High-Level Hessian-Based Image Processing with the Frangi Neuron
Abstract
1. Introduction
2. Materials and Methods
2.1. Frangi Filter
| Algorithm 1: Frangi filtering algorithm. |
 |
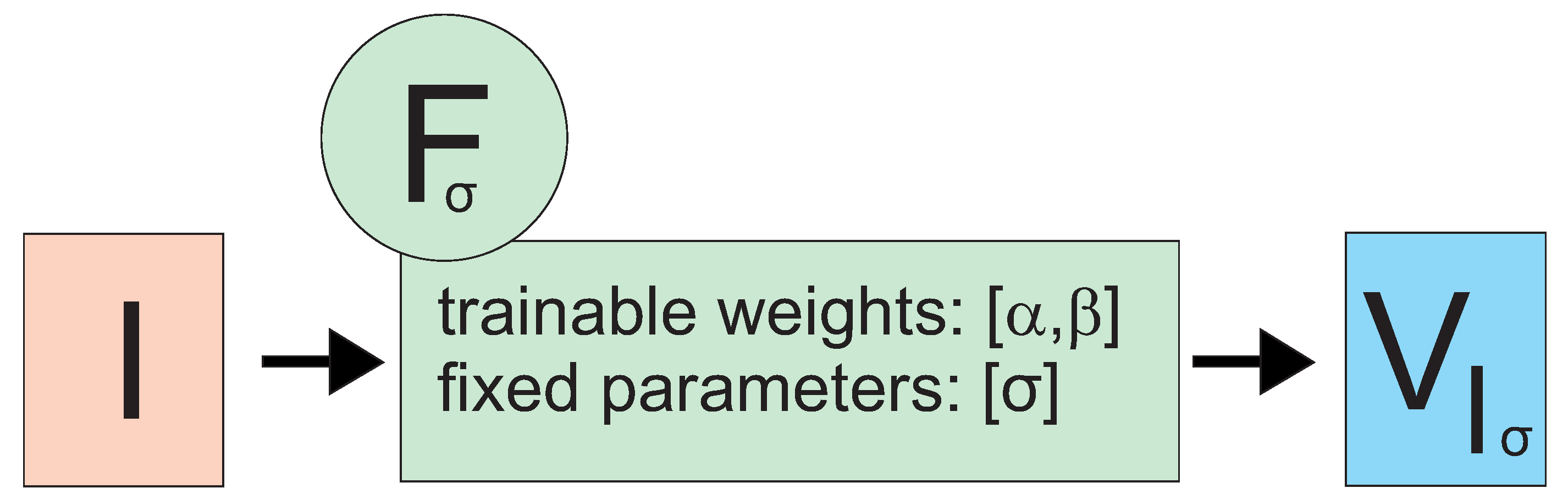
2.2. Frangi Neuron
2.3. Example Application: Network Using Frangi Neurons for Image Segmentation (Frangi Network)
2.4. Datasets
2.4.1. Retinopathy (Retina)
2.4.2. X-ray Coronary Artery Angiography (Coronary Artery)
2.4.3. Brain MRA (Brain)
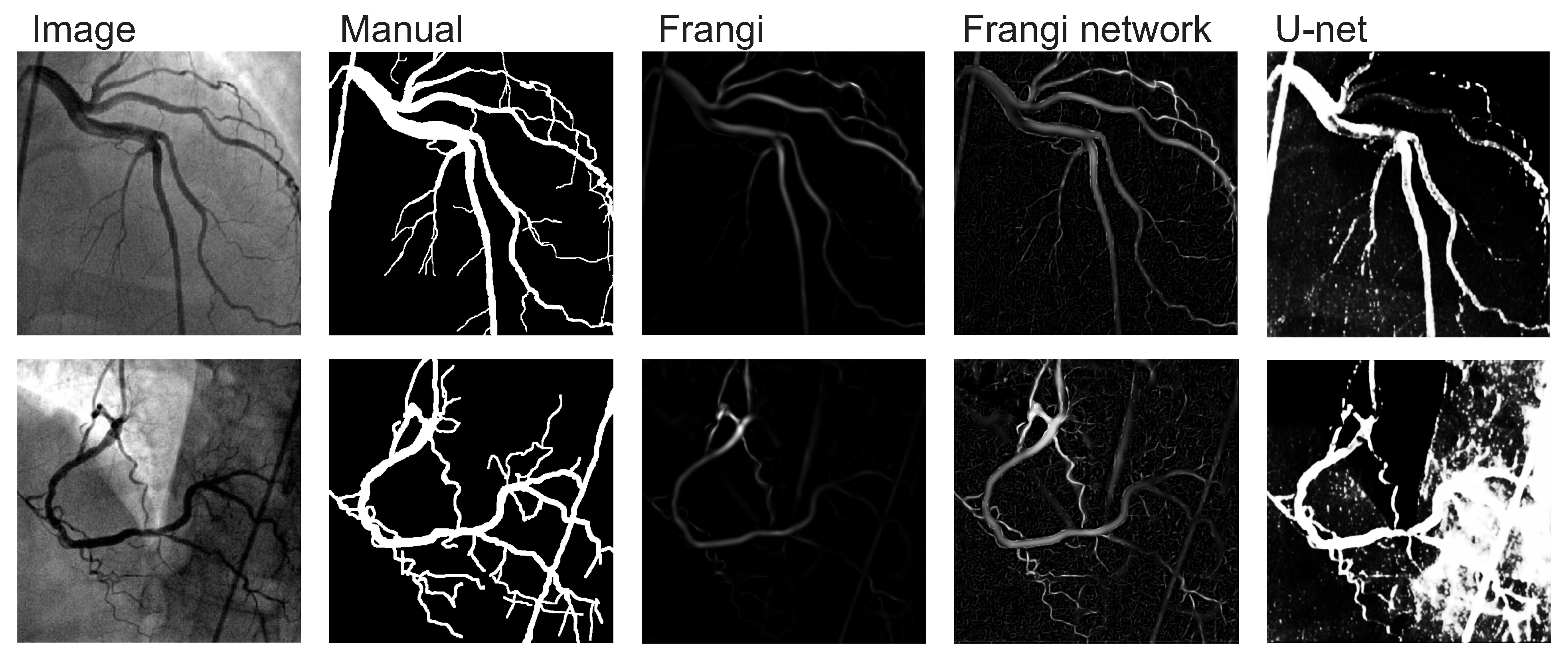
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lamy, J.; Merveille, O.; Kerautret, B.; Passat, N. A Benchmark Framework for Multi-Region Analysis of Vesselness Filters. IEEE Trans. Med. Imaging 2022, 41, 3649–3662. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Yoon, H.; Lee, J.; Yoo, S. Semi-automatic Labeling and Training Strategy for Deep Learning-based Facial Wrinkle Detection. In Proceedings of the 2022 IEEE 35th International Symposium on Computer-Based Medical Systems (CBMS), Shenzhen, China, 21–23 July 2022; pp. 383–388. [Google Scholar] [CrossRef]
- Covaciu, A.G.; Florea, C.; Szolga, L.A. Microscopic Images Analysis for Saliva Ferning Prediction. In Proceedings of the 2020 International Symposium on Fundamentals of Electrical Engineering (ISFEE), Bucharest, Romania, 5–7 November 2020; pp. 1–5. [Google Scholar] [CrossRef]
- Yin, P.; Cai, H.; Wu, Q. DF-Net: Deep fusion network for multi-source vessel segmentation. Inf. Fusion 2022, 78, 199–208. [Google Scholar] [CrossRef]
- Ali, O.; Muhammad, N.; Jadoon, Z.; Kazmi, B.M.; Muzamil, N.; Mahmood, Z. A Comparative Study of Automatic Vessel Segmentation Algorithms. In Proceedings of the 2020 3rd International Conference on Computing, Mathematics and Engineering Technologies (iCoMET), Sukkur, Pakistan, 29–30 January 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Zamani, M.; Salkim, E.; Saeed, S.R.; Demosthenous, A. A Fast and Reliable Three-Dimensional Centerline Tracing: Application to Virtual Cochlear Implant Surgery. IEEE Access 2020, 8, 167757–167766. [Google Scholar] [CrossRef]
- Rodrigues, E.O.; Conci, A.; Liatsis, P. ELEMENT: Multi-Modal Retinal Vessel Segmentation Based on a Coupled Region Growing and Machine Learning Approach. IEEE J. Biomed. Health Inform. 2020, 24, 3507–3519. [Google Scholar] [CrossRef] [PubMed]
