Appraising the Effects of Metabolic Traits on the Risk of Glaucoma: A Mendelian Randomization Study
Abstract
:1. Introduction
2. Methods
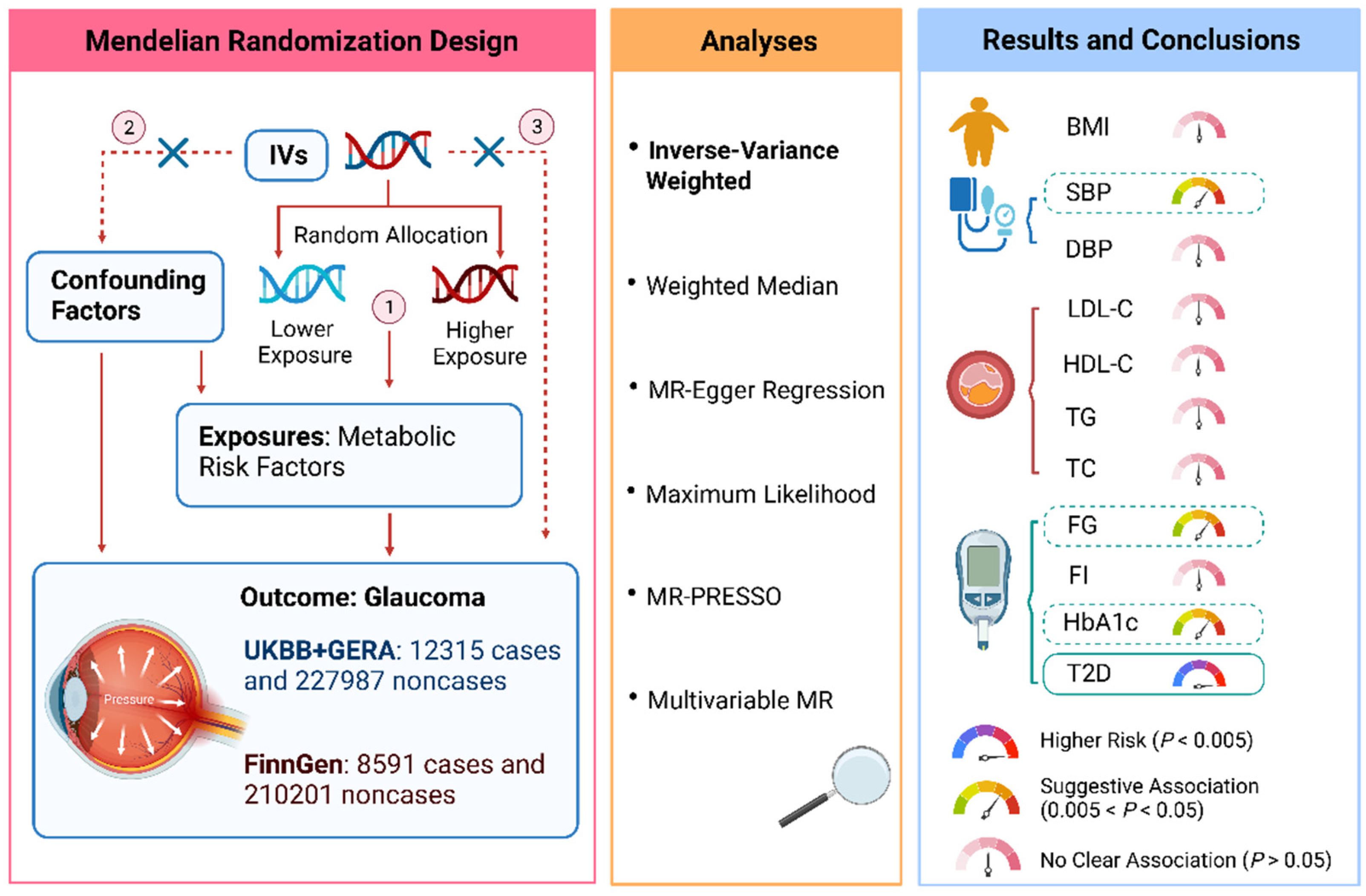
2.1. Study Design
2.2. Selection of the Genetic Instruments
2.3. Data Source for Glaucoma
2.4. Statistical Analyses
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Tham, Y.C.; Li, X.; Wong, T.Y.; Quigley, H.A.; Aung, T.; Cheng, C.Y. Global prevalence of glaucoma and projections of glaucoma burden through 2040: A systematic review and meta-analysis. Ophthalmology 2014, 121, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- Quigley, H.A. Glaucoma. Lancet 2011, 377, 1367–1377. [Google Scholar] [CrossRef] [PubMed]
- Gumus, M.; Babacan, S.N.; Demir, Y.; Sert, Y.; Koca, I.; Gulcin, I. Discovery of sulfadrug-pyrrole conjugates as carbonic anhydrase and acetylcholinesterase inhibitors. Arch. Pharm. 2022, 355, e2100242. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Cho, J.; Kim, M.H.; Friedman, D.S.; Guallar, E. Diabetes, fasting glucose, and the risk of glaucoma: A meta-analysis. Ophthalmology 2015, 122, 72–78. [Google Scholar] [CrossRef]
- Zhao, Y.X.; Chen, X.W. Diabetes and risk of glaucoma: Systematic review and a Meta-analysis of prospective cohort studies. Int. J. Ophthalmol. 2017, 10, 1430–1435. [Google Scholar] [CrossRef]
- Pertl, L.; Mossbock, G.; Wedrich, A.; Weger, M.; Konigsbrugge, O.; Silbernagel, G.; Posch, F. Triglycerides and Open Angle Glaucoma—A Meta-analysis with meta-regression. Sci. Rep. 2017, 7, 7829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Posch-Pertl, L.; Michelitsch, M.; Wagner, G.; Wildner, B.; Silbernagel, G.; Pregartner, G.; Wedrich, A. Cholesterol and glaucoma: A systematic review and meta-analysis. Acta Ophthalmol. 2022, 100, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Bao, X. Hyperlipidemia, Blood Lipid Level, and the Risk of Glaucoma: A Meta-Analysis. Investig. Ophthalmol. Vis. Sci. 2019, 60, 1028–1043. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; Cho, J.; Kim, M.H.; Guallar, E. The association of blood pressure and primary open-angle glaucoma: A meta-analysis. Am. J. Ophthalmol. 2014, 158, 615–627.e9. [Google Scholar] [CrossRef]
- Nislawati, R.; Taufik Fadillah Zainal, A.; Ismail, A.; Waspodo, N.; Kasim, F.; Gunawan, A. Role of hypertension as a risk factor for open-angle glaucoma: A systematic review and meta-analysis. BMJ Open Ophthalmol. 2021, 6, e000798. [Google Scholar] [CrossRef]
- Ko, F.; Boland, M.V.; Gupta, P.; Gadkaree, S.K.; Vitale, S.; Guallar, E.; Zhao, D.; Friedman, D.S. Diabetes, Triglyceride Levels, and Other Risk Factors for Glaucoma in the National Health and Nutrition Examination Survey 2005–2008. Investig. Ophthalmol. Vis. Sci. 2016, 57, 2152–2157. [Google Scholar] [CrossRef]
