Abstract
Gliomas, primary brain tumors arising from glial cells, can be effectively identified using Magnetic Resonance Imaging (MRI), a widely employed diagnostic tool in clinical settings. Accurate glioma segmentation, which is crucial for diagnosis and surgical intervention, can be achieved by integrating multiple MRI modalities that offer complementary information. However, limited access to multiple modalities in certain clinical contexts often results in suboptimal performance of glioma segmentation methods. This study introduces a novel generalized knowledge distillation framework designed to transfer multimodal knowledge from a teacher model to a unimodal student model via two distinct distillation strategies: segmentation graph distillation and cascade region attention distillation. The former enables the student to replicate the teacher’s softened output, whereas the latter facilitates extraction and learning of region feature information at various levels within the teacher model. Our evaluation of the proposed distillation strategies using the BraTS 2018 dataset confirms their superior performance in unimodal segmentation contexts compared with existing methods.
1. Introduction
Gliomas are a common type of brain tumor originating from glial cells in the brain [1]. They are classified into four grades based on their malignancy, with grades I and II being low-grade and grades III and IV being high-grade. Low-grade gliomas are less dangerous, while high-grade gliomas are heterogeneous and aggressive [2]. Overall, gliomas have become the most prevalent and deadly brain tumor disease.
Currently, magnetic resonance imaging (MRI) technology is usually used for glioma examination. Compared with other auxiliary imaging methods [3], MRI provides clear anatomical structures and presents high-quality images without skull artifacts [4]. Four MRI modalities, namely T1, T1ce, T2, and Flair [5], are commonly used for glioma diseases. Different MRI modalities can accentuate and describe different tissues. Figure 1a–d show imaging results of these four modalities, and (e) shows the segmentation results labeled by experts, where red indicates a region of necrosis and non-enhancing tumor (NCR/NET), yellow indicates an enhancing tumor (ET) region, and green indicates a peritumoral edema (ED) region [6]. To evaluate glioma image segmentation, these three regions are typically combined into three nested subregions: the enhancing tumor (ET), tumor core (TC, including ET and NCR/NET), and the whole tumor (WT, including ET, NCR/NET and ED) [5].
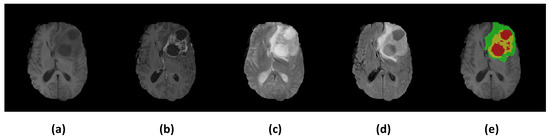
Figure 1.
Illustrations of multimodal MR images from the BraTS 2018 dataset [5,7]. (a) T1; (b) T1ce; (c) T2; (d) Flair; (e) Ground truth, in which red indicates NCR/NET regions, yellow indicates ET regions, and green indicates ED regions.
In general, the accurate segmentation of glioma images is crucial for clinical diagnosis and effective surgical planning. In recent years, deep learning has gained popularity in medical image segmentation due to its promising performance. Because different image modalities contain different tissue structure information, most previous work [8,9,10,11,12,13,14,15,16] has fused multiple modalities to significantly improve segmentation accuracy. Using multiple modalities for joint learning typically leads to very good results, but optimal performance during inference requires the use of the complete set of modalities; otherwise, the performance may be significantly compromised. However, in clinical settings, it is often difficult to obtain complete multimodal datasets. In most cases, only one modality can be collected for segmentation at the time of inference, due to broken scanners, limited numbers, patients’ allergies to certain contrast agents, and the unavailability of acquired MRI modalities [17].
Having multiple modalities available during training but most of them missing during inference can result in poor segmentation accuracy. There are several mainstream approaches to solving the problem of missing modalities at inference time. The first approach to the problem involves trying to synthesize the missing modalities to complete the set of modalities during inference time. Van Tulder and de Bruijne et al. [18] suggested that, when dealing with missing modalities in medical image classification, accuracy can be enhanced by using synthetic data to substitute for the missing modalities. Jog et al. [19] provided a random forest image synthesis approach to synthesize the missing modalities. In addition, Ben-Cohen et al. [20] generated simulated PET data using input CT data. The synthesized PET data can be used to reduce false positives when detecting liver lesions. Similarly, Yu et al. [21] generated Flair modality using a 3D conditional Generative Adversarial Network to help improve brain tumor segmentation from the single modality of T1. However, such methods are computationally cumbersome and resource-intensive. They require a generative model to be trained for each missing modality. Furthermore, such image synthesis-based methods also face great challenges in the scenario of unimodal segmentation, because it is very difficult to recover other modalities when only one modality is available.