- Chakour, E.; Mrad, Y.; Mansouri, A.; Elloumi, Y.; Bedoui, M.; Andaloussi, I.; Ahaitouf, A. Blood vessel segmentation of retinal fundus images using dynamic preprocessing and mathematical morphology. In Proceedings of the 2022 8th International Conference on Control, Decision and Information Technologies (CoDIT), Istanbul, Turkey, 17–20 May 2022; Volume 1, pp. 1473–1478. [Google Scholar] [CrossRef]
- Mahapatra, S.; Agrawal, S.; Mishro, P.K.; Pachori, R.B. A novel framework for retinal vessel segmentation using optimal improved frangi filter and adaptive weighted spatial FCM. Comput. Biol. Med. 2022, 147, 105770. [Google Scholar] [CrossRef]
- Usman, I.; Almejalli, K.A. Intelligent Automated Detection of Microaneurysms in Fundus Images Using Feature-Set Tuning. IEEE Access 2020, 8, 65187–65196. [Google Scholar] [CrossRef]
- Rodrigues, E.O.; Rodrigues, L.O.; Machado, J.H.P.; Casanova, D.; Teixeira, M.; Oliva, J.T.; Bernardes, G.; Liatsis, P. Local-Sensitive Connectivity Filter (LS-CF): A Post-Processing Unsupervised Improvement of the Frangi, Hessian and Vesselness Filters for Multimodal Vessel Segmentation. J. Imaging 2022, 8, 291. [Google Scholar] [CrossRef]
- Tian, F.; Li, Y.; Wang, J.; Chen, W. Blood Vessel Segmentation of Fundus Retinal Images Based on Improved Frangi and Mathematical Morphology. Comput. Math. Methods Med. 2021, 2021, 4761517. [Google Scholar] [CrossRef]
- Longo, A.; Morscher, S.; Najafababdi, J.M.; Jüstel, D.; Zakian, C.; Ntziachristos, V. Assessment of hessian-based Frangi vesselness filter in optoacoustic imaging. Photoacoustics 2020, 20, 100200. [Google Scholar] [CrossRef]
- Yang, J.; Huang, M.; Fu, J.; Lou, C.; Feng, C. Frangi based multi-scale level sets for retinal vascular segmentation. Comput. Methods Programs Biomed. 2020, 197, 105752. [Google Scholar] [CrossRef]
- Challoob, M.; Gao, Y.; Busch, A.; Nikzad, M. Separable Paravector Orientation Tensors for Enhancing Retinal Vessels. IEEE Trans. Med. Imaging 2022, 42, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Chen, W.; Luo, H.; Tan, Y.; Liu, M.; Wang, Y. Neuron image segmentation via learning deep features and enhancing weak neuronal structures. IEEE J. Biomed. Health Inform. 2020, 25, 1634–1645. [Google Scholar] [CrossRef] [PubMed]
