EFMviz: A COBRA Toolbox Extension to Visualize Elementary Flux Modes in Genome-Scale Metabolic Models
Abstract
:1. Introduction
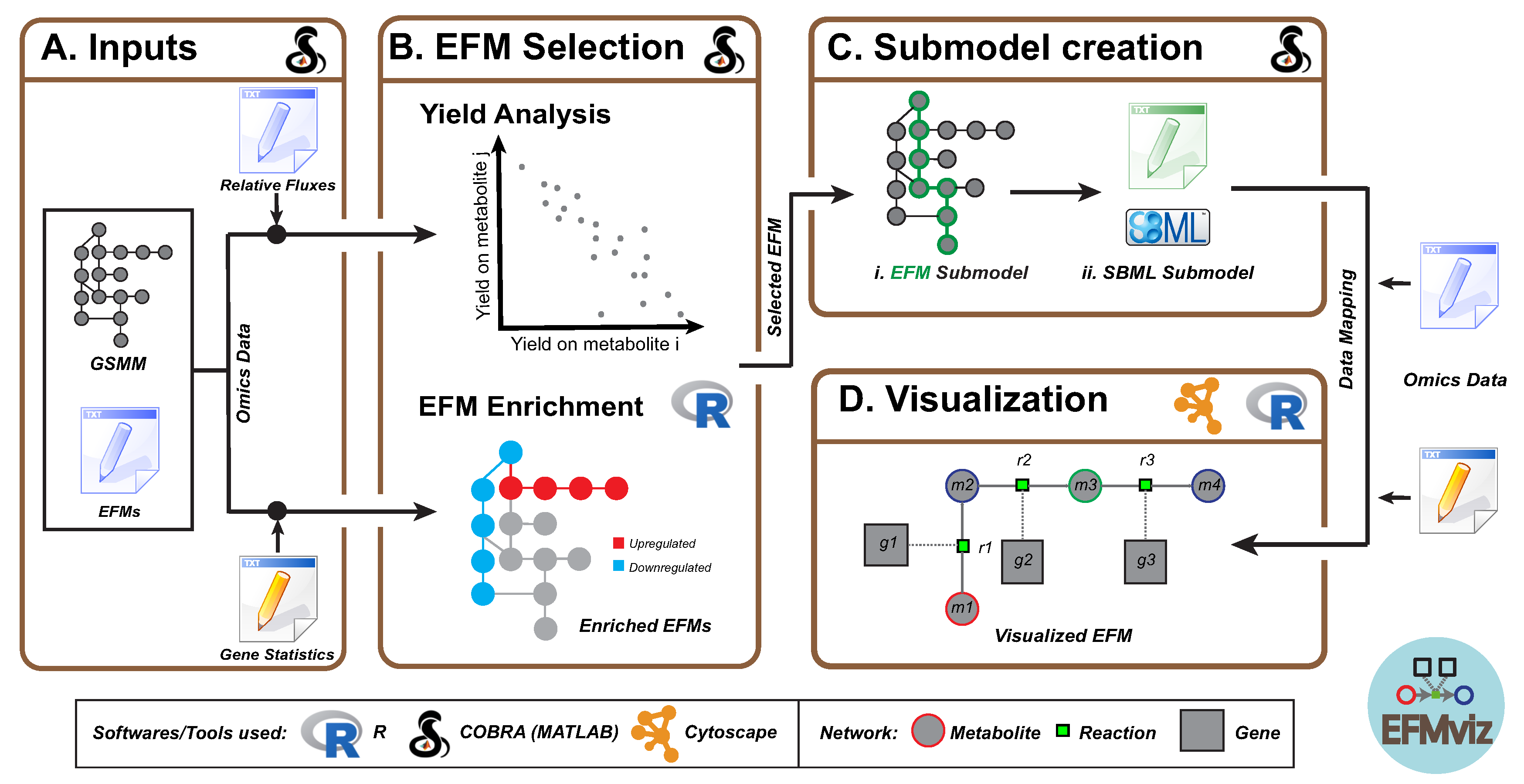
2. Materials and Methods
2.1. Inputs
2.2. EFM Selection
2.2.1. Yield Analysis
2.2.2. EFM Enrichment
2.3. Submodel Creation
2.4. Visualization
3. Results
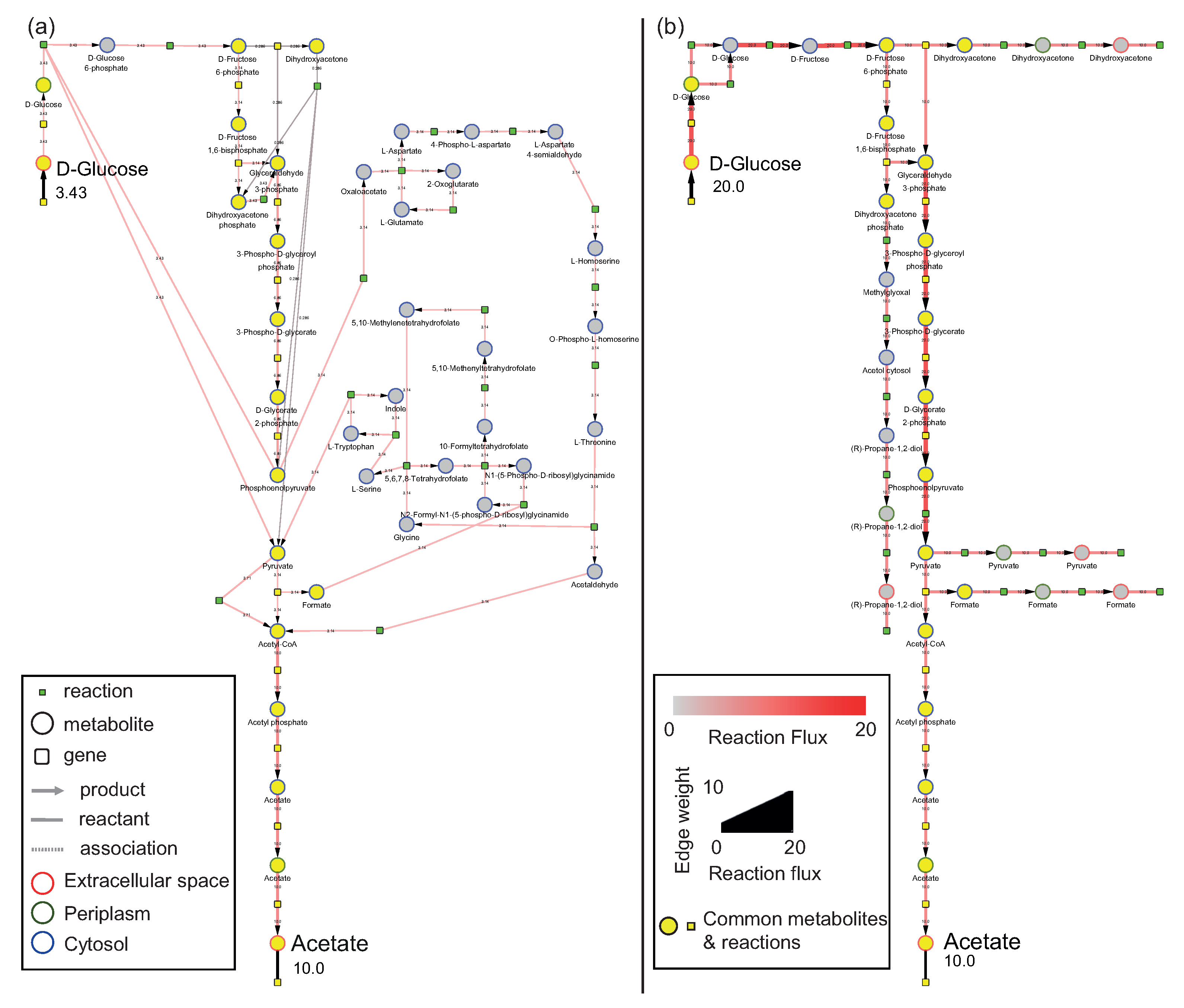
3.1. Use Case 1: Visualizing Elementary Flux Modes from the E. coli Model
3.1.1. Model
3.1.2. Analysis
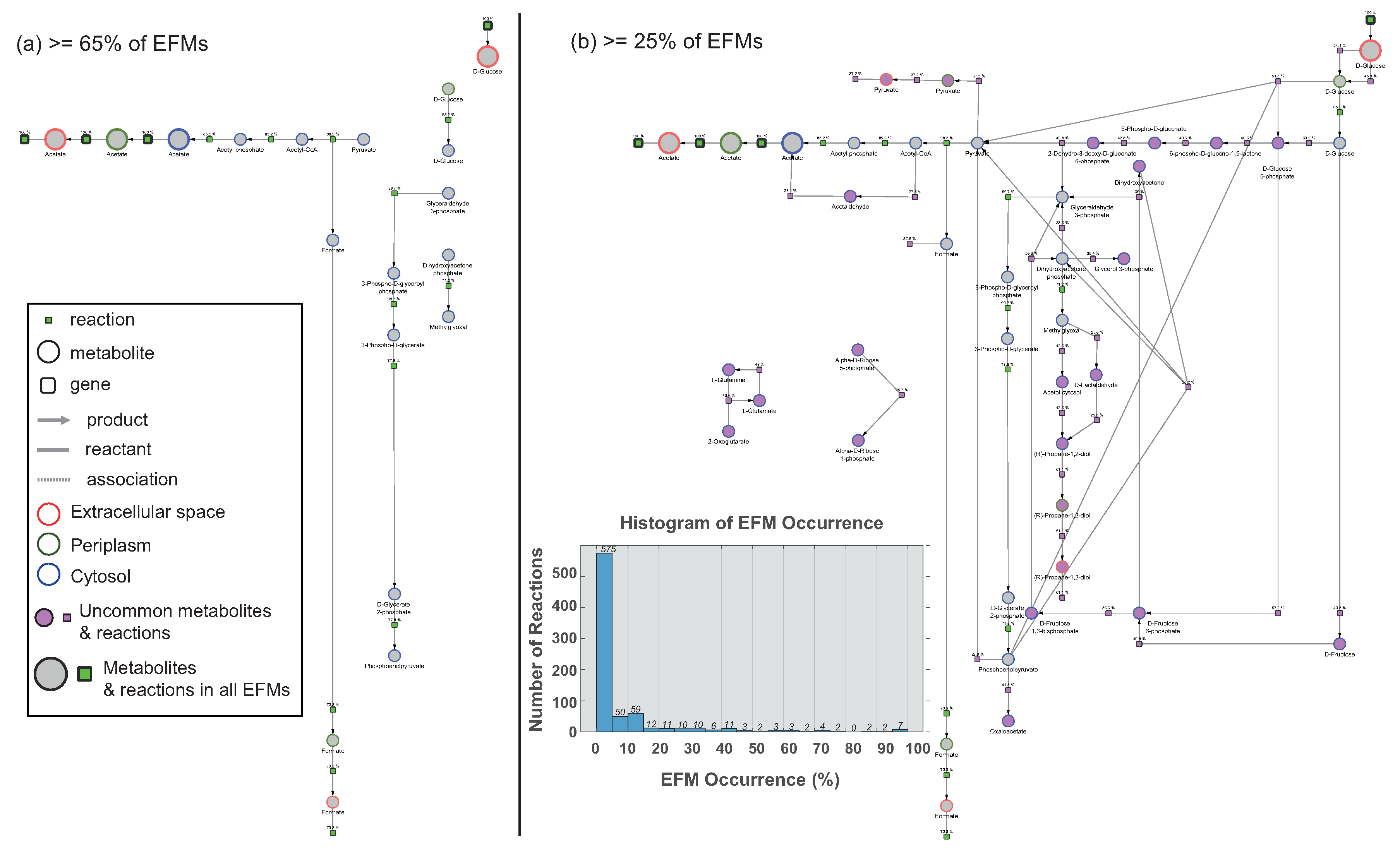
Comparison of EFMs Using Preserved Visual Arrangement
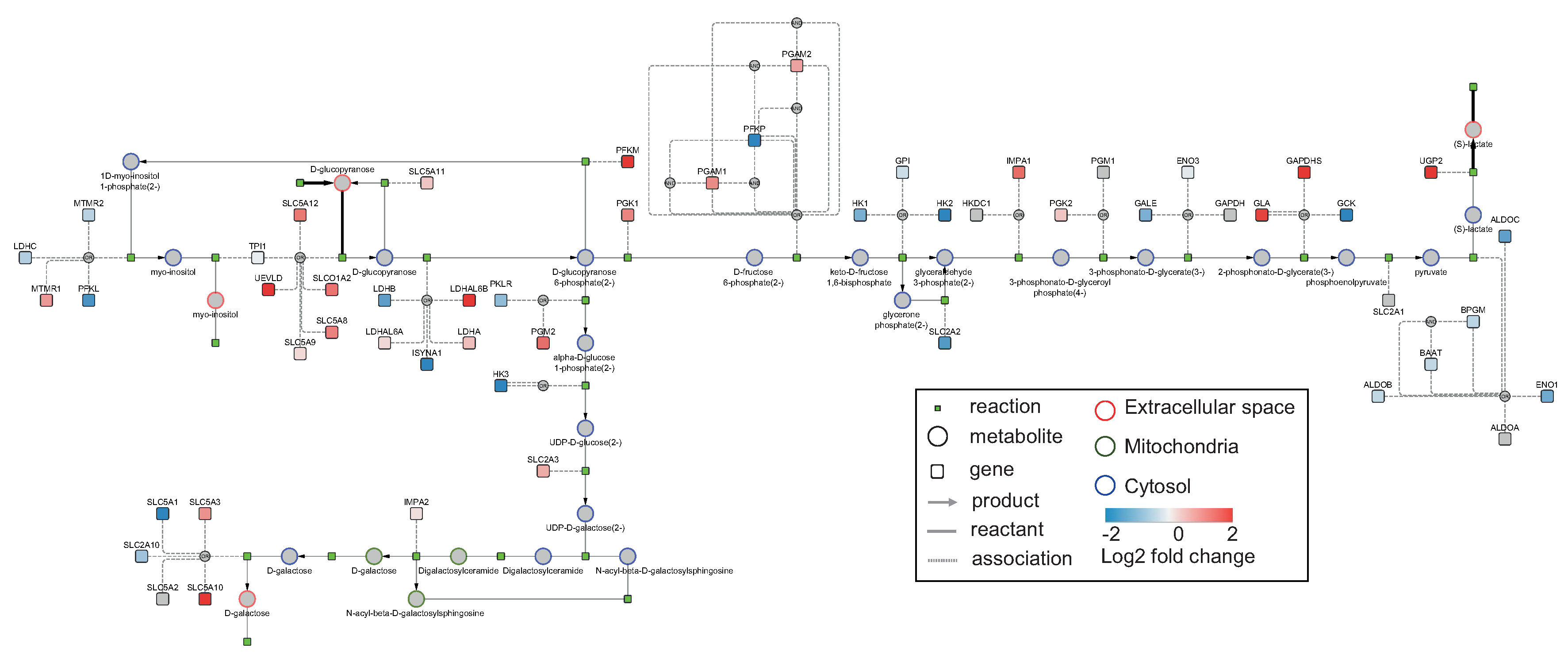
3.2. Use Case 2: Visualizing Elementary Flux Modes from the Human Model
3.2.1. Data
3.2.2. Model
3.2.3. Analysis
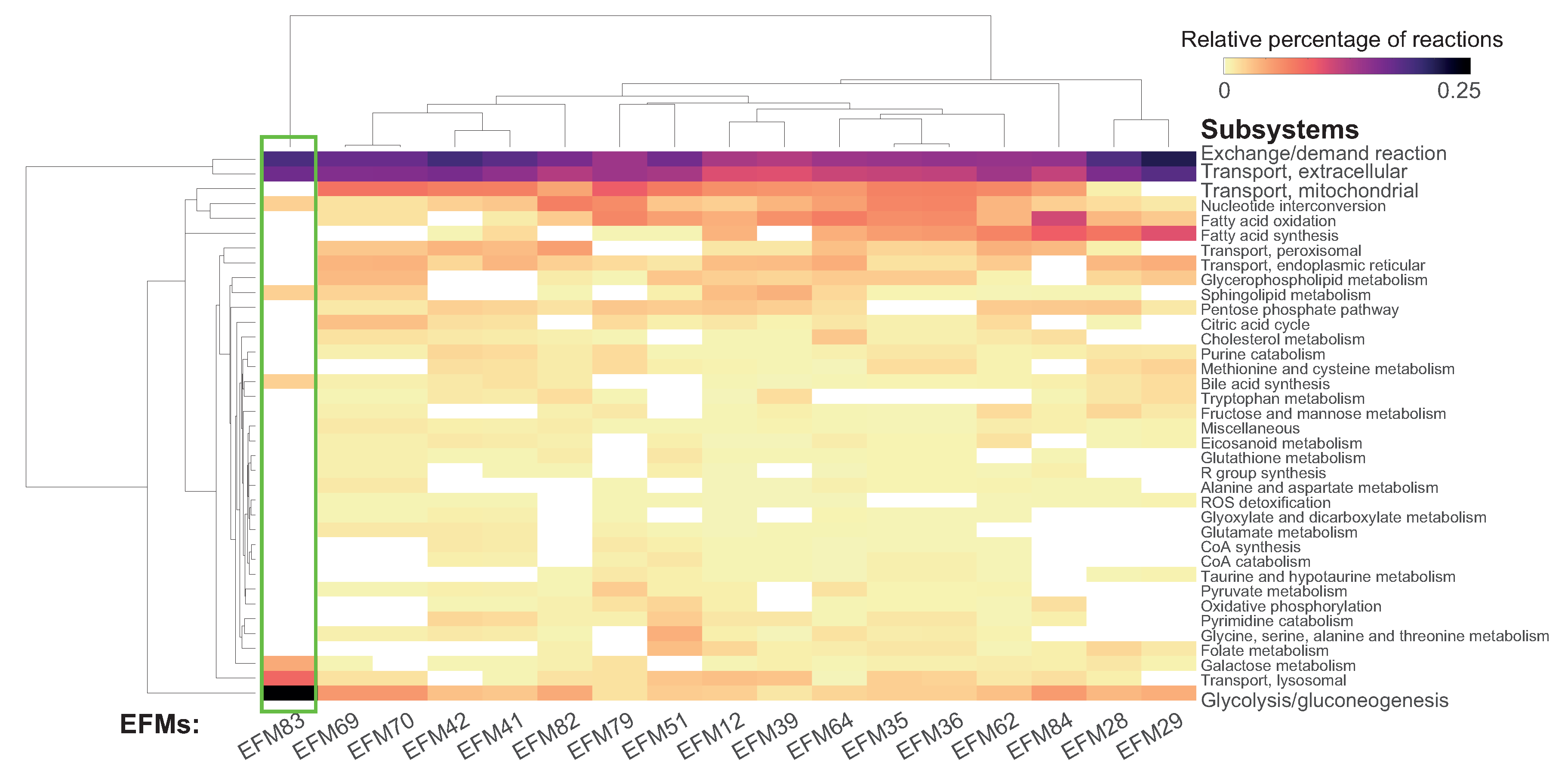
Comparing EFMs Using Subsystem Occurrence
3.3. Assessing Overlap between EFMs: Backbone Identification
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Robinson, J.L.; Nielsen, J. Integrative analysis of human omics data using biomolecular networks. Mol. BioSyst. 2016, 12, 2953–2964. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.; Dandekar, T.; Fell, D.A.; Schuster, S.; Dandekar, T.; Fell, D.; Schuster, S.; Dandekar, T.; Fell, D.; Schuster, S.; et al. Detection of elementary flux modes in biochemical networks: A promising tool for pathway analysis and metabolic engineering. Trends Biotechnol. 1999, 17, 53–60. [Google Scholar] [CrossRef]
- Schuster, S.; Fell, D.A.; Dandekar, T. A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nat. Biotechnol. 2000, 18, 326. [Google Scholar] [CrossRef]