The second approach is to learn a common modality-invariant latent representation space that allows any combinatorial subset of available modalities as input during inference time. The HeMIS [22] proposed by Havaei et al. first trains a feature encoder for each modality from the input modality to the latent space, and then the mean and variance of the feature maps for all modalities are calculated and combined through concatenation in the latent space. Finally, the concatenated mean and variance feature maps are fed to the decoder for training to obtain segmentation maps. Inspired by HeMIS, Dorent et al. proposed U-HVED [23]. U-HVED trains different feature extractors for each modality to extract features and then constructs a shared representation space by modeling the Gaussian distribution of the features. U-HVED outperforms HeMIS on the BraTS 2018 dataset. In [24], the RS-Net, a regression-segmentation 3D CNN, creates a shared representation of all modalities and can generate missing modalities. It consists of three modules. The first module generates intermediate latent representations from all modality data, and the second module uses the latent representations and existing modalities to synthesise the missing modalities. The third module takes the generated latent representation as input and outputs a segmentation map. Ideally, obtaining a single model can handle various combinations of missing modalities [17]. However, when only one modality is available for inference, this second approach does not perform as well as models trained with only a specific modality.
Additionally, Hu et al. [25] proposed a segmentation framework, KD-Net, based on a generalized knowledge distillation strategy [26] to solve the aforementioned missing modality problem in glioma. KD-Net employed the KL divergence loss to incentivize the student’s latent space to resemble the teacher’s and applied a distillation loss between the outputs of the teacher and student. This method transfers knowledge from the multimodal teacher network to the unimodal student network. However, their framework does not extract the features of different layers to learn rich knowledge, and this may waste the network’s learning ability.
While earlier approaches offer versatility in dealing with different missing modality scenarios, they tend to fall short in delivering accurate segmentation results when only a single modality is available, particularly in clinical settings. Considering this drawback of the above works, in this paper, we propose a novel framework based on a generalized knowledge distillation strategy that combines distillation and privileged information [26,27,28]. Privileged information is specific information that the teacher model can access during training, while the student model cannot directly access this information during training and can only obtain it by distillation learning from the teacher model. In our framework, the multimodal privileged information in the multimodal teacher model is distilled and transferred to the unimodal student model, improving the segmentation accuracy of the unimodal model. We designed two distillation modules to distill privileged information: the cascade region attention distillation module and the segmentation graph distillation module. The cascade region attention distillation module extracts the features that need to be learned in the three layers of the teacher model by using the WT, TC, and ET subregion label masks and then forces the student’s region features to resemble the teacher’s related region features, to avoid redundant learning and enable more information to be obtained. The segmentation graph distillation module encourages the student model to imitate the softened output of the teacher model to learn segmentation capabilities. In real clinical scenarios, doctors usually diagnose and grade gliomas based on their subregions (WT, ET, TC), so we should focus more on the information in the subregions. Therefore, unlike other knowledge distillation methods, we distill information in a targeted manner based on the information that doctors need in real situations. The distillation strategy is accurate and efficient, which significantly improves the segmentation accuracy of the unimodal model. We conducted experiments on the BraTS 2018 dataset in order to demonstrate our proposed framework’s effectiveness and superior performance in unimodal scenarios.
Overall, our main contributions are:
- We propose a knowledge distillation framework that can fully extract the knowledge of the multimodal teacher model and transfer it to the unimodal student model, improving the segmentation performance of the student model.
- We devise a novel Cascade Region Attention Distillation (CRAD) module to construct feature region similarity through label masks, distill the region feature information of the teacher model and transfer it to the student model, and improve the segmentation accuracy of the student model.