- Ghavami, S.; Bayat, M.; Fatemi, M.; Alizad, A. Quantification of morphological features in non-contrast-enhanced ultrasound microvasculature imaging. IEEE Access 2020, 8, 18925–18937. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Frangi, A.F.; Niessen, W.J.; Vincken, K.L.; Viergever, M.A. Multiscale vessel enhancement filtering. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI’98, Cambridge, MA, USA, 11–13 October 1998; Wells, W.M., Colchester, A., Delp, S., Eds.; Springer: Berlin/Heidelberg, Germany, 1998; pp. 130–137. [Google Scholar]
- Bukenya, F.; Bai, L.; Kiweewa, A. A Review of Blood Vessel Segmentation Techniques. In Proceedings of the 2018 1st International Conference on Computer Applications Information Security (ICCAIS), Riyadh, Saudi Arabia, 4–6 April 2018; pp. 1–10. [Google Scholar] [CrossRef]
- Lamy, J.; Merveille, O.; Kerautret, B.; Passat, N.; Vacavant, A. Vesselness Filters: A Survey with Benchmarks Applied to Liver Imaging. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; pp. 3528–3535. [Google Scholar] [CrossRef]
- Khattak, D.; Khaliq, A.; Jalil, A.; Shahid, M. A robust technique based on VLM and Frangi filter for retinal vessel extraction and denoising. PLoS ONE 2018, 13, e0203418. [Google Scholar] [CrossRef]
- Jiang, J.; Wang, D.; Song, Y.; Sachdev, P.S.; Wen, W. Computer-aided extraction of select MRI markers of cerebral small vessel disease: A systematic review. NeuroImage 2022, 261, 119528. [Google Scholar] [CrossRef]
- Tran, N.C.; Wang, J.C. A Survey of Finger Vein Recognition. In Proceedings of the 2021 9th International Conference on Orange Technology (ICOT), Tainan, Taiwan, 16–17 December 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Ng, C.C.; Yap, M.H.; Costen, N.; Li, B. Automatic Wrinkle Detection Using Hybrid Hessian Filter. In Proceedings of the Computer Vision—ACCV 2014, Singapore, 1–5 November 2014; Cremers, D., Reid, I., Saito, H., Yang, M.H., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 609–622. [Google Scholar]
- Kang, X.; Zhang, E. A universal defect detection approach for various types of fabrics based on the Elo-rating algorithm of the integral image. Text. Res. J. 2019, 89, 004051751984063. [Google Scholar] [CrossRef]
- Survarachakan, S.; Pelanis, E.; Khan, Z.A.; Kumar, R.P.; Edwin, B.; Lindseth, F. Effects of Enhancement on Deep Learning Based Hepatic Vessel Segmentation. Electronics 2021, 10, 1165. [Google Scholar] [CrossRef]
