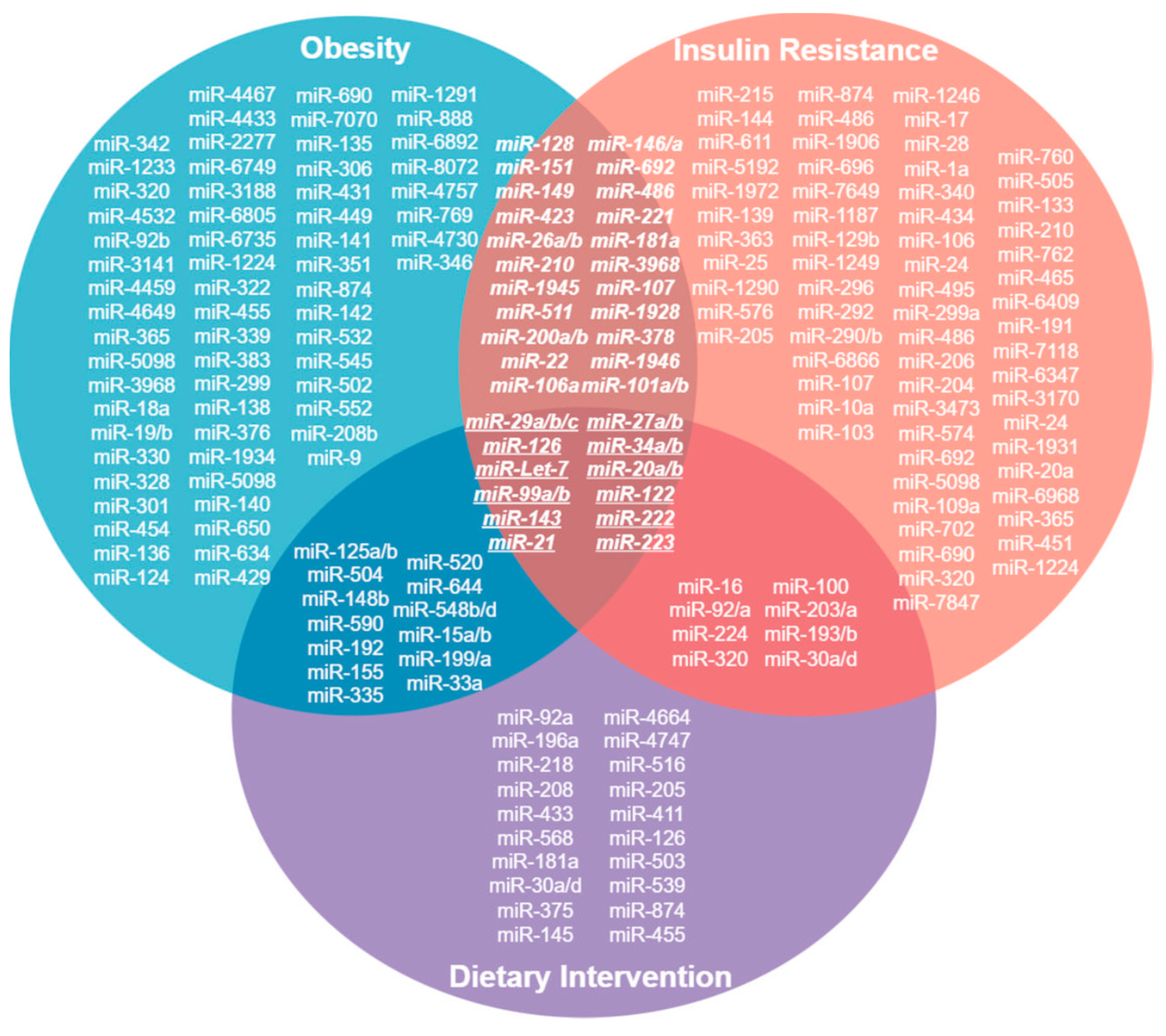
6. miRNAs Related to Obesity, Insulin Resistance and Diet
7. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Bartel, D.P. Metazoan microRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandão, B.B.; Kahn, C.R. Extracellular miRNAs: From biomarkers to mediators of physiology and disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef] [PubMed]
- Safdar, A.; Tarnopolsky, M.A. Exosomes as mediators of the systemic adaptations to endurance exercise. Cold Spring Harb. Perspect. Med. 2018, 8, a029827. [Google Scholar] [CrossRef] [PubMed]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Hannon, G.J. Small RNA sorting: Matchmaking for Argonautes. Nat. Rev. Genet. 2011, 12, 19–31. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of exosome composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef]
- Vallabhajosyula, P.; Korutla, L.; Habertheuer, A.; Yu, M.; Rostami, S.; Yuan, C.X.; Reddy, S.; Liu, C.; Korutla, V.; Koeberlein, B.; et al. Tissue-specific exosome biomarkers for noninvasively monitoring immunologic rejection of transplanted tissue. J. Clin. Investig. 2017, 127, 1375–1391. [Google Scholar] [CrossRef]
- Jankovičová, J.; Sečová, P.; Michalková, K.; Antalíková, J. Tetraspanins, more than markers of extracellular vesicles in reproduction. Int. J. Mol. Sci. 2020, 21, 7568. [Google Scholar] [CrossRef]
- Horibe, S.; Tanahashi, T.; Kawauchi, S.; Murakami, Y.; Rikitake, Y. Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer 2018, 18, 47. [Google Scholar] [CrossRef]
- Lai, C.P.; Mardini, O.; Ericsson, M.; Prabhakar, S.; Maguire, C.A.; Chen, J.W.; Tannous, B.A.; Breakefield, X.O. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano 2014, 8, 483–494. [Google Scholar] [CrossRef]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef]
- Tang, Y.; Yang, L.J.; Liu, H.; Song, Y.J.; Yang, Q.Q.; Liu, Y.; Qian, S.W.; Tang, Q.Q. Exosomal miR-27b-3p secreted by visceral adipocytes contributes to endothelial inflammation and atherogenesis. Cell Rep. 2023, 42, 111948. [Google Scholar] [CrossRef] [PubMed]
- Aas, V.; Øvstebø, R.; Brusletto, B.S.; Aspelin, T.; Trøseid, A.M.S.; Qureshi, S.; Eid, D.S.O.; Olstad, O.K.; Nyman, T.A.; Haug, K.B.F. Distinct microRNA and protein profiles of extracellular vesicles secreted from myotubes from morbidly obese donors with type 2 diabetes in response to electrical pulse stimulation. Front. Physiol. 2023, 14, 1143966. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell 2017, 171, 372–384.e12. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Hu, L.; Gui, W.; Xiao, L.; Wang, W.; Xia, J.; Fan, H.; Li, Z.; Zhu, Q.; Hou, X.; et al. Hepatocyte TGF-β signaling inhibiting WAT browning to promote NAFLD and obesity is associated with let-7b-5p. Hepatol. Commun. 2022, 6, 1301–1321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Yao, X.; Teng, Y.; Zhao, T.; Lin, L.; Li, Y.; Shang, H.; Jin, Y.; Jin, Q. Adipocytes-derived exosomal microRNA-1224 inhibits M2 macrophage polarization in obesity-induced adipose tissue inflammation via MSI2-mediated Wnt/β-catenin axis. Mol. Nutr. Food Res. 2022, 66, e2100889. [Google Scholar] [CrossRef] [PubMed]
- Castaño, C.; Novials, A.; Párrizas, M. Exosomes from short-term high-fat or high-sucrose fed mice induce hepatic steatosis through different pathways. Cells 2022, 12, 169. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Chen, J.; Ren, Y.; Fan, L.; Xiang, W.; He, X. Exosomal miR-122 promotes adipogenesis and aggravates obesity through the VDR/SREBF1 axis. Obesity 2022, 30, 666–679. [Google Scholar] [CrossRef]
- Phu, T.A.; Ng, M.; Vu, N.K.; Bouchareychas, L.; Raffai, R.L. IL-4 polarized human macrophage exosomes control cardiometabolic inflammation and diabetes in obesity. Mol. Ther. 2022, 30, 2274–2297. [Google Scholar] [CrossRef]
- Ying, W.; Gao, H.; Dos Reis, F.C.G.; Bandyopadhyay, G.; Ofrecio, J.M.; Luo, Z.; Ji, Y.; Jin, Z.; Ly, C.; Olefsky, J.M. MiR-690, an exosomal-derived miRNA from M2-polarized macrophages, improves insulin sensitivity in obese mice. Cell Metab. 2021, 33, 781–790.e5. [Google Scholar] [CrossRef]
- Xia, S.F.; Jiang, Y.Y.; Qiu, Y.Y.; Huang, W.; Wang, J. Role of diets and exercise in ameliorating obesity-related hepatic steatosis: Insights at the microRNA-dependent thyroid hormone synthesis and action. Life Sci. 2020, 242, 117182. [Google Scholar] [CrossRef]
- Dang, S.; Leng, Y.; Wang, Z.; Xiao, X.; Zhang, X.; Wen, T.; Gong, H.; Hong, A.; Ma, Y. Exosomal transfer of obesity adipose tissue for decreased miR-141-3p mediate insulin resistance of hepatocytes. Int. J. Biol. Sci. 2019, 15, 351–368. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Hui, X.; Hoo, R.L.C.; Ye, D.; Chan, C.Y.C.; Feng, T.; Wang, Y.; Lam, K.S.L.; Xu, A. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J. Clin. Investig. 2019, 129, 834–849. [Google Scholar] [CrossRef] [PubMed]
- Castaño, C.; Kalko, S.; Novials, A.; Párrizas, M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 12158–12163. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, Q.; Xiao, X.; Wu, C.; Gao, R.; Peng, C.; Li, D.; Zhang, W.; Du, T.; Wang, Y.; et al. miR-1934, downregulated in obesity, protects against low-grade inflammation in adipocytes. Mol. Cell. Endocrinol. 2016, 428, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Heianza, Y.; Xue, Q.; Rood, J.; Bray, G.; Sacks, F.; Qi, L. Circulating thrifty microRNA is related to insulin sensitivity, adiposity, and energy metabolism in adults with overweight and obesity: The POUNDS Lost trial. Am. J. Clin. Nutr. 2023, 117, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Cabiati, M.; Randazzo, E.; Guiducci, L.; Falleni, A.; Cecchettini, A.; Casieri, V.; Federico, G.; Del Ry, S. Evaluation of exosomal coding and non-coding RNA signature in obese adolescents. Int. J. Mol. Sci. 2022, 24, 139. [Google Scholar] [CrossRef]