- We validate our framework with extensive experiments on the BraTS 2018 dataset, demonstrating the effectiveness of the proposed distillation framework and achieving state-of-the-art segmentation performance in unimodal scenarios.
2. Methodology
Our proposed knowledge distillation framework for unimodal glioma segmentation is shown in Figure 2. The framework has a multimodal glioma segmentation network (teacher model) and a unimodal glioma segmentation network (student model), with identical U-shaped network architecture. Our proposed framework aims to enhance the segmentation accuracy of the unimodal student model by leveraging the rich and complete modal information from the teacher model during training.
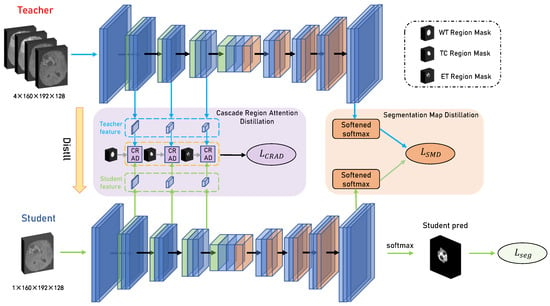
Figure 2.
The presentation of our proposed distillation framework. Two networks, one for the teacher model and the other for the student model, are vertically depicted in the framework. The teacher model uses a combination of four modalities as input, and the student model uses one modality as input. The distillation process is separated into two modules, i.e., the Cascade Region Attention Distillation (CRAD) module and the Segmentation Graph Distillation (SMD) module.
The segmentation loss was first used to train the teacher model. Because the input of the teacher model was the complete four-modal data, the modal information can be fully utilized, resulting in high segmentation accuracy and good results. Then, the student model was trained using three parts: cascade region attention distillation, segmentation graph distillation, and segmentation loss.
The cascade region attention distillation module transferred the intermediate feature information of the teacher model by constructing region feature similarity through label masks. Then, the segmentation graph distillation module encouraged the student model to learn segmentation skills by mimicking the output of the last layer of the teacher model. Finally, the segmentation loss was added to make the segmentation output of the student model as similar as possible to the segmentation labels. Benefiting from this architecture, the student model performed the segmentation task during inference time without requiring other modalities. Each module is described in detail below.
2.1. Segmentation Map Distillation
Knowledge distillation [29] usually distills knowledge from the teacher model and transfers it to the student model. Typically, this is accomplished by calculating the difference between the outputs of their final layers. Inspired by the above knowledge distillation method, we followed the previous work on knowledge distillation of medical semantic segmentation [30,31,32] to construct the segmentation map distillation (SMD) module.
Specifically, to train the student model’s segmentation ability, we aimed to minimize the difference between its output feature segmentation map at the final layer and that of the teacher model. The segmentation graph distillation loss function is as follows:
where is the Kullback–Leibler (KL) divergence function, which can measure the difference between the distributions of the two datasets. and are the values of the segmentation map adjusted by and , where is the output segmentation map of the last layer of the teacher model, and is the output segmentation map of the student model. i equals the number of classes. The temperature parameter is a hyperparameter that adjusts the softness of the probability distribution. When , the softmax function is standard. Increasing the value of results in a softer probability distribution, providing more information.
Note that the KL divergence function is asymmetric; is not equal to . was used to let the teacher model guide the student model for training. This module is shown as the orange area in Figure 2.
2.2. Cascade Region Attention Distillation
In our network architecture, we have encoder and decoder modules, and the encoder part is mainly responsible for extracting data features. Generally, the shallow layers of the encoder extract low-level image features, including color, texture, and edges. When the receptive field of the convolution layer is small, the overlapping area of the receptive field is also small, which can ensure that the network captures more details. As the network’s depth increases, the convolution layer’s receptive field gradually increases, and the overlapping area of the receptive field becomes larger, so it will extract more global and high-level semantic features. As a result, the features extracted by the encoder increase gradually from low- to high-level.