- Carlson, R.; Srienc, F. FundamentalEscherichia coli biochemical pathways for biomass and energy production: Identification of reactions. Biotechnol. Bioeng. 2004, 85, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Trinh, C.T.; Wlaschin, A.; Srienc, F. Elementary mode analysis: A useful metabolic pathway analysis tool for characterizing cellular metabolism. Appl. Microbiol. Biotechnol. 2009, 81, 813–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlson, R. Metabolic systems cost-benefit analysis for interpreting network structure and regulation. Bioinformatics 2007, 23, 2202. [Google Scholar] [CrossRef] [Green Version]
- Trinh, C.T.; Srienc, F. Metabolic engineering of Escherichia coli for efficient conversion of glycerol to ethanol. Appl. Environ. Microbiol. 2009, 75, 6696–6705. [Google Scholar] [CrossRef] [Green Version]
- Trinh, C.T.; Unrean, P.; Srienc, F. Minimal Escherichia coli cell for the most efficient production of ethanol from hexoses and pentoses. Appl. Environ. Microbiol. 2008, 74, 3634–3643. [Google Scholar] [CrossRef] [Green Version]
- Klamt, S.; Gilles, E.D. Minimal cut sets in biochemical reaction networks. Bioinformatics 2004, 20, 226–234. [Google Scholar] [CrossRef]
- Stelling, J.; Klamt, S.; Bettenbrock, K.; Schuster, S.; Gilles, E.D. Metabolic network structure determines key aspects of functionality and regulation. Nature 2002, 420, 190–193. [Google Scholar] [CrossRef]
- Gebauer, J.; Schuster, S.; de Figueiredo, L.F.; Kaleta, C. Detecting and investigating substrate cycles in a genome-scale human metabolic network. FEBS J. 2012, 279, 3192–3202. [Google Scholar] [CrossRef]