- Cheung, N.; Wong, T.Y. Obesity and eye diseases. Surv. Ophthalmol. 2007, 52, 180–195. [Google Scholar] [CrossRef] [Green Version]
- Laville, V.; Kang, J.H.; Cousins, C.C.; Iglesias, A.I.; Nagy, R.; Cooke Bailey, J.N.; Igo, R.P., Jr.; Song, Y.E.; Chasman, D.I.; Christen, W.G.; et al. Genetic Correlations Between Diabetes and Glaucoma: An Analysis of Continuous and Dichotomous Phenotypes. Am. J. Ophthalmol. 2019, 206, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Pulit, S.L.; Stoneman, C.; Morris, A.P.; Wood, A.R.; Glastonbury, C.A.; Tyrrell, J.; Yengo, L.; Ferreira, T.; Marouli, E.; Ji, Y.; et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 2019, 28, 166–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evangelou, E.; Warren, H.R.; Mosen-Ansorena, D.; Mifsud, B.; Pazoki, R.; Gao, H.; Ntritsos, G.; Dimou, N.; Cabrera, C.P.; Karaman, I.; et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet. 2018, 50, 1412–1425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willer, C.J.; Schmidt, E.M.; Sengupta, S.; Peloso, G.M.; Gustafsson, S.; Kanoni, S.; Ganna, A.; Chen, J.; Buchkovich, M.L.; Mora, S.; et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013, 45, 1274–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Spracklen, C.N.; Marenne, G.; Varshney, A.; Corbin, L.J.; Luan, J.; Willems, S.M.; Wu, Y.; Zhang, X.; Horikoshi, M.; et al. The trans-ancestral genomic architecture of glycemic traits. Nat. Genet. 2021, 53, 840–860. [Google Scholar] [CrossRef]
- Xue, A.; Wu, Y.; Zhu, Z.; Zhang, F.; Kemper, K.E.; Zheng, Z.; Yengo, L.; Lloyd-Jones, L.R.; Sidorenko, J.; Wu, Y.; et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat. Commun. 2018, 9, 2941. [Google Scholar] [CrossRef] [Green Version]
- Clarke, L.; Zheng-Bradley, X.; Smith, R.; Kulesha, E.; Xiao, C.; Toneva, I.; Vaughan, B.; Preuss, D.; Leinonen, R.; Shumway, M.; et al. The 1000 Genomes Project: Data management and community access. Nat. Methods 2012, 9, 459–462. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Chen, S.; Qu, Z.; Wang, K.; Xie, X.; Cui, H. Genetic Liability to Sedentary Behavior in Relation to Stroke, Its Subtypes and Neurodegenerative Diseases: A Mendelian Randomization Study. Front. Aging Neurosci. 2021, 13, 757388. [Google Scholar] [CrossRef]
- Choquet, H.; Paylakhi, S.; Kneeland, S.C.; Thai, K.K.; Hoffmann, T.J.; Yin, J.; Kvale, M.N.; Banda, Y.; Tolman, N.G.; Williams, P.A.; et al. A multiethnic genome-wide association study of primary open-angle glaucoma identifies novel risk loci. Nat. Commun. 2018, 9, 2278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Track Football Consortium. FinnGen Documentation of R5 Release. 2021. Available online: https://finngen.gitbook.io/documentation/ (accessed on 18 August 2021).