Additionally, we noticed that, for the glioma segmentation task, the expert labeling results are often evaluated based on three nested subregions: the enhancing tumor (ET), the tumor core (TC), and the whole tumor (WT). Among these three subregions, the WT region is the largest and covers the whole tumor, while the ET region is the smallest. Moreover, in real clinical scenarios, when doctors manually segment glioma MR images, they would typically first determine the overall lesion area (WT), and then further divide the internal subregions of the lesion (TC, ET). Therefore, inspired by doctors to obtain information from the outside to the inside when segmenting gliomas, we proposed a novel distillation module named Cascade Region Attention Distillation (CRAD). The CRAD module combines the encoder’s pattern of extracting features from low-level to high-level with the three nested subregions of the lesion, effectively mines the feature information in the multiple layers of the teacher model, and uses this information to guide student model training.
To this end, the ground truth labels were first binarized according to the three subregions of WT, TC, and ET to obtain three binary label masks. The formula is as follows:
where i is the index of the label mask, denoting three label masks (WT, TC, ET), respectively, and j is the index of the pixel. It can be seen that the value of the pixel inside the region is set to 1, while the value of the pixel outside the region is 0. This allows us to selectively extract feature information from the model feature map using label masks, keeping the feature information inside the region and discarding the unimportant information outside the region.
The above shows that the encoder has a small receptive field in the shallow layer, usually capturing low-level image features. Therefore, the WT label was used as a mask with the largest valid region (pixel value of 1) to extract feature maps in the shallow layers, preserving as much feature information as possible. In the deeper layers, a label mask with a smaller valid region was used to extract global semantic information. Then, we calculated the similarity of the corresponding region information between teachers and students. In this way, our student model can not only avoid the interference of redundant features but also efficiently learn useful feature information from the teacher model.
The CRAD module’s architecture is depicted in Figure 3. In detail, the feature maps are extracted from the three layers of the encoder respectively, and the sizes are , , . First, the ground truth labels were binarized according to the WT, TC, and ET to obtain three binary label masks , and resize these three label masks as , , . Then, given a binary label mask , we multiplied it channel-by-channel with the feature map of the corresponding size to calculate the feature region vector . The element-wise multiplication formula is as follows:
where is the index of the label mask and the corresponding feature map, is the size of the label mask, j represents pixel indices, and denotes the number of pixels with a pixel value of 1 in the label mask i. Then, given the feature vectors and of a certain layer of the teacher and student models, the cosine similarity between the two feature vectors was computed to encourage the student model to learn from the teacher model’s feature information. The loss function of this distillation module is defined as:
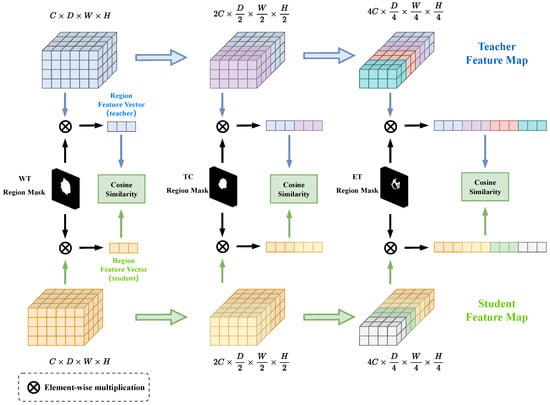
Figure 3.
The presentation of the Cascade Region Attention Distillation (CRAD) module. The CRAD module takes in multilayered feature maps from the teacher and student models as input and then multiplies them with the corresponding label masks to obtain the regional feature vectors. The cascade regional distillation loss is obtained by calculating the cosine similarity between the regional feature vectors of teachers and students in the end.
We concatenated the losses calculated by the three layers of the network together to obtain an overall loss to efficiently train the student model.