- Klamt, S.; Stelling, J. Combinatorial Complexity of Pathway Analysis in Metabolic Networks. Mol. Biol. Rep. 2002, 29, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Gagneur, J.; Klamt, S. Computation of elementary modes: A unifying framework and the new binary approach. BMC Bioinform. 2004, 5, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schilling, C.H.; Letscher, D.; Palsson, B.Ø. Theory for the Systemic Definition of Metabolic Pathways and their use in Interpreting Metabolic Function from a Pathway-Oriented Perspective. J. Theor. Biol. 2000, 203, 229–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeung, M.; Thiele, I.; Palsson, B.Ø. Estimation of the number of extreme pathways for metabolic networks. BMC Bioinform. 2007, 8, 363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croes, D.; Couche, F.; Wodak, S.J.; van Helden, J. Inferring meaningful pathways in weighted metabolic networks. J. Mol. Biol. 2006, 356, 222–236. [Google Scholar] [CrossRef] [PubMed]
- Blum, T.; Kohlbacher, O. Using Atom Mapping Rules for an Improved Detection of Relevant Routes in Weighted Metabolic Networks. J. Comput. Biol. 2008, 15, 565–576. [Google Scholar] [CrossRef]
- de Figueiredo, L.F.; Podhorski, A.; Rubio, A.; Kaleta, C.; Beasley, J.E.; Schuster, S.; Planes, F.J. Computing the shortest elementary flux modes in genome-scale metabolic networks. Bioinformatics 2009, 25, 3158–3165. [Google Scholar] [CrossRef]
- Machado, D.; Soons, Z.; Patil, K.R.; Ferreira, E.C.; Rocha, I. Random sampling of elementary flux modes in large-scale metabolic networks. Bioinformatics 2012, 28, i515–i521. [Google Scholar] [CrossRef] [Green Version]