- Zhang, J.W.; Pan, H.T. microRNA profiles of serum exosomes derived from children with nonalcoholic fatty liver. Genes Genom. 2022, 44, 879–888. [Google Scholar] [CrossRef]
- Eirin, A.; Meng, Y.; Zhu, X.Y.; Li, Y.; Saadiq, I.M.; Jordan, K.L.; Tang, H.; Lerman, A.; Van Wijnen, A.J.; Lerman, L.O. The micro-RNA cargo of extracellular vesicles released by human adipose tissue-derived mesenchymal stem cells is modified by obesity. Front. Cell Dev. Biol. 2021, 9, 660851. [Google Scholar] [CrossRef]
- Al-Rawaf, H.A. Circulating microRNAs and adipokines as markers of metabolic syndrome in adolescents with obesity. Clin. Nutr. 2019, 38, 2231–2238. [Google Scholar] [CrossRef]
- Cannataro, R.; Caroleo, M.C.; Fazio, A.; La Torre, C.; Plastina, P.; Gallelli, L.; Lauria, G.; Cione, E. Ketogenic diet and microRNAs linked to antioxidant biochemical homeostasis. Antioxidants 2019, 8, 269. [Google Scholar] [CrossRef]
- Tang, D.; Bai, S.; Li, X.; Yao, M.; Gong, Y.; Hou, Y.; Li, J.; Yang, D. Improvement of microvascular endothelial dysfunction induced by exercise and diet is associated with microRNA-126 in obese adolescents. Microvasc. Res. 2019, 123, 86–91. [Google Scholar] [CrossRef]
- Parr, E.B.; Camera, D.M.; Burke, L.M.; Phillips, S.M.; Coffey, V.G.; Hawley, J.A. Circulating microRNA responses between ‘high’ and ‘low’ responders to a 16-Wk diet and exercise weight loss intervention. PLoS ONE 2016, 11, e0152545. [Google Scholar] [CrossRef]
- Tabet, F.; Torres, L.F.C.; Ong, K.L.; Shrestha, S.; Choteau, S.A.; Barter, P.J.; Clifton, P.; Rye, K.A. High-density lipoprotein-associated miR-223 is altered after diet-induced weight loss in overweight and obese males. PLoS ONE 2016, 11, e0151061. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Cardona-Alvarado, M.I.; Mercader, J.M.; Moreno-Navarrete, J.M.; Moreno, M.; Sabater, M.; Fuentes-Batllevell, N.; Ramírez-Chávez, E.; Ricart, W.; Molina-Torres, J.; et al. Circulating profiling reveals the effect of a polyunsaturated fatty acid-enriched diet on common microRNAs. J. Nutr. Biochem. 2015, 26, 1095–1101. [Google Scholar] [CrossRef]
- Pescador, N.; Pérez-Barba, M.; Ibarra, J.M.; Corbatón, A.; Martínez-Larrad, M.T.; Serrano-Ríos, M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS ONE 2013, 8, e77251. [Google Scholar] [CrossRef]
- Ortega, F.J.; Mercader, J.M.; Catalán, V.; Moreno-Navarrete, J.M.; Pueyo, N.; Sabater, M.; Gómez-Ambrosi, J.; Anglada, R.; Fernández-Formoso, J.A.; Ricart, W.; et al. Targeting the circulating microRNA signature of obesity. Clin. Chem. 2013, 59, 781–792. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Ye, Y.; Li, D.; Liu, Y.; Lee, E.; Zhang, M.; Dai, X.; Zhang, X.; Wang, S.; et al. Pancreatic β cells control glucose homeostasis via the secretion of exosomal miR-29 family. J. Extracell. Vesicles 2021, 10, e12055. [Google Scholar] [CrossRef]
- Wen, Z.; Li, J.; Fu, Y.; Zheng, Y.; Ma, M.; Wang, C. Hypertrophic adipocyte–derived exosomal miR-802-5p contributes to insulin resistance in cardiac myocytes through targeting HSP60. Obesity 2020, 28, 1932–1940. [Google Scholar] [CrossRef]
- Tian, F.; Tang, P.; Sun, Z.; Zhang, R.; Zhu, D.; He, J.; Liao, J.; Wan, Q.; Shen, J. miR-210 in exosomes derived from macrophages under high glucose promotes mouse diabetic obesity pathogenesis by suppressing NDUFA4 expression. J. Diabetes Res. 2020, 2020, 6894684. [Google Scholar] [CrossRef]
- Su, T.; Xiao, Y.; Xiao, Y.; Guo, Q.; Li, C.; Huang, Y.; Luo, X. Bone marrow mesenchymal stem cells-derived exosomal MiR-29b-3p regulates aging-associated insulin resistance. ACS Nano 2019, 13, 2450–2462. [Google Scholar] [CrossRef]
- Xiong, H.; Liu, W.; Song, J.; Gu, X.; Luo, S.; Lu, Z.; Hao, H.; Xiao, X. Adipose tissue macrophage-derived exosomal miR-210-5p in modulating insulin sensitivity in rats born small for gestational age with catch-up growth. Transl. Pediatr. 2023, 12, 587–599. [Google Scholar] [CrossRef]
- Hong, Y.; Wu, J.; Yu, S.; Hui, M.; Lin, S. Serum-derived exosomal microRNAs in lipid metabolism in polycystic ovary syndrome. Reprod. Sci. 2022, 29, 2625–2635. [Google Scholar] [CrossRef]
- Wang, Y.; Li, M.; Chen, L.; Bian, H.; Chen, X.; Zheng, H.; Yang, P.; Chen, Q.; Xu, H. Natural killer cell-derived exosomal miR-1249-3p attenuates insulin resistance and inflammation in mouse models of type 2 diabetes. Signal Transduct. Target. Ther. 2021, 6, 409. [Google Scholar] [CrossRef]
- Li, L.; Zuo, H.; Huang, X.; Shen, T.; Tang, W.; Zhang, X.; An, T.; Dou, L.; Li, J. Bone marrow macrophage-derived exosomal miR-143-5p contributes to insulin resistance in hepatocytes by repressing MKP5. Cell Prolif. 2021, 54, e13140. [Google Scholar] [CrossRef]
- Jalabert, A.; Reininger, L.; Berger, E.; Coute, Y.; Meugnier, E.; Forterre, A.; Errazuriz-Cerda, E.; Geloen, A.; Aouadi, M.; Bouzakri, K.; et al. Profiling of ob/ob mice skeletal muscle exosome-like vesicles demonstrates combined action of miRNAs, proteins and lipids to modulate lipid homeostasis in recipient cells. Sci. Rep. 2021, 11, 21626. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhou, Y.; Shi, Y.; Zhang, Y.; Liu, K.; Liang, R.; Sun, P.; Chang, X.; Tang, W.; Zhang, Y.; et al. Expression of miRNA-29 in pancreatic β cells promotes inflammation and diabetes via TRAF3. Cell Rep. 2021, 34, 108576. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Song, H.; Shuo, L.; Wang, L.; Xie, P.; Li, W.; Liu, J.; Tong, Y.; Zhang, C.Y.; Jiang, X.; et al. Gonadal white adipose tissue-derived exosomal MiR-222 promotes obesity-associated insulin resistance. Aging 2020, 12, 22719–22743. [Google Scholar] [CrossRef]