- Ballerini, L.; Lovreglio, R.; del C. Valdés Hernández, M.; Gonzalez-Castro, V.; Maniega, S.M.; Pellegrini, E.; Bastin, M.E.; Deary, I.J.; Wardlaw, J.M. Application of the Ordered Logit Model to Optimising Frangi Filter Parameters for Segmentation of Perivascular Spaces. Procedia Comput. Sci. 2016, 90, 61–67. [Google Scholar] [CrossRef][Green Version]
- Fu, W.; Breininger, K.; Schaffert, R.; Ravikumar, N.; Würfl, T.; Fujimoto, J.; Moult, E.; Maier, A. Frangi-Net. In Proceedings of the Bildverarbeitung für die Medizin 2018; Maier, A., Deserno, T.M., Handels, H., Maier-Hein, K.H., Palm, C., Tolxdorff, T., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 341–346. [Google Scholar]
- Zhao, G.; Zhao, H. One-Shot Image Segmentation with U-Net. J. Phys. Conf. Ser. 2021, 1848, 012113. [Google Scholar] [CrossRef]
- Shaban, A.; Bansal, S.; Liu, Z.; Essa, I.; Boots, B. One-shot learning for semantic segmentation. arXiv 2017, arXiv:1709.03410. [Google Scholar]
- Sun, Z.; Rooke, E.; Charton, J.; He, Y.; Lu, J.; Baek, S. ZerNet: Convolutional Neural Networks on Arbitrary Surfaces Via Zernike Local Tangent Space Estimation. Comput. Graph. Forum 2020, 39, 204–216. [Google Scholar] [CrossRef]
- Theodoridis, T.; Loumponias, K.; Vretos, N.; Daras, P. Zernike Pooling: Generalizing Average Pooling Using Zernike Moments. IEEE Access 2021, 9, 121128–121136. [Google Scholar] [CrossRef]
- Wang, S.; Lv, L.T.; Yang, H.C.; Lu, D. Zernike-CNNs for image preprocessing and classification in printed register detection. Multimed. Tools Appl. 2021, 80, 32409–32421. [Google Scholar] [CrossRef]
- Liu, J.; Li, P.; Tang, X.; Li, J.; Chen, J. Research on improved convolutional wavelet neural network. Sci. Rep. 2021, 11, 17941. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Kingma, D.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the ICLR 2015. Ithaca, NY. arXiv 2015, arXiv:1412.6980. [Google Scholar]
- Ma, J.; Chen, J.; Ng, M.; Huang, R.; Li, Y.; Li, C.; Yang, X.; Martel, A.L. Loss odyssey in medical image segmentation. Med. Image Anal. 2021, 71, 102035. [Google Scholar] [CrossRef]
- Murugesan, B.; Liu, B.; Galdran, A.; Ayed, I.B.; Dolz, J. Calibrating segmentation networks with margin-based label smoothing. Med Image Anal. 2023, 87, 102826. [Google Scholar] [CrossRef]
- Staal, J.; Abramoff, M.; Niemeijer, M.; Viergever, M.; van Ginneken, B. Ridge based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef]