- Kelk, S.M.; Olivier, B.G.; Stougie, L.; Bruggeman, F.J. Optimal flux spaces of genome-scale stoichiometric models are determined by a few subnetworks. Sci. Rep. 2012, 2, 580. [Google Scholar] [CrossRef]
- Rezola, A.; Pey, J.; de Figueiredo, L.F.; Podhorski, A.; Schuster, S.; Rubio, A.; Planes, F.J. Selection of human tissue-specific elementary flux modes using gene expression data. Bioinformatics 2013, 29, 2009–2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunt, K.A.; Folsom, J.P.; Taffs, R.L.; Carlson, R.P. Complete enumeration of elementary flux modes through scalable demand-based subnetwork definition. Bioinformatics 2014, 30, 1569–1578. [Google Scholar] [CrossRef] [PubMed]
- Gerstl, M.P.; Ruckerbauer, D.E.; Mattanovich, D.; Jungreuthmayer, C.; Zanghellini, J. Metabolomics integrated elementary flux mode analysis in large metabolic networks. Sci. Rep. 2015, 5, 8930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rezola, A.; Pey, J.; Rubio, Á.; Planes, F.J. In-Silico Prediction of Key Metabolic Differences between Two Non-Small Cell Lung Cancer Subtypes. PLoS ONE 2014, 9, e103998. [Google Scholar] [CrossRef] [PubMed]
- Orth, J.D.; Thiele, I.; Palsson, B.Ø. What is flux balance analysis? Nat. Biotechnol. 2010, 28, 245–248. [Google Scholar] [CrossRef]
- Rowe, E.; Palsson, B.O.; King, Z.A. Escher-FBA: A web application for interactive flux balance analysis. BMC Syst. Biol. 2018, 12, 84. [Google Scholar] [CrossRef]