- Liu, T.; Sun, Y.C.; Cheng, P.; Shao, H.G. Adipose tissue macrophage-derived exosomal miR-29a regulates obesity-associated insulin resistance. Biochem. Biophys. Res. Commun. 2019, 515, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Jalabert, A.; Vial, G.; Guay, C.; Wiklander, O.P.B.; Nordin, J.Z.; Aswad, H.; Forterre, A.; Meugnier, E.; Pesenti, S.; Regazzi, R.; et al. Exosome-like vesicles released from lipid-induced insulin-resistant muscles modulate gene expression and proliferation of beta recipient cells in mice. Diabetologia 2016, 59, 1049–1058. [Google Scholar] [CrossRef]
- Infante-Menéndez, J.; López-Pastor, A.R.; González-Illanes, T.; González-López, P.; Huertas-Lárez, R.; Rey, E.; González-Rodríguez, Á.; García-Monzón, C.; Patil, N.P.; De Céniga, M.V.; et al. Increased let-7d-5p in non-alcoholic fatty liver promotes insulin resistance and is a potential blood biomarker for diagnosis. Liver Int. 2023, 43, 1714–1728. [Google Scholar] [CrossRef]
- Ye, Z.; Cheng, M.; Fan, L.; Ma, J.; Zhang, Y.; Gu, P.; Xie, Y.; You, X.; Zhou, M.; Wang, B.; et al. Plasma microRNA expression profiles associated with zinc exposure and type 2 diabetes mellitus: Exploring potential role of miR-144-3p in zinc-induced insulin resistance. Environ. Int. 2023, 172, 107807. [Google Scholar] [CrossRef]
- Ali, H.S.; Kamel, M.M.; Agwa, S.H.A.; Hakeem, M.S.A.; Meteini, M.S.E.; Matboli, M. Analysis of mRNA-miRNA-lncRNA differential expression in prediabetes/type 2 diabetes mellitus patients as potential players in insulin resistance. Front. Endocrinol. 2023, 14, 1131171. [Google Scholar] [CrossRef]
- Brandão-Lima, P.N.; De Carvalho, G.B.; Payolla, T.B.; Sarti, F.M.; Fisberg, R.M.; Malcomson, F.C.; Mathers, J.C.; Rogero, M.M. Circulating microRNAs showed specific responses according to metabolic syndrome components and sex of adults from a population-based study. Metabolites 2022, 13, 2. [Google Scholar] [CrossRef]
- Byun, J.S.; Lee, H.Y.; Tian, J.; Moon, J.S.; Choi, J.; Lee, S.H.; Kim, Y.G.; Yi, H.S. Effect of salivary exosomal miR-25-3p on periodontitis with insulin resistance. Front. Immunol. 2022, 12, 775046. [Google Scholar] [CrossRef]
- Mantilla-Escalante, D.C.; De Las Hazas, M.C.L.; Crespo, M.C.; Martín-Hernández, R.; Tomé-Carneiro, J.; Del Pozo-Acebo, L.; Salas-Salvadó, J.; Bulló, M.; Dávalos, A. Mediterranean diet enriched in extra-virgin olive oil or nuts modulates circulating exosomal non-coding RNAs. Eur. J. Nutr. 2021, 60, 4279–4293. [Google Scholar] [CrossRef]
- Sardu, C.; Modugno, P.; Castellano, G.; Scisciola, L.; Barbieri, M.; Petrella, L.; Fanelli, M.; Macchia, G.; Caradonna, E.; Massetti, M.; et al. Atherosclerotic plaque fissuration and clinical outcomes in pre-diabetics vs. normoglycemics patients affected by asymptomatic significant carotid artery stenosis at 2 years of follow-up: Role of microRNAs modulation: The ATIMIR study. Biomedicines 2021, 9, 401. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Du, H.; Wei, S.; Feng, L.; Li, J.; Yao, F.; Zhang, M.; Hatch, G.M.; Chen, L. Adipocyte-derived exosomal MiR-27a induces insulin resistance in skeletal muscle through repression of PPARγ. Theranostics 2018, 8, 2171–2188. [Google Scholar] [CrossRef] [PubMed]
- Párrizas, M.; Brugnara, L.; Esteban, Y.; González-Franquesa, A.; Canivell, S.; Murillo, S.; Gordillo-Bastidas, E.; Cussó, R.; Cadefau, J.A.; García-Roves, P.M.; et al. Circulating miR-192 and miR-193b are markers of prediabetes and are modulated by an exercise intervention. J. Clin. Endocrinol. Metab. 2015, 100, E407–E415. [Google Scholar] [CrossRef]
- Nunez Lopez, Y.O.; Garufi, G.; Seyhan, A.A. Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Mol. Biosyst. 2016, 13, 106–121. [Google Scholar] [CrossRef]
- Dandona, P.; Aljada, A.; Bandyopadhyay, A. Inflammation: The link between insulin resistance, obesity and diabetes. Trends Immunol. 2004, 25, 4–7. [Google Scholar] [CrossRef]
- Yamada, K.; Takizawa, S.; Ohgaku, Y.; Asami, T.; Furuya, K.; Yamamoto, K.; Takahashi, F.; Hamajima, C.; Inaba, C.; Endo, K.; et al. MicroRNA 16-5p is upregulated in calorie-restricted mice and modulates inflammatory cytokines of macrophages. Gene 2020, 725, 144191. [Google Scholar] [CrossRef] [PubMed]
- Manning, P.; Munasinghe, P.E.; Papannarao, J.B.; Gray, A.R.; Sutherland, W.; Katare, R. Acute weight loss restores dysregulated circulating microRNAs in individuals who are obese. J. Clin. Endocrinol. Metab. 2019, 104, 1239–1248. [Google Scholar] [CrossRef] [PubMed]
- Deiuliis, J.A.; Syed, R.; Duggineni, D.; Rutsky, J.; Rengasamy, P.; Zhang, J.; Huang, K.; Needleman, B.; Mikami, D.; Perry, K.; et al. Visceral adipose microRNA 223 is upregulated in human and murine obesity and modulates the inflammatory phenotype of macrophages. PLoS ONE 2016, 11, e0165962. [Google Scholar] [CrossRef]
- Ramzan, F.; Mitchell, C.J.; Milan, A.M.; Schierding, W.; Zeng, N.; Sharma, P.; Mitchell, S.M.; D’Souza, R.F.; Knowles, S.O.; Roy, N.C.; et al. Comprehensive profiling of the circulatory miRNAome response to a high protein diet in elderly men: A potential role in inflammatory response modulation. Mol. Nutr. Food Res. 2019, 63, e1800811. [Google Scholar] [CrossRef] [PubMed]
- Cannataro, R.; Perri, M.; Gallelli, L.; Caroleo, M.C.; De Sarro, G.; Cione, E. Ketogenic diet acts on body remodeling and microRNAs expression profile. MicroRNA 2019, 8, 116–126. [Google Scholar] [CrossRef]
- Hernández-Alonso, P.; Giardina, S.; Salas-Salvadó, J.; Arcelin, P.; Bulló, M. Chronic pistachio intake modulates circulating microRNAs related to glucose metabolism and insulin resistance in prediabetic subjects. Eur. J. Nutr. 2017, 56, 2181–2191. [Google Scholar] [CrossRef]
- Lu, H.; Buchan, R.J.; Cook, S.A. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc. Res. 2010, 86, 410–420. [Google Scholar] [CrossRef]