- Sharma, S.; Bhattacharya, M.; Sharma, G. Optimized Coronary Artery Segmentation Using Frangi Filter and Anisotropic Diffusion Filtering. In Proceedings of the 2013 International Symposium on Computational and Business Intelligence, New Delhi, India, 24–26 August 2013; pp. 261–264. [Google Scholar] [CrossRef]
- Avadiappan, S.; Payabvash, S.; Morrison, M.A.; Jakary, A.; Hess, C.P.; Lupo, J.M. A Fully Automated Method for Segmenting Arteries and Quantifying Vessel Radii on Magnetic Resonance Angiography Images of Varying Projection Thickness. Front. Neurosci. 2020, 14, 537. [Google Scholar] [CrossRef]
- Bock, S.; Weiß, M. A proof of local convergence for the Adam optimizer. In Proceedings of the 2019 International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019; pp. 1–8. [Google Scholar]
- Jalalian, A.; Mashohor, S.; Mahmud, R.; Karasfi, B.; Iqbal Saripan, M.; Ramli, A.R. Computer-assisted diagnosis system for breast cancer in computed tomography laser mammography (CTLM). J. Digit. Imaging 2017, 30, 796–811. [Google Scholar] [CrossRef] [PubMed]
- Van Ginneken, B.; Heimann, T.; Styner, M. 3D segmentation in the clinic: A grand challenge. In Proceedings of the MICCAI Workshop on 3D Segmentation in the Clinic: A Grand Challenge, Brisbane, Australia, 29 October–2 November 2007; Volume 1, pp. 7–15. [Google Scholar]
- Yeghiazaryan, V.; Voiculescu, I. Family of boundary overlap metrics for the evaluation of medical image segmentation. J. Med. Imaging 2018, 5, 015006. [Google Scholar] [CrossRef] [PubMed]
- Meng, C.; Sun, K.; Guan, S.; Wang, Q.; Zong, R.; Liu, L. Multiscale dense convolutional neural network for DSA cerebrovascular segmentation. Neurocomputing 2020, 373, 123–134. [Google Scholar] [CrossRef]
- Lu, Y.; Zheng, K.; Li, W.; Wang, Y.; Harrison, A.P.; Lin, C.; Wang, S.; Xiao, J.; Lu, L.; Kuo, C.F.; et al. Contour transformer network for one-shot segmentation of anatomical structures. IEEE Trans. Med. Imaging 2020, 40, 2672–2684. [Google Scholar] [CrossRef] [PubMed]
- Zhao, A.; Balakrishnan, G.; Durand, F.; Guttag, J.V.; Dalca, A.V. Data augmentation using learned transformations for one-shot medical image segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–17 June 2019; pp. 8543–8553. [Google Scholar]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P. Microsoft COCO: Common Objects in Context. In Computer Vision—ECCV 2014. ECCV 2014; Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2014; Volume 8693. [Google Scholar] [CrossRef]