- König, M.; Holzhütter, H.G. Fluxviz—Cytoscape plug-in for visualization of flux distributions in networks. In Genome Informatics 2010; Imperial College Press: London, UK, 2010; pp. 96–103. [Google Scholar] [CrossRef]
- Hoppe, A.; Hoffmann, S.; Gerasch, A.; Gille, C.; Holzhütter, H.G. FASIMU: Flexible software for flux-balance computation series in large metabolic networks. BMC Bioinform. 2011, 12, 28. [Google Scholar] [CrossRef] [Green Version]
- Junker, B.; Klukas, C.; Schreiber, F. VANTED: A system for advanced data analysis and visualization in the context of biological networks. BMC Bioinform. 2006, 7, 109. [Google Scholar] [CrossRef] [Green Version]
- Kostromins, A.; Stalidzans, E. Paint4Net: COBRA Toolbox extension for visualization of stoichiometric models of metabolism. Biosystems 2012, 109, 233–239. [Google Scholar] [CrossRef]
- Heirendt, L.; Arreckx, S.; Pfau, T.; Mendoza, S.N.; Richelle, A.; Heinken, A.; Haraldsdóttir, H.S.; Wachowiak, J.; Keating, S.M.; Vlasov, V.; et al. Creation and analysis of biochemical constraint-based models: The COBRA Toolbox v3.0. arXiv 2017, arXiv:1710.04038. [Google Scholar] [CrossRef] [Green Version]
- Klamt, S.; Saez-Rodriguez, J.; Gilles, E.D. Structural and functional analysis of cellular networks with CellNetAnalyzer. BMC Syst. Biol. 2007, 1, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, P.A.; Papin, J.A. MetDraw: Automated visualization of genome-scale metabolic network reconstructions and high-throughput data. Bioinformatics 2014, 30, 1327–1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noronha, A.; Daníelsdóttir, A.D.; Gawron, P.; Jóhannsson, F.; Jónsdóttir, S.; Jarlsson, S.; Gunnarsson, J.P.; Brynjólfsson, S.; Schneider, R.; Thiele, I.; et