- Druz, A.; Chen, Y.C.; Guha, R.; Betenbaugh, M.; Martin, S.E.; Shiloach, J. Large-scale screening identifies a novel microRNA, miR-15a-3p, which induces apoptosis in human cancer cell lines. RNA Biol. 2013, 10, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.Y.; Hu, B.; Hu, X.B.; Zhong, J.; Feng, S.D.; Qin, L.; Liu, G.; Wen, G.B.; Liao, D.F. MiRNA-21 reverses high glucose and high insulin induced insulin resistance in 3T3-L1 adipocytes through targeting phosphatase and tensin homologue. Exp. Clin. Endocrinol. Diabetes 2012, 120, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Poy, M.N.; Hausser, J.; Trajkovski, M.; Braun, M.; Collins, S.; Rorsman, P.; Zavolan, M.; Stoffel, M. miR-375 maintains normal pancreatic α- and β-cell mass. Proc. Natl. Acad. Sci. USA 2009, 106, 5813–5818. [Google Scholar] [CrossRef] [PubMed]
- Seclaman, E.; Balacescu, L.; Balacescu, O.; Bejinar, C.; Udrescu, M.; Marian, C.; Sirbu, I.O.; Anghel, A. MicroRNAs mediate liver transcriptome changes upon soy diet intervention in mice. J. Cell. Mol. Med. 2019, 23, 2263–2267. [Google Scholar] [CrossRef] [PubMed]
- Adi, N.; Adi, J.; Lassance-Soares, R.M.; Kurlansky, P.; Yu, H.; Webster, K.A. High protein/fish oil diet prevents hepatic steatosis in NONcNZO10 mice; association with diet/genetics-regulated micro-RNAs. J. Diabetes Metab. 2016, 7, 676. [Google Scholar] [CrossRef]
- Wang, H.; Shao, Y.; Yuan, F.; Feng, H.; Li, N.; Zhang, H.; Wu, C.; Liu, Z. Fish oil feeding modulates the expression of hepatic microRNAs in a western-style diet-induced nonalcoholic fatty liver disease rat model. BioMed Res. Int. 2017, 2017, 2503847. [Google Scholar] [CrossRef]
- Improta Caria, A.C.; Nonaka, C.K.V.; Pereira, C.S.; Soares, M.B.P.; Macambira, S.G.; Souza, B.S.F. Exercise training-induced changes in micrornas: Beneficial regulatory effects in hypertension, type 2 diabetes, and obesity. Int. J. Mol. Sci. 2018, 19, 3608. [Google Scholar] [CrossRef]
- Ryu, H.S.; Park, S.Y.; Ma, D.; Zhang, J.; Lee, W. The induction of microRNA targeting IRS-1 is involved in the development of insulin resistance under conditions of mitochondrial dysfunction in hepatocytes. PLoS ONE 2011, 6, e17343. [Google Scholar] [CrossRef]
- Hubal, M.J.; Nadler, E.P.; Ferrante, S.C.; Barberio, M.D.; Suh, J.H.; Wang, J.; Dohm, G.L.; Pories, W.J.; Mietus-Snyder, M.; Freishtat, R.J. Circulating adipocyte-derived exosomal MicroRNAs associated with decreased insulin resistance after gastric bypass. Obesity 2017, 25, 102–110. [Google Scholar] [CrossRef]
| Author | Study Population | Study Design | Dietary Intervention/Treatment | miRNAs Regulated | Significant Target Genes | Metabolic Effect | Tissue | Exosomes |
|---|---|---|---|---|---|---|---|---|
| In vitro studies | ||||||||
| Tang et al. [14] | Gonadal adipocytes from WT and ob/ob mice Human umbilical vein endothelial cells ApoE-/- mice. | In vitro study. | In animal model WT and ob/ob mice: Group 1. Normal diet Group 2. High-fat diet. In vitro model: TNF-α stimulation. In animal model ApoE-/- mice: Western diet. | ↑ miR-221, miR-22, miR-27b, miR-27a, miR-342, miR-34a-5p, miR-101b-3p, miR-22-3p, miR-222-3p | ↓ VCAM1, ICAM1, MCP1, P65, PPARα | Proinflammatory pathways Endothelial inflammation Adipogenesis Atherogenesis. | Cell culture supernatant Serum. | Yes |
| Aas et al. [15] | Human myotubes with obesity (BMI 45 ± 5 kg/m2) and diabetes (HbA1c 6.7 ± 1.2%). | In vitro study. | Electro pulsed stimulation. | ↑ miR-1233-5p, miR-320b, miR4532, miR-92b-5p, miR-3141, miR-4459, miR-4649-5p, miR-4467, miR-4433b-3p, miR-2277-5p, miR-6749-5p, miR-3188, miR-6805-5p, miR6735-5p | ↑ GANAB, VCL, SYNPO2, TFRC, WARS, PLEC, EEF2, MAP4, SND1, RPS15A, ZYX, CNDP2, MAP1A, RNH1, DES, GSN, FAM129B, PLOD3, AARS ↓ S100A7, COL6A1, PVR, CLSTN1, SEMA7A, RERPING1, CSF1, BMP1, APOE, SDF4 | Receptors of the serine/threonine protein kinase pathway Ubiquitin proteasome system Guanine nucleotide exchange factor. | Cell culture supernatant. | Yes |
| Ying et al. [16] | 3T3-L1 adipocytes WT mice with obesity WT mice without obesity Primary hepatocytes of WT mice Primary myocytes of WT mice. | In vitro study Animal model. | Stimulus: miR-223 Obese mice: High-fat diet. Nonobese mice: Normal diet + adipose tissue exosomal miRNAs Stimulus with glucagon and insulin. | ↑ miR-223, miR-155, miR-181, miR-149, miR-210, miR-1945 ↓ miR-3968, miR-692, miR-365, miR-7070, miR-692, miR-5098, miR-1928, miR-690, miR-7054, miR-682, miR-1946, miR-511 | ↓ PPARγ, GLUT4, ERα, AKT | Insulin signaling pathway. | Cell culture supernatant. | Yes |
| In vivo studies in animal models | ||||||||
| Zhao et al. [17] | TGF-β receptor II (Tgfbr2)-deficient C57BL/6 mice C57BL/6 mice expressing TGF-β receptor II (Tgfbr2 flox/flox) AML-12 cell line Humans with obesity (BMI ≥ 30) and hepatic steatosis Nonobese and healthy humans. | Animal model In vitro study Cross-sectional clinical study. | High-fat diet (60% of L) both groups of mice Normal chow diet (10.2% of L) both groups of mice 4 groups at the end 16 weeks 2 more groups on β3-AR agonist stimulation 1 mg/kg/day 7 days 2 more groups with exposure to temperature of 4 °C. | ↑ Let-7b-5p | ↓ CD36, FATP1, FABP1, ATP5A, COX5B, UCP1, DIO2, PRDM16, PGC-1A | Thermogenesis Mitochondrial oxidative phosphorylation. | Cell culture supernatant Serum. | Yes |
| Zhang et al. [18] | Mice with obesity Mice without obesity KO mice for miR-1224 Primary adipocytes. | Animal model In vitro study. | Animal model: High-fat diet Standard diet. | ↑ miR-1224 | ↓ MSI2 | Wnt/β-catenin pathway Inhibition of M2 macrophage polarization Inflammation. | Plasma Cell culture supernatant. | Yes |