In summary, the WT label mask was used to extract the region feature information of the feature map in the shallow layer, the TC label mask was used to extract the feature map’s information in the middle layer, and the deepest region feature information was extracted by the ET label mask. It can be seen that as the encoder layer gets deeper, the valid region of the used label mask gets smaller. This design was based on the fact that during feature extraction, the encoder identified the more obvious outer lesion regions (WT) using low-level image features (color, texture, edges) extracted at a shallow layer, and further delineated the lesion regions (TC, ET) using high-level semantic information learned at a deeper layer. Therefore, we used different label masks in different layers, which can extract feature information accurately and effectively, and transfer more useful knowledge to the student model. In the experimental section, we further demonstrate the rationality and effectiveness of the order in which the label masks were used by this distillation module.
2.3. Objective Function
As shown in Figure 2, the two distillation loss functions mentioned above were combined to train the student model end-to-end, with the total loss function defined as follows:
where is the segmentation loss function, which makes the output segmentation map similar to the ground truth, and is composed of both the cross-entropy loss function and the Dice loss function. The hyperparameters and range between 0 and 1. Based on the experimental results, was set to 0.75, and was set to 0.9.
The teacher model was pre-trained using segmentation loss, which combined the cross-entropy and Dice loss functions, as the first step in training our framework. This ensured that the well-trained teacher model could provide high-quality feature maps and soft labels. After pre-training, the teacher model was not updated, and the student model was trained using the above loss function (Equation (6)), guided by the teacher model.
Experiments showed that our method can effectively improve the segmentation performance of the unimodal student model and solve the missing modality problem.
3. Experimental Results
3.1. Dataset
We evaluated the proposed framework’s performance using the BraTS 2018 dataset [5,7] from the Multimodal Brain Tumor Segmentation Challenge. It contains MR images from a total of 285 patients: 210 high-grade gliomas and 75 low-grade gliomas. Four modalities, namely T1, T2, T1ce, and Flair, are present in each MR image. The label for each image segmentation includes three subregions: the enhancing tumor (ET), the peritumoral edema (ED), and the necrotic and non-enhancing tumor core (NCR/NET). To evaluate the segmentation results, these three subregions are organized into three nested subregions: the enhancing tumor (ET), the tumor core (TC), and the whole tumor (WT). All images were normalized and cropped to .
3.2. Implementation Details
All experiments were implemented using PyTorch, a deep learning framework supported by the PyTorch Foundation, and run on a 40GB NVIDIA TESLA A100 GPU, maufactured in State of California, USA. We adopted U-net as the network architecture for the teacher and student models.
During model training, the objective loss function parameters were set as and . In addition, the input image size was . The Adam optimizer was utilized, and the initial learning rate was set to . The learning rate was multiplied by the formula every epoch and gradually decreased. Note that the number of epochs was 200. We performed three-fold cross-validation on the BraTS 2018 dataset.
3.3. Evaluation Metric
The segmentation performance of three nested subregions of glioma is usually evaluated using the Dice similarity coefficient (DSC) [33]. A higher DSC indicates better segmentation performance. The formula for the Dice similarity coefficient is as follows:
where is the segmentation result of the model, and is the ground truth label. We used the Dice similarity coefficient to judge the similarity between the model output and the ground truth. The DSC effectively measures the segmentation performance of the model.
3.4. Results and Analysis
In our experiments, we use a combination of four MR modalities to train the teacher model. The training of the unimodal student model is subsequently guided by the trained teacher model.
Model comparison: We compare our framework with four methods to verify the superiority and effectiveness of the framework. U-net [34] is used as a benchmark for all methods. Because the basic settings and datasets are the same, we directly reference their results to compare.
First, we compared our framework with two well-known frameworks, U-HeMIS [22] and HVED [23]. These are multimodal segmentation methods that can handle various combinations of possible missing modes. Here we compared the segmentation performance in a unimodal scenario. As shown in Table 1, we observed that U-HeMIS and HVED are not robust enough. They gave poor results when applied to unimodality. The performance of our framework far outperformed these two methods. For example, in T1 modality, the DSC score of both methods on ET was only around , while our method achieved a DSC score close to , making it more useful in clinical practice.

Table 1.
Comparison of the proposed method with U-HeMIS [22] and U-HVED [23] methods on three nested sub-regions (ET, TC, WT). DSC is used as an evaluation metric. Existing modalities are marked with •, while missing modalities are marked with ∘.