al. ReconMap: An interactive visualization of human metabolism. Bioinformatics 2016, 33, btw667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose, T.; Mazat, J.P.; Rose, T.D.; Mazat, J.P. FluxVisualizer, a Software to Visualize Fluxes through Metabolic Networks. Processes 2018, 6, 39. [Google Scholar] [CrossRef] [Green Version]
- Chazalviel, M.; Frainay, C.; Poupin, N.; Vinson, F.; Merlet, B.; Gloaguen, Y.; Cottret, L.; Jourdan, F. MetExploreViz: Web component for interactive metabolic network visualization. Bioinformatics 2018, 34, 312–313. [Google Scholar] [CrossRef]
- Noronha, A.; Vilaça, P.; Rocha, M. An integrated network visualization framework towards metabolic engineering applications. BMC Bioinform. 2014, 15, 420. [Google Scholar] [CrossRef] [Green Version]
- Feist, A.M.; Henry, C.S.; Reed, J.L.; Krummenacker, M.; Joyce, A.R.; Karp, P.D.; Broadbelt, L.J.; Hatzimanikatis, V.; Palsson, B.Ø. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol. Syst. Biol. 2007, 3, 121. [Google Scholar] [CrossRef]
- Swainston, N.; Smallbone, K.; Hefzi, H.; Dobson, P.D.; Brewer, J.; Hanscho, M.; Zielinski, D.C.; Ang, K.S.; Gardiner, N.J.; Gutierrez, J.M.; et al. Recon 2.2: From reconstruction to model of human metabolism. Metabolomics 2016, 12, 109. [Google Scholar] [CrossRef]
- Thiele, I.; Palsson, B.Ø. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010, 5, 93–121. [Google Scholar] [CrossRef] [Green Version]