- Kirbas, C.; Quek, F. Vessel extraction techniques and algorithms: A survey. In Proceedings of the Third IEEE Symposium on Bioinformatics and Bioengineering, 2003, Bethesda, MD, USA, 12 March 2003; pp. 238–245. [Google Scholar] [CrossRef]
- Ciecholewski, M.; Kassjański, M. Computational Methods for Liver Vessel Segmentation in Medical Imaging: A Review. Sensors 2021, 21, 2027. [Google Scholar] [CrossRef]
- Kirbas, C.; Quek, F. A review of vessel extraction techniques and algorithms. ACM Comput. Surv. (CSUR) 2004, 36, 81–121. [Google Scholar] [CrossRef]
- Li, H.; Tang, Z.; Nan, Y.; Yang, G. Human treelike tubular structure segmentation: A comprehensive review and future perspectives. Comput. Biol. Med. 2022, 151, 106241. [Google Scholar] [CrossRef]
- Spuhler, C.; Harders, M.; Székely, G. Fast and robust extraction of centerlines in 3D tubular structures using a scattered-snakelet approach. In Proceedings of the Medical Imaging 2006 Conference, San Diego, CA, USA, 13–16 February 2006; Volume 6144, pp. 1295–1302. [Google Scholar]
- Wu, X.; Cui, M.; Gao, Y.; Sun, D.; Ma, H.; Zhang, E.; Xie, Y.; Zaki, N.; Qin, W. Tubular Structure-Aware Convolutional Neural Networks for Organ at Risks Segmentation in Cervical Cancer Radiotherapy. In Proceedings of the International Workshop on Computational Mathematics Modeling in Cancer Analysis, Singapore, 18 September 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 131–140. [Google Scholar]
- Smistad, E.; Elster, A.C.; Lindseth, F. GPU accelerated segmentation and centerline extraction of tubular structures from medical images. Int. J. Comput. Assist. Radiol. Surg. 2014, 9, 561–575. [Google Scholar] [CrossRef]
- Bauer, C.; Pock, T.; Bischof, H.; Beichel, R. Airway tree reconstruction based on tube detection. In Proceedings of the Second International Workshop on Pulmonary Image Analysis, London, UK, 20 September 2009; pp. 203–213. [Google Scholar]



| Frangi Network | Frangi | U-Net | |
|---|---|---|---|
| Retina | 0.8684 | 0.7883 | 0.7769 |
| Coronary artery | 0.9117 | 0.9110 | 0.7264 |
| Brain | 0.9397 | 0.9297 | 0.5720 |
| Exec time [S] | 0.160 | 1.187 | 0.143 |
| Sensitivity | Specificity | IoU | VOE | F1-Score/Dice | |
|---|---|---|---|---|---|
| Retina | 0.6648 | 0.9557 | 0.4251 | 0.5749 | 0.5966 |
| Coronary artery | 0.9891 | 0.9143 | 0.4842 | 0.5158 | 0.6526 |
| Brain | 0