In Table 2, our method is compared with KD-Net [25], a framework for knowledge distillation used for unimodal segmentation, and the state-of-the-art method ACN [35]. Compared with KD-Net, our proposed knowledge distillation framework can learn rich knowledge of different layers, and the mean DSC scores increased by , , and on the four modalities (Flair, T1, T1ce and T2), respectively. Moreover, we observed that our framework outperformed the state-of-the-art method ACN in unimodal scenarios. The mean DSC scores increased by , , and on the four modalities (Flair, T1, T1ce and T2) respectively.

Table 2.
Comparison of the proposed method with the knowledge distillation framework KD-Net [25] and the state-of-the-art method ACN [35] (DSC%).
Experiments demonstrated the effectiveness and superiority of the knowledge distillation strategy proposed in our method.
Qualitative results: To highlight the effectiveness of our framework, Figure 4 shows the qualitative results of our framework in a unimodal scenario. Compared with the ground truth, we can observe that inference for each modality as input produces acceptable segmentation results. This shows that our method effectively learns the knowledge of other modalities from the teacher network, complementing the missing information.
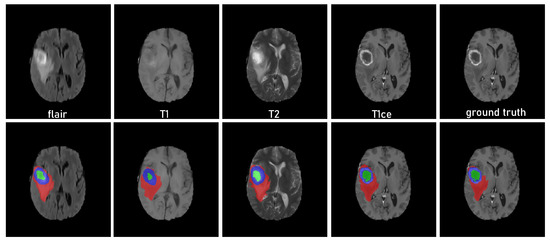
Figure 4.
Qualitative results of the framework obtained on the brain tumor dataset [5] using unimodality as input. The green indicates NCR/NET regions, blue indicates ET regions, and red indicates ED regions.
Ablation study: In this section, we conducted experiments to test how well the proposed distillation strategy works. We first evaluated the contributions of each proposed distillation module (SMD and CRAD). We chose Flair as the only available modality. First, we built a baseline model using only the segmentation loss . We then gradually added each proposed module. As shown in Table 3, we observed that both the SMD and CRAD modules effectively extracted additional helpful knowledge from the teacher model, improving the segmentation performance of the baseline network. This demonstrated the effectiveness of the proposed distillation module.

Table 3.
Contribution of each module in the unimodal (Flair) scenario (DSC%).
In addition, we explored the rationality of the order in which label masks are used in the cascade region attention distillation module. We first trained the model using the segmentation loss and the cascade region attention distillation loss . Then, we exchanged the order in which the label masks were used, let the ET label mask with a small valid region (pixel value of 1) extract the region feature information of the feature map in the shallow layer, and let the WT label mask with the largest valid region extract the region features in the deep layer; hence, we obtained the distillation loss to train the model. As shown in Table 4, we observed that the segmentation performance of the model trained using was significantly better than that using , with an increase of . The experimental results demonstrated the effectiveness and rationality of using label masks with large valid regions to extract features in the shallow layer and label masks with smaller valid regions in the deeper layer.

Table 4.
Exploration of CRAD loss in the unimodal (Flair) scenario (DSC%).
4. Conclusions
In this study, we presented a novel generalized knowledge distillation framework to overcome the limitations of missing modalities in glioma segmentation, particularly in unimodal scenarios. Our framework successfully extracted rich knowledge from a multimodal segmentation model and transferred it to a unimodal segmentation model, enhancing its performance. We introduced two knowledge distillation strategies—segmentation map distillation and cascade region attention distillation—to effectively transfer multimodal knowledge from the teacher model. The segmentation map distillation strategy enabled the student model to mimic the teacher’s output and acquire segmentation capabilities. In contrast, the cascade region attention distillation strategy employed label masks to concentrate on local features and allowed the student model to focus on essential knowledge without being distracted by superfluous feature information.
Notably, our proposed framework required less training effort than alternative methods and demonstrated superior segmentation performance in unimodal scenarios. When applied to the BraTS 2018 dataset in a unimodal inference context, our framework outperformed existing approaches, highlighting its effectiveness.