- Pey, J.; Villar, J.A.; Tobalina, L.; Rezola, A.; García, J.M.; Beasley, J.E.; Planes, F.J. TreeEFM: Calculating elementary flux modes using linear optimization in a tree-based algorithm. Bioinformatics 2015, 31, 897–904. [Google Scholar] [CrossRef] [Green Version]
- Song, H.S.; Ramkrishna, D. Reduction of a set of elementary modes using yield analysis. Biotechnol. Bioeng. 2009, 102, 554–568. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Väremo, L.; Nielsen, J.; Nookaew, I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013, 41, 4378–4391. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- König, M.; Drager, A.; Holzhutter, H.G. CySBML: A Cytoscape plugin for SBML. Bioinformatics 2012, 28, 2402–2403. [Google Scholar] [CrossRef] [Green Version]
- Novère, N.L.; Hucka, M.; Mi, H.; Moodie, S.; Schreiber, F.; Sorokin, A.; Demir, E.; Wegner, K.; Aladjem, M.I.; Wimalaratne, S.M.; et al. The Systems Biology Graphical Notation. Nat. Biotechnol. 2009, 27, 735–741. [Google Scholar] [CrossRef]
- yWorks GmbH. yFiles Layout Algorithms for Cytoscape app; yWorks GmbH: Tübingen, Germany, 2018. [Google Scholar]
- Ono, K.; Muetze, T.; Kolishovski, G.; Shannon, P.; Demchak, B. CyREST: Turbocharging Cytoscape Access for External Tools via a RESTful API. F1000Research 2015, 4, 478. [Google Scholar] [CrossRef]
- Pratt, D.; Chen, J.; Welker, D.; Rivas, R.; Pillich, R.; Rynkov, V.; Ono, K.; Miello, C.; Hicks, L.; Szalma, S.; et al. NDEx, the network data exchange. Cell Syst. 2015, 1, 302–305. [Google Scholar] [CrossRef] [Green Version]
- Eiteman, M.A.; Altman, E. Overcoming acetate in Escherichia coli recombinant protein fermentations. Trends Biotechnol. 2006, 24, 530–536. [Google Scholar] [CrossRef]