| Castaño et al. [19] | Mice with obesity Mice with hepatic steatosis Mice without obesity or hepatic steatosis. | Animal model. | Model with obesity: High-fat diet Model with hepatic steatosis: Normal diet Chow + sucrose in water Control model: Normal diet Chow 15 weeks. | ↑ miR-22-3p, miR-122-5p, miR-223-3p, miR-34a-5p, miR-378a-3p, miR-101-3p, miR-107-3p, miR-19-3p, miR-200b-3p, miR-322-5p ↓ miR-199a-5p, miR-23b-3p, miR-143-3p | ↓ PTGDS, GGT1, HK3, PLKP, GM13882, PKM2, GOT1, DLD, G6PC, ENO3 ↑ SLC2A5, GPI1, ALDOC, | Hepatic steatosis Gluconeogenesis De novo lipogenesis Insulin resistance. | Serum Hepatocytes. | Yes |
| Huang et al. [20] | Mice with obesity. | Animal model. | High-fat diet. | ↑ miR-122 | ↓ VDR, SREBF1, PPARγ, ADIPOQ, LPL | Adipogenesis. | Cell culture supernatant | Yes |
| Phu et al. [21] | Obese mice Human THP1 monocytes 3T3-L1 adipocytes WT mice with obesity Ldlr-/- mice. | Animal model In vitro study. | High-fat diet 6 weeks In WT mice with obesity and Ldlr-/-: Intraperitoneal THP1-IL-4 exosomes. | ↑ miR-21, miR-99a, miR-146b, miR-378a ↓ miR-33 | ↑ PPARγ, GLUT4, UCP1, OXPHOS | Lipophagy Mitochondrial activity Oxidative phosphorylation Beiging Hepatic steatosis Glucose metabolism Insulin resistance. | Plasma Cell culture supernatant. | Yes |
| Ying et al. [22] | Mice with obesity 3T3-L1 adipocytes L6 myotubes Primary hepatocytes. | Animal model In vitro study. | High-fat diet 8 weeks + treatment with M2-type macrophage exosomes 4 weeks. | ↑ miR-690, miR-7070, miR-365, miR-5098, miR-3968 | ↓ NADK, BMDM, RAW264, PPARγ, YM1, MRC1, INOS, TNFA. IL1B, CC12, IFBG | Citrate cycle Pyruvate metabolism Glycolysis Gluconeogenesis JAK STAT pathway signaling Insulin pathway signaling mTOR pathway signaling. | Cell culture supernatant. | Yes |
| Xia et al. [23] | C57BL/6J Mice. | Animal model. | High-fat diet for 10 weeks, then divided into 5 groups: High-fat diet Low fat diet (10% L) Energy-restricted diet (low-fat diet with 70% energy restriction) Quercetin-enriched diet (0.005% quercetin) Diet with exercise For 7 weeks. | Liver: ↑ miR-28, miR-382, miR-146 Thyroid: ↓ miR-200a, miR-339, miR-383, miR-146 | ↑ TRB, SREBP1C, FASN, ACC1, SCD1, CD36, FABP, G6PC, PCK1, FAGF21, PGC1α ↓ NIS, NRF2, NQO1, HO-1, DIO1, ATP5C1, COX7C | Energy metabolism Lipid metabolism Thyroid gland function. | Liver Thyroid. | No |
| Dang et al. [24] | B6 WT Mice B6 ob/ob mice AML12 cells. | Animal model In vitro study. | Animal model 3 groups: B6 WT control B6 WT with obesity, high-fat diet. B6 ob/ob 3 months. In vitro study: Stimulus with exosomes from each of the groups. 48 h. | ↑ 151-5p, miR-299a-5p, miR-135b-5p, miR-15b-3p, miR-306-5p, miR-431-5p, miR-449a-5p ↓ miR-141-3p, miR-351-5p, miR-874-3p | ↓ STAT3, PPP1R3B, SLC2A2, GSK3B, PTEN, PIK3CB, IRS2, SLC2A4, RPS6KA1, PRKAA2, PTPN1, NFKB1, FOXO1, MAPK8, AKT3 | Insulin resistance. | Adipose tissue Cell culture supernatant. | Yes |
| Pan et al. [25] | C57BL/6J WT mice with and without obesity KO mice for miR-34a. | Animal model. | WT and KO mice with obesity: high-fat diet. WT and KO mice without obesity: standard chow diet 16 weeks. | ↑ miR-34a | ↑ TNFα, IL6, IL1B, INOS, MCP1 ↓ FIZZ1, YM1, ARG1, IL10, KLF4 | Proinflammatory cytokines Glucose intolerance Insulin resistance. | Adipose tissue Cell culture supernatant. | Yes |
| Castaño et al. [26] | C57BL/6J mice Group 1 with obesity Group 2 without obesity. | Animal model In vitro study. | High-fat diet Chow control diet 15 weeks. | ↑ miR-192, miR-122, miR-27a ↓ miR-375 | ↓ PPARα, PPARγ, CD36, FADS1, PPARD, LIPE, PNPLA2 ↑ PLIN2, CPT1α, FGF21, CCL2, TNF | Adipogenesis Lipogenesis Fatty acid oxidation Inflammation Insulin resistance Dyslipidemia. | Plasma. | Yes |
| Liu et al. [27] | C57BL/6J mice 3T3-L1 adipocytes Human adipocytes: Group 1: nonobese BMI ≤ 25 kg/m2 Group 2: overweight and obese BMI > 25 kg/m2. | Animal model In vitro study. | Animal study: Group 1: Control diet with 15% L Group 2: High-fat diet with 45% L 8 weeks. In vitro study on 3T3-L1 adipocytes stimulated with or without 50 ng/mL TNFα. | ↓ miR-1934 | ↑ TNF-α, IL-6, IL-1β, CD11c, MCP-1 | Inflammatory state. | Adipose tissue Serum. | No |
| Clinical studies | ||||||||
| Heianza et al. [28] | Humans with overweight and obesity (BMI 32.7 ± 3.8 kg/m2). | Randomized controlled clinical trial (N = 495): Group 1 = 124 Group 2 = 113 Group 3 = 116 Group 4 = 92. | 4 types of energy restricted diet: Group 1. Low fat and average protein (20% L, 15% P, 65% HC). Group 2. Low fat and high protein (20% L, 25% P, 55% HC). Group 3. High in fat and average protein (40% L, 15% P, 45% HC) Group 4. High fat and high protein (40% L, 25% P, 35% HC) + 90 min of moderate exercise a week 6 months. | ↑ miR-128-1-5p | ↓ LCT, R3HDM1, PRDM, PPARγ, C1A, PPARα | Energy expenditure Glucose metabolism Insulin resistance. | Plasma. | No |
| Cabiati et al. [29] | Adolescents with obesity BMI 29.5 ± 0.8 kg/m2 Adolescents without obesity BMI 21.1 ± 0.6 kg/m2. | Cross-sectional clinical study (N = 44), (22 per group). | No treatment. | ↓ miR-223-5p, 33a-3p, miR-181a-5p, miR-199-5p | ↓ CHD9, PTEN, MTMR12, TBL1X, CPOX, ACOT9 | Lipid metabolism Inflammatory state Insulin resistance. | Plasma. | Yes |
| Zhang and Pan [30] | Adolescents with obesity and no non-alcoholic fatty liver disease Adolescents with obesity and non-alcoholic fatty liver disease. | Cross-sectional clinical trial (N = 10), (5 per group). | No treatment. | ↑ miR-122-5p, miR-27a, miR-335-5p | ↓ WNT10B, PPARγ, SREBP-1C | Hepatic lipid metabolism Adipocyte differentiation. | Serum. | Yes |
| Eirin et al. [31] | Vascular stromal adipose cells from abdominal subcutaneous fat tissue of two groups: Humans with obesity (BMI ≥ 30 kg/m2) Humans without obesity (BMI ≤ 25 kg/m2) Proximal tubule epithelial cells (HK2). | Cross-sectional clinical study In vitro study (N = 10), (5 per group). | HK2 stimulation: a model of ischemic kidney injury with TNF-α 10 ng/mL and antimycin A (AMA). | ↑ miR-1291, miR-888-5p, miR-6892, miR-222-5p, miR-8072, miR-4757-5p, miR-769-5p, miR-4730 ↓ miR-346, miR-650, miR-634, miR-429, miR-136-3p, miR-222-3p, miR-124b-3p, miR-454-5p, miR-552, miR-208b-3p, miR-9-3p, miR-548-5p, miR-20a-5p, miR-545-5p, miR-455-3p, miR-146a-3p | ↑ NFK-β, PP38, MAPK ↓ WNT1 | MAPK pathway Apoptosis NF-κβ signaling pathway. | Cell culture supernatant. | Yes |