Future work will investigate the potential for further development of our framework, including exploring more efficient modality fusion methods for the teacher model to address potential missing modality issues during training and examining the mapping relationships between different region masks to enhance segmentation performance. These advancements will further improve our framework and significantly impact real-world clinical practice.
Author Contributions
Conceptualization, F.X. and X.W.; methodology, F.X., C.S. and X.W.; software, F.X. and C.S.; validation, F.X. and C.S.; writing—original draft preparation, F.X.; writing—review and editing, X.W. and C.S.; visualization, F.X.; supervision, X.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are publicly available. The data can be found at: https://www.kaggle.com/datasets/sanglequang/brats2018?select=MICCAI_BraTS_2018_Data_Training (accessed on 28 February 2023).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Claus, E.B.; Walsh, K.M.; Wiencke, J.K.; Molinaro, A.M.; Wiemels, J.L.; Schildkraut, J.M.; Bondy, M.L.; Berger, M.; Jenkins, R.; Wrensch, M. Survival and low-grade glioma: The emergence of genetic information. Neurosurg. Focus 2015, 38, E6. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Yang, B.; Xiao, Y.; Liu, S.; Liu, M.; Yin, L.; Zheng, W. Iterative reconstruction of low-dose CT based on differential sparse. Biomed. Signal Process. Control 2023, 79, 104204. [Google Scholar] [CrossRef]
- Yan, J.; Chen, S.; Zhang, Y.; Li, X. Neural architecture search for compressed sensing magnetic resonance image reconstruction. Comput. Med. Imaging Graph. 2020, 85, 101784. [Google Scholar] [CrossRef] [PubMed]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; Hong, Y.; Liu, J. Aggregation-and-Attention Network for brain tumor segmentation. BMC Med. Imaging 2021, 21, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.M.; Larochelle, H. Brain tumor segmentation with deep neural networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef]
- Zhou, C.; Ding, C.; Lu, Z.; Wang, X.; Tao, D. One-pass multi-task convolutional neural networks for efficient brain tumor segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, 16–20 September 2018; pp. 637–645. [Google Scholar]
- Wang, Y.; Zhang, Y.; Hou, F.; Liu, Y.; Tian, J.; Zhong, C.; Zhang, Y.; He, Z. Modality-pairing learning for brain tumor segmentation. In Proceedings of the International MICCAI Brainlesion Workshop, Lima, Peru, 4 October 2020; pp. 230–240. [Google Scholar]
- Maier, O.; Menze, B.H.; von der Gablentz, J.; Häni, L.; Heinrich, M.P.; Liebrand, M.; Winzeck, S.; Basit, A.; Bentley, P.; Chen, L.; et al. ISLES 2015—A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Med. Image Anal. 2017, 35, 250–269. [Google Scholar] [CrossRef]
- Dolz, J.; Gopinath, K.; Yuan, J.; Lombaert, H.; Desrosiers, C.; Ayed, I.B. HyperDense-Net: A hyper-densely connected CNN for multi-modal image segmentation. IEEE Trans. Med. Imaging 2018, 38, 1116–1126. [Google Scholar] [CrossRef]
- Tseng, K.L.; Lin, Y.L.; Hsu, W.; Huang, C.Y. Joint sequence learning and cross-modality convolution for 3D biomedical segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 6393–6400. [Google Scholar]
- Yu, B.; Zhou, L.; Wang, L.; Yang, W.; Yang, M.; Bourgeat, P.; Fripp, J. Learning sample-adaptive intensity lookup table for brain tumor segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; pp. 216–226. [Google Scholar]
- Jia, H.; Xia, Y.; Cai, W.; Huang, H. Learning high-resolution and efficient non-local features for brain glioma segmentation in MR images. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; pp. 480–490. [Google Scholar]