- Clomburg, J.M.; Gonzalez, R. Biofuel production in Escherichia coli: The role of metabolic engineering and synthetic biology. Appl. Microbiol. Biotechnol. 2010, 86, 419–434. [Google Scholar] [CrossRef]
- Nakano, K.; Rischke, M.; Sato, S.; Märkl, H. Influence of acetic acid on the growth of Escherichia coli K12 during high-cell-density cultivation in a dialysis reactor. Appl. Microbiol. Biotechnol. 1997, 48, 597–601. [Google Scholar] [CrossRef]
- Contiero, J.; Beatty, C.; Kumari, S.; DeSanti, C.L.; Strohl, W.R.; Wolfe, A. Effects of mutations in acetate metabolism on high-cell-density growth of Escherichia coli. J. Ind. Microbiol. Biotechnol. 2000, 24, 421–430. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CGAN. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarathy, C.; Kutmon, M.; Lenz, M.; Adriaens, M.E.; Evelo, C.T.; Arts, I.C.W. EFMviz: A COBRA Toolbox Extension to Visualize Elementary Flux Modes in Genome-Scale Metabolic Models. Metabolites 2020, 10, 66. https://doi.org/10.3390/metabo10020066
Sarathy C, Kutmon M, Lenz M, Adriaens ME, Evelo CT, Arts ICW. EFMviz: A COBRA Toolbox Extension to Visualize Elementary Flux Modes in Genome-Scale Metabolic Models. Metabolites. 2020; 10(2):66. https://doi.org/10.3390/metabo10020066
Chicago/Turabian StyleSarathy, Chaitra, Martina Kutmon, Michael Lenz, Michiel E. Adriaens, Chris T. Evelo, and Ilja C.W. Arts. 2020. "EFMviz: A COBRA Toolbox Extension to Visualize Elementary Flux Modes in Genome-Scale Metabolic Models" Metabolites 10, no. 2: 66. https://doi.org/10.3390/metabo10020066