- Bowden, J.; Davey Smith, G.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgess, S.; Thompson, S.G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 2017, 32, 377–389. [Google Scholar] [CrossRef] [Green Version]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018, 7, e34408. [Google Scholar] [CrossRef] [PubMed]
- Verbanck, M.; Chen, C.Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Multivariable Mendelian randomization: The use of pleiotropic genetic variants to estimate causal effects. Am. J. Epidemiol. 2015, 181, 251–260. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Walter, S.; Melles, R.B.; Glymour, M.M.; Jorgenson, E. Diabetes Pathology and Risk of Primary Open-Angle Glaucoma: Evaluating Causal Mechanisms by Using Genetic Information. Am. J. Epidemiol. 2016, 183, 147–155. [Google Scholar] [CrossRef] [Green Version]
- Kanamori, A.; Nakamura, M.; Mukuno, H.; Maeda, H.; Negi, A. Diabetes has an additive effect on neural apoptosis in rat retina with chronically elevated intraocular pressure. Curr. Eye Res. 2004, 28, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, M.; Kanamori, A.; Negi, A. Diabetes mellitus as a risk factor for glaucomatous optic neuropathy. Ophthalmologica 2005, 219, 1–10. [Google Scholar] [CrossRef]
- Aschard, H.; Kang, J.H.; Iglesias, A.I.; Hysi, P.; Cooke Bailey, J.N.; Khawaja, A.P.; Allingham, R.R.; Ashley-Koch, A.; Lee, R.K.; Moroi, S.E.; et al. Genetic correlations between intraocular pressure, blood pressure and primary open-angle glaucoma: A multi-cohort analysis. Eur. J. Hum. Genet. 2017, 25, 1261–1267. [Google Scholar] [CrossRef]
- He, Z.; Vingrys, A.J.; Armitage, J.A.; Bui, B.V. The role of blood pressure in glaucoma. Clin. Exp. Optom. 2011, 94, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Konieczka, K.; Liu, X.; Chen, M.; Yao, K.; Wang, K.; Flammer, J. Role of ocular blood flow in normal tension glaucoma. Adv. Ophthalmol. Pract. Res. 2022, 2, 100036. [Google Scholar] [CrossRef]
- Marshall, H.; Mullany, S.; Qassim, A.; Siggs, O.; Hassall, M.; Ridge, B.; Nguyen, T.; Awadalla, M.; Andrew, N.H.; Healey, P.R.; et al. Cardiovascular Disease Predicts Structural and Functional Progression in Early Glaucoma. Ophthalmology 2021, 128, 58–69. [Google Scholar] [CrossRef]
- Ramdas, W.D.; Wolfs, R.C.; Hofman, A.; de Jong, P.T.; Vingerling, J.R.; Jansonius, N.M. Lifestyle and risk of developing open-angle glaucoma: The Rotterdam study. Arch. Ophthalmol. 2011, 129, 767–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.; Zhu, X.; Luo, W.; Jiang, B.; Lin, Q.; Tang, M.; Li, X.; Xie, L. The Causal Association Between Obesity and Primary Open-Angle Glaucoma: A Two-Sample Mendelian Randomization Study. Front. Genet. 2022, 13, 835524. [Google Scholar] [CrossRef] [PubMed]