- Chen, C.; Dou, Q.; Jin, Y.; Liu, Q.; Heng, P.A. Learning with privileged multimodal knowledge for unimodal segmentation. IEEE Trans. Med. Imaging 2021, 41, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Tulder, G.V.; Bruijne, M.D. Why does synthesized data improve multi-sequence classification? In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 531–538. [Google Scholar]
- Jog, A.; Carass, A.; Roy, S.; Pham, D.L.; Prince, J.L. Random forest regression for magnetic resonance image synthesis. Med. Image Anal. 2017, 35, 475–488. [Google Scholar] [CrossRef]
- Ben-Cohen, A.; Klang, E.; Raskin, S.P.; Soffer, S.; Ben-Haim, S.; Konen, E.; Amitai, M.M.; Greenspan, H. Cross-modality synthesis from CT to PET using FCN and GAN networks for improved automated lesion detection. Eng. Appl. Artif. Intell. 2019, 78, 186–194. [Google Scholar] [CrossRef]
- Yu, B.; Zhou, L.; Wang, L.; Fripp, J.; Bourgeat, P. 3D cGAN based cross-modality MR image synthesis for brain tumor segmentation. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 626–630. [Google Scholar]
- Havaei, M.; Guizard, N.; Chapados, N.; Bengio, Y. Hemis: Hetero-modal image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; pp. 469–477. [Google Scholar]
- Dorent, R.; Joutard, S.; Modat, M.; Ourselin, S.; Vercauteren, T. Hetero-modal variational encoder-decoder for joint modality completion and segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; pp. 74–82. [Google Scholar]
- Mehta, R.; Arbel, T. RS-Net: Regression-segmentation 3D CNN for synthesis of full resolution missing brain MRI in the presence of tumours. In Proceedings of the International Workshop on Simulation and Synthesis in Medical Imaging, Granada, Spain, 16 September 2018; pp. 119–129. [Google Scholar]
- Hu, M.; Maillard, M.; Zhang, Y.; Ciceri, T.; La Barbera, G.; Bloch, I.; Gori, P. Knowledge distillation from multi-modal to mono-modal segmentation networks. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 772–781. [Google Scholar]
- Lopez-Paz, D.; Bottou, L.; Schölkopf, B.; Vapnik, V. Unifying distillation and privileged information. arXiv 2015, arXiv:1511.03643. [Google Scholar]
- Vapnik, V.; Vashist, A. A new learning paradigm: Learning using privileged information. Neural Netw. 2009, 22, 544–557. [Google Scholar] [CrossRef]
- Vapnik, V.; Izmailov, R. Learning using privileged information: Similarity control and knowledge transfer. J. Mach. Learn. Res. 2015, 16, 2023–2049. [Google Scholar]
- Hinton, G.; Vinyals, O.; Dean, J. Distilling the knowledge in a neural network. arXiv 2015, arXiv:1503.02531. [Google Scholar]
- He, T.; Shen, C.; Tian, Z.; Gong, D.; Sun, C.; Yan, Y. Knowledge adaptation for efficient semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 578–587. [Google Scholar]
- Liu, Y.; Chen, K.; Liu, C.; Qin, Z.; Luo, Z.; Wang, J. Structured knowledge distillation for semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 2604–2613. [Google Scholar]
- Qin, D.; Bu, J.J.; Liu, Z.; Shen, X.; Zhou, S.; Gu, J.J.; Wang, Z.H.; Wu, L.; Dai, H.F. Efficient medical image segmentation based on knowledge distillation. IEEE Trans. Med. Imaging 2021, 40, 3820–3831. [Google Scholar] [CrossRef]
- Zou, K.H.; Warfield, S.K.; Bharatha, A.; Tempany, C.M.; Kaus, M.R.; Haker, S.J.; Wells III, W.M.; Jolesz, F.A.; Kikinis, R. Statistical validation of image segmentation quality based on a spatial overlap index1: Scientific reports. Acad. Radiol. 2004, 11, 178–189. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Wang, Y.; Zhang, Y.; Liu, Y.; Lin, Z.; Tian, J.; Zhong, C.; Shi, Z.; Fan, J.; He, Z. Acn: Adversarial co-training network for brain tumor segmentation with missing modalities. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 October–1 November 2021; pp. 410–420. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).