| Al-Rawaf [32] | Humans Adolescents Group 1: Obese (BMI 26.7 ± 2.91 kg/m2) Group 2: Overweight (BMI 21.9 ± 5.7 kg/m2) Group 3: Normal weight (BMI 17.4 ± 4.3 kg/m2). | Cross-sectional clinical trial (N = 250): Group 1 = 100 Group 2 = 100 Group 3 = 50. | No treatment. | ↑ miR-142-3p, miR-140-5p, miR-22, miR-143, miR-130 ↓ miR-532-5p, miR-423-5p, miR-520c-3p, miR-146a, miR-15a | ↓ P38, MAPK | Lipid metabolism. | Plasma. | No |
| Cannataro et al. [33] | Humans with obesity (BMI 46 ± 10 kg/m2) separated by sex. | Controlled clinical trial (N = 36): Women = 18 Men = 18. | Hypocaloric ketogenic diet with 2 phases: Phase 1: −300 kcal, 30 g HC, 57% L, 37% P. Phase 2: −200 kcal, 120 g HC, 44% L, 32% P 3 weeks each phase. | ↑ miR-504-5p ↓ miR-Let7-5p, miR-143-3p, miR-30a-5p, miR-502-5p, miR-590-5p, miR-644a, miR-148b-3p, miR-26a-5p, miR-520, miR-548bmi-3p | ↓ ALDH, CS, DLAT, GPI, HK, PFKM, BPGM, ACACA, CRKL, GYS, IRS2 Y 4, MAPK, NRAS, PHKA, PRKAA, PPARγ, AKT, HRAS, KRAS, ACSL, ALDH, MCAT, HADHA, HMGCS, LPL, RXRB, SCD, FADS, SLC, ATP, OX, NDUFA, NFKBIA, HIF, HSL, FHIT, TP53, VEGFA, MDM2, TFF1, TCEA1, DRD1 | Glycolysis, gluconeogenesis and citrate cycle Insulin signaling pathway Fatty acid metabolism PPAR signaling pathway mTOR, amino acids and cytokine signaling. | Serum. | No |
| Tang et al. [34] | Humans: Group 1: adolescents with obesity (BMI 33.2 ± 4.23 kg/m2) Group 2: adolescents without obesity (BMI 23.21 ± 4.23 kg/m2). | Controlled clinical study (N = 67): Group 1 = 57 Group 2 = 10. | Group 1: Restrictive diet (20–25 Kcal/day) + exercise program of 50 min, 5 days a week. Group 2: Control diet + sedentary lifestyle 6 weeks. | ↑ miR-126 | ↓ SPRED-1, PI3KR2, CXCL12 | Angiogenesis PI3K-eNOs pathway. | Serum. | No |
| Parr et al. [35] | Humans: Overweight and obese (BMI 27–40 kg/m2). | Controlled clinical study (N = 89): Diet 1 = 32 Diet 2 = 29 Diet 3 control = 28. | Energy restriction diet (<250 Kcal) Diet 1: high protein and high HC (~30% P, 55% HC, 15% L) Diet 2: high protein and moderate HC (~30% P, 40% HC, 30% L) Diet 3 control: low protein and high HC (15% P, 55% CH, 30% L) + exercise with energy expenditure of ~250 Kcal 16 weeks. | ↓ miR-221, miR-223, miR-140, miR-935, miR-448, miR-310, miR-263 | ↓ ARL15, CROT, CRTC3, FNDC5, SOCS7, STRADE, CYP7A1, PIK3R1, HDAC4, IGF1R, PLA2G6, SORBS, HMGB1, MEF2D, PHIP, PPARγ | Lipid metabolism and β fatty acid oxidation. Regulation of energy metabolism Insulin metabolism Low grade inflammation. | Serum. | No |
| Tabet et al. [36] | Humans: Group 1 High protein diet: Obesity (BMI 32.7 ± 4.2 kg/m2) Group 2 Normal-protein diet: Obesity (BMI 32.6 ± 4.4 kg/m2). | Controlled clinical trial (N = 47): Group 1 = 20 Group 2 = 27. | Group 1: High protein diet Group 2: Normal protein diet 12 weeks. | ↑ miR-223 | ↓ GLUT4 | Glucose metabolism. | Serum. | No |
| Ortega et al. [37] | Humans: BMI 30–35 kg/m2. | Clinical study First phase (N = 10) Second validation phase (N = 30). | Isocaloric diet: 55–60% HC, 15% P, <30% L (<10% saturated fats, 10-15% monounsaturated fats, 10% polyunsaturated fats; 5–8% omega-6, 1–2% omega-3) 15 g almonds and 15 g walnuts; for 8 weeks. | ↑ miR-328, miR-330-3p, miR-221, miR-125-5p ↓ miR-192, miR-486-5p, miR-19b, miR-106a, miR-18a, miR-130b | Not mentioned | Inflammatory state Lipid metabolism. | Plasma. | No |
| Pescador et al. [38] | Humans: Group 1: Obesity BMI 42.7 ± 4.67 kg/m2 Group 2: Control BMI 22.7 ± 2.43 kg/m2 Group 3: DM2 BMI 24.8 ± 1.49 kg/m2 Group 4: OB-DM2 BMI 33.3 ± 3.86 kg/m2. | Cross-sectional clinical trial: Group 1 = 20 Group 2 = 20 Group 3 = 13 Group 4 = 16. | No treatment. | ↑miR-138, miR-15b, miR-376a | ↓ P85a, PIK3R1 | Hepatic triglyceride storage Apoptosis Adipogenesis. | Serum. | No |
| Ortega et al. [39] | Humans: Group 1: Obesity BMI 42.9 ± 5.9 kg/m2 Group 2: Obesity BMI 32.4 ± 3.8 kg/m2. | Cross-sectional and longitudinal clinical trial First phase identification = 32 Group 1 = 6 Second phase validation = 102 Group 2 = 9. | Group 1: Bariatric surgery Group 2: Energy restrictive diet. | ↑ miR-142-3p, miR-140-5p, miR-222 ↓ miR-221, miR-130b, miR-423-5p, miR-15a, miR-520-3p | ↑ TGFRB1, LIFR, VEGFA | JAK-STAT and MAPK pathway Adipocyte development Energy expenditure Apoptosis Angiogenesis. | Plasma. | No |
| Author | Study Population | Study Design | Dietary Intervention/Treatment | miRNAs Regulated | Significant Target Genes | Metabolic Effect | Tissue | Exosomes |
| In vitro studies | ||||||||
| Li et al. [40] | Mouse pancreatic cells MIN6 3T3-L1 cells C57BL/6J WT control mice C57BL/6J Lep ob (ob) mice Humans with DM2 with overweight and obesity (BMI > 25 kg/m2). | In vitro study Animal model Cross-sectional clinical study. | Animal model High fat diet (20% CH, 20% P, 60% L) + injections with exosomes from control mice 6 weeks. | ↑ miR-29a, miR-29b, miR-29c | ↓ MCTL, STXLA, PI3KRL, P85α | Hepatic glucose production Glucose homeostasis | Cell culture supernatant Plasma. | Yes |
| Wen et al. [41] | 3T3-L1 adipocytes Rat ventricular myocytes. | In vitro study. | Stimulus with palmitate Stimulus with insulin. | ↑ miR-802-5p | ↓ HSP60 | Oxidative stress Insulin resistance | Cell culture supernatant | Yes |
| Tian et al. [42] | Mouse macrophages RAW264.7 3T3-L1 adipocytes C57BL/6J WT mice C56BL/6J KO for miR-210-/- mice. | In vitro study Animal model. | In vitro study: Stimulus with exosomes (2 µg) of macrophages 24 h. Animal model 3 groups: WT control normal diet 2. WT with obesity, high fat diet 3. KO with obesity and diabetes high fat diet + streptozotocin 7 weeks. | ↑ miR-210 | ↓ NDUFA4 | Mitochondrial dysfunction Insulin resistance | Cell culture supernatant Serum. | Yes |