| Exposure or Outcome | Unit | Participants Included in Analysis | Adjustments | IVs a | PMID |
|---|---|---|---|---|---|
| BMI | SD of BMI | 806,834 European-descent individuals | Age, age square, sex, and 1–5 PCs | 613 | 30239722 |
| Systolic blood pressure | 10 mmHg | 757,601 European-descent individuals | Sex, age, age square, BMI, genotyping chips | 227 | 30224653 |
| Diastolic blood pressure | 10 mmHg | 757,602 European-descent individuals | Sex, age, age square, BMI, genotyping chips | 292 | 30224653 |
| LDL cholesterol | SD of LDL cholesterol | 188,578 individuals of multiancestries (90% European) | Age, age square, sex | 80 | 24097068 |
| HDL cholesterol | SD of HDL cholesterol | 188,578 individuals of multiancestries (90% European) | Age, age square, sex | 87 | 24097068 |
| Triglyceride | SD of Triglyceride | 188,578 individuals of multiancestries (90% European) | Age, age square, sex | 55 | 24097068 |
| Total cholesterol | SD of Total cholesterol | 188,578 individuals of multiancestries (90% European) | Age, age square, sex | 86 | 24097068 |
| Fasting glucose | mmol/L | 200,622 European-descent individuals | BMI, study-specific covariates, and PCs | 69 | 34059833 |
| Fasting insulin | pmol/L | 151,013 European-descent individuals | BMI, study-specific covariates, and PCs | 36 | 34059833 |
| Hemoglobin A1c | 1% | 146,806 European-descent individuals | study-specific covariates and PCs | 76 | 34059833 |
| Type 2 diabetes | 1-log unit odds of type 2 diabetes | 62,892 type 2 diabetes cases and 596,424 controls of European ancestry | Age, sex, and 20 PCs | 135 | 30054458 |
| Glaucoma (UKBB + GERA) | — | 12,315 glaucoma cases and 227,987 noncases of multiancestries (89% European) | Age, sex, and ancestry PCs | — | 29891935 |
| Glaucoma (FinnGen) | — | 8591 glaucoma cases and 210,201 noncases of European descent | Age, sex, 10 PCs, and genotyping batch | — | — |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, K.; Yang, F.; Liu, X.; Lin, X.; Yin, H.; Tang, Q.; Jiang, L.; Yao, K. Appraising the Effects of Metabolic Traits on the Risk of Glaucoma: A Mendelian Randomization Study. Metabolites 2023, 13, 109. https://doi.org/10.3390/metabo13010109
Wang K, Yang F, Liu X, Lin X, Yin H, Tang Q, Jiang L, Yao K. Appraising the Effects of Metabolic Traits on the Risk of Glaucoma: A Mendelian Randomization Study. Metabolites. 2023; 13(1):109. https://doi.org/10.3390/metabo13010109
Chicago/Turabian StyleWang, Kai, Fangkun Yang, Xin Liu, Xueqi Lin, Houfa Yin, Qiaomei Tang, Li Jiang, and Ke Yao. 2023. "Appraising the Effects of Metabolic Traits on the Risk of Glaucoma: A Mendelian Randomization Study" Metabolites 13, no. 1: 109. https://doi.org/10.3390/metabo13010109