| Su et al. [43] | Bone marrow mesenchymal stem cells from 3- and 18-month-old mice 3T3-L1 adipocytes C2C12 myocytes Primary hepatocytes C57BL/6J WT mice. | In vitro study Animal model. | In vitro study: Stimulation with bone marrow mesenchymal stem cell exosomes from 3- and 18-month-old rats to adipocytes, myocytes and hepatocytes. 12 h. Animal model: Exosomes from bone marrow mesenchymal stem cells of 3- and 18-month-old rats 7 days. | ↑ miR-29b-3p, miR-17-50, miR-762, miR-465b-5p, miR-221-3p, miR-6409, miR-151-5p, miR-191-5p, miR-99b-5p, miR-7118-5p, miR-6347, miR-290b-3p, miR-let-7a-5p, miR-3170-5p, miR486-5p, miR-107-3p, miR-486-5p, miR-107-3p, miR-24-3p, miR-1931, miR-92b-3p, miR-20a-5p, miR-6968-5p ↓ miR-190a-3p, miR-702-5p | ↓ SIRT1 | Insulin resistance | Cell culture supernatant. | Yes |
| Ying et al. [16] | 3T3-L1 adipocytes WT mice without obesity WT mice with obesity KO mice for miR-155 L6 myocytes cells. | In vivo study Animal model. | Stimulus with macrophages transfected with miR-223 Treatment with macrophage exosomes from adipose tissue. KO mice for miR-155: High fat diet 20 weeks. | ↑ miR-155, miR-181a, miR-149, miR-210, miR-1945 ↓ miR-3968, miR-692, miR-365, miR-7070, miR-5098, miR-1928, miR-690, miR-7054, miR-682, miR-1946, miR-511 | ↓ PPARγ, GLUT4 | Insulin resistance Glucose homeostasis | Cell culture supernatant. | Yes |
| In vivo studies in animal models | ||||||||
| Xiong et al. [44] | Small rats by gestational age (SGA) Control term rats Primary hepatocytes 3T3-L1 adipocytes L6 myocytes. | Animal model In vitro study. | Pregnant rats Control group: Standard diet ad libitum. SGA rats group: Energy restriction (50% of normal intake). | ↑ miR-210 | ↓ SIDT2 | Insulin resistance Autophagy Lipid metabolism Inflammation | Cell culture supernatant. | Yes |
| Hong et al. [45] | Polycystic ovary syndrome mice with insulin resistance 3T3-L1 adipocytes. | Animal model In vitro study. | High-fat diet + dehydroepiandrosterone. | ↓ miR-20b-5p, miR-106a-5p | Not mentioned | Lipid metabolism Adipocyte differentiation | Serum. | Yes |
| Wang et al. [46] | Mice with DM2 Control mice without DM2 3T3-L1 adipocytes AML12 cells. | Animal model In vitro study. | DM2 model: High-fat diet + streptozocin. Non-DM2 model: Normal Chow diet. 3T3-L1 and AML12 stimulation: NK cell exosomes from high-fat diet and control diet. | ↑ miR-1906, miR-696, miR-7649-5p, miR-1187, miR-129b-5p, miR1249-3p ↓ miR-1249, miR-296-5p, miR292-3p, miR-290b-3p, miR-6866 | ↓ PAKT, PPARγ, GLUT4 | Insulin sensitivity Inflammation | Cell culture supernatant. | Yes |
| Li et al. [47] | WT C57BL/6K mice with and without obesity Hep1-6 cells HEK293 cells. | Animal model In vitro study. | Mice with obesity: High fat diet (45% of L). Mice without obesity: Normal diet Chow. HEP1-6 stimulation: miR-143-5p mimics. HEK293 cell stimulation: miR-143-5p mimics and pmirGLO-MKP5. | ↑ miR-143-5p | ↓ DUSP10 | Decreased AKT and GSK phosphorylation Glycogen synthesis | Culture medium supernatant. | Yes |
| Jalabert et al. [48] | Leptin-deficient C57BL/6 mice (ob/ob) C57BL/6 WT control mice 3T3-L1 adipocytes C2C12 muscle cells. | Animal model In vitro study. | Normal chow diet (57% HC, 25% L, 18% P) 12 weeks 3T3-L1 cells incubated with exosomes from animal models C2C12 cells incubated with exosomes from animal models. | ↑ miR-1a-3p, miR-101a-3p, miR-340-5p, miR-434-5p, miR-106-b, miR-146a-5p, miR-24-3p, miR-200-3p, miR-203-3p ↓ miR-224-5p, miR-29b-5p, miR-495-3p, miR-434-3p, miR-299a-5p | ↓ PPARγ, PPARα, CPT1, CPT2, CD36, ABCA1, HMGCR, CIDEC, FABP4, INSR, IGF1R | Lipid metabolism TNF-β pathway WNT pathway Proteolysis Thyroid hormone signaling Adrenergic signaling in muscle Insulin resistance | Cell culture supernatant. | Yes |
| Sun et al. [49] | db/m mice db/db mice Primary pancreatic islets. | Animal model In vitro study. | Normal diet control High fat diet 4 weeks. | ↑ miR-29a-5p, miR-30a-5p, miR378a-3p, miR-203-3p, miR-486a-3p, miR-206-3p, miR-1a-3p, miR-30a-3p, miR-145a-5p, miR-192-5p, miR-146a-5p ↓ miR-Let-7-5p, miR-204-3p, miR-3473b, miR-193b-5p, miR-574-5p, miR-423-5p, miR-128-3p, miR-760-3p, miR-505-5p | ↓ TRAF3, CXCL10, AHSG, P2RX1, KNG1, NOS2, CXCL17 | Glucose intolerance Insulin resistance Inflammatory response | Cell culture supernatant. | Yes |
| Li et al. [50] | C57BL/6J mice With obesity-related insulin resistance Without obesity or insulin resistance. | Animal model | High fat diet Normal chow diet (control) 8 weeks. | ↑ miR-222 | ↓ IRS1 | Insulin resistance | Serum Gonadal adipose tissue. | Yes |
| Liu et al. [51] | C56BL/6J Mice With obesity Without obesity 3T3-L1 adipose tissue L6 myocytes Primary hepatocytes. | Animal model In vitro study. | Mice with obesity: High-fat diet (60% L, 20% P, 20% HC). Mice without obesity: Normal chow diet (control) 3 months. Stimulus in vitro study: miR-29a mimic. | ↑ miR-29a | ↓ PPARγ | Insulin resistance | Serum Adipose tissue. | Yes |
| Jalabert et al. [52] | C57BL/6J mice Primary pancreatic islets MIN6B1 cell line C2C12 myoblasts 3T3-L1 preadipocytes. | Animal model In vitro study. | Animal model: Group 1. Standard chow diet (57% HC, 25% L, 18% P). Group 2. Standard chow diet enriched with 20% palm oil. 16 weeks. In vitro study: Stimulation with skeletal muscle exosomes from animal model (group 1 and group 2). | miR-146a, miR-92a, miR-16 | ↓ PTCH1, HMGA1, WDR26, PDPK1, MXD1, RAP1A, RHOB, CCNE2, GPR137C, ATXN3 | Adipogenesis and cell differentiation Cell cycle MAPK signaling pathway PI3K/Akt signaling pathway Insulin resistance | Tissue (muscle conditioning medium). | Yes |
| Clinical studies | ||||||||
| Infante-Menéndez et al. [53] | Humans with non-alcoholic hepatic steatosis Humans without non-alcoholic hepatic steatosis ApoE-/- mice with hepatic steatosis C57BL/6J WT mice without hepatic steatosis Huh7 cells. | Cross-sectional clinical study: Humans with non-alcoholic hepatic steatosis = 30 Humans without non-alcoholic hepatic steatosis = 21 Animal model In vitro study. | Animal model Steatosis mice: high fat diet Control mice: standard diet 13 weeks Huh7 cell stimulation with oleic acid and palmitic acid. | ↑ miR-Let-7d, miR-34-5p ↓ miR-26b-5p, | ↓ AKT, IGF1, INSR, IGF1R | Insulin resistance Cell proliferation Cell differentiation | Plasma Liver tissue. | Yes |
| Ye et al. [54] | Humans with newly diagnosed DM2 Healthy human controls HepG2 cells. | Prospective clinical study: Humans with newly diagnosed DM2 = 9 Healthy human controls = 9 Validation phase = 161 In vitro study. | In vitro study: Zinc sulfate stimulation from 0 to 140 µM. 24 h, then insulin stimulation Transfected with miR-144-3p mimic, 200 nM miR-144-3p inhibitor or negative control with Lipofectamine + zinc sulfate 18 h. | ↑ miR-215-5p, miR-144-3p | ↓ NRF2 | Insulin resistance Oxidative stress TGF-β pathway | Plasma. | No |
| Ali et al. [55] | Humans with DM2 Humans with prediabetes Healthy humans (control). | Cross-sectional clinical study: Humans with DM2 = 66 Humans with prediabetes = 49 Healthy humans (control) = 45. | No treatment. | ↑ miR-611, miR-5192, miR-1976 | ↑ CHUK, TMEM173 | Inflammation Insulin resistance INF pathway activation | Serum. | No |
| Brandão-Lima et al. [56] | Humans with metabolic syndrome. | Cross-sectional clinical study (N = 192): Men = 87 Women = 105. | No treatment. | ↑ miR-122, miR-Let-7c, miR-15a, miR-222, miR-146a, miR-miR-30d ↓ miR-139, miR-16, miR-363, miR-486 | ↓ IGF2BP, RFX6, IL10, CCL3, PDK4, SIRT4, AKAT3, VEGFA, IKBKB, CDK8, FOXO1, BCL2, MARK1, UCP3, PPARγ, C1A ↑ SOCS1/4, PIK3R1, CDK8, MAP3K2, IGF1, TRAF6, PRKAR2A | Inflammatory state Cytokine signaling PKA activation Stress response Insulin signaling Energy metabolism | Plasma. | Yes |
| Byun et al. [57] | Humans with overweight BMI 26.5 ± 2.9 kg/m2 and DM2 Humans without overweight and DM2 C57BL/6 mice Submandibular lymph node cells. | Cross-sectional clinical study (N = 60): Overweight humans BMI 26.5 ± 2.9 kg/m2 and DM2 = 30 Humans without overweight or DM2 = 30 Animal model In vitro study. | Animal model: Group 1: high fat diet Group 2: normal chow diet 12 weeks. In vitro study: Phorbol myristate acetate. | ↑ miR-92-3p, miR-25-3p, miR-1290, miR-576-5p, miR-221-3p, miR-205-5p, miR-7847-3p, miR-320b, miR-25-3p, miR-451a, miR-130b-3p, miR-210-3p, miR-874-3p, miR-486-5p | ↑ IL17A, IL17F ↓ RORC | Cell differentiation Inflammatory response Insulin resistance | Saliva Cell culture supernatant. | Yes |
| Mantilla-Escalante et al. [58] | Overweight and obese older adults BMI 29 ± 4 kg/m2. | Longitudinal clinical study (N = 150): Group 1 = 50 Group 2 = 50 Group 3 = 50. | Group 1: Mediterranean diet + extra virgin olive oil ≥ 4 tablespoons per day. Group 2: Mediterranean diet + nuts ≥ 3 servings per day. Group 3: Low-fat diet Duration: 1 year. | ↓ miR-222-3p, mir-185-5p, miR-27a-3p, miR-21-5p, miR-29c-3p, miR-34b-5p, miR-320b, miR-107, miR-20b-5p, miR-20a-3p, miR-1246, miR-106a-5p, miR-23a-3p, miR-28-5p, miR-215, miR-21, miR-34, miR-103, miR-151, miR-22, miR-671-5p, miR-200c-3p, miR-193a-3p, miR-381-3p, miR-10b-3p, miR-24-3p, miR-122-5p, miR-16-5p, miR-28-5p, miR-195-50, miR-15a-5p, miR-26a-5p ↑ miR-215-5p, miR_369-3p, miR-10a-5p, miR-210-3p, miR-215-5p | ↓ AKT1, CDK1, MYC, BHLH, PTEN, PLK1, TP53, MAPK1, CCND1, AMPK, FOXO, P53, HIF1 | Insulin resistance Tumor suppression Cell proliferation and growth | Plasma. | Yes |
| Sardu et al. [59] | Overweight and obese humans with carotid artery stenosis Group 1: normal glycaemia Group 2: prediabetes without metformin treatment Group 3: prediabetes with metformin treatment. | Longitudinal clinical study (N = 234): Group 1 = 125 Group 2 = 73 Group 3 = 36. | Carotid revascularization surgery. | ↑ miR-24, miR-27, miR-100, miR-126, miR-133 | Not mentioned | Glucose metabolism Atherosclerosis | Serum. | Yes |
| Yu et al. [60] | Children with and without obesity Mice with obesity KO for miR-27a db/db mice C2C12 cells. | Cross-sectional clinical study (N = 90): Children with obesity = 45 Children without obesity = 45 Animal model In vitro study. | Animal model: High-fat diet Low fat diet 12 weeks. In vitro study: Stimulus with palmitate-treated 3T3-L1 adipocyte culture medium. | ↑ miR-27a | ↓ IRS1, GLUT4, PPARγ | Insulin resistance | Serum. | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Gómez, K.G.; Avila-Nava, A.; González-Salazar, L.E.; Noriega, L.G.; Serralde-Zúñiga, A.E.; Guizar-Heredia, R.; Medina-Vera, I.; Gutiérrez-Solis, A.L.; Torres, N.; Tovar, A.R.; et al. Modulation of MicroRNAs and Exosomal MicroRNAs after Dietary Interventions for Obesity and Insulin Resistance: A Narrative Review. Metabolites 2023, 13, 1190. https://doi.org/10.3390/metabo13121190
Hernández-Gómez KG, Avila-Nava A, González-Salazar LE, Noriega LG, Serralde-Zúñiga AE, Guizar-Heredia R, Medina-Vera I, Gutiérrez-Solis AL, Torres N, Tovar AR, et al. Modulation of MicroRNAs and Exosomal MicroRNAs after Dietary Interventions for Obesity and Insulin Resistance: A Narrative Review. Metabolites. 2023; 13(12):1190. https://doi.org/10.3390/metabo13121190
Chicago/Turabian StyleHernández-Gómez, Karla G., Azalia Avila-Nava, Luis E. González-Salazar, Lilia G. Noriega, Aurora E. Serralde-Zúñiga, Rocio Guizar-Heredia, Isabel Medina-Vera, Ana Ligia Gutiérrez-Solis, Nimbe Torres, Armando R. Tovar, and et al. 2023. "Modulation of MicroRNAs and Exosomal MicroRNAs after Dietary Interventions for Obesity and Insulin Resistance: A Narrative Review" Metabolites 13, no. 12: 1190. https://doi.org/10.3390/metabo13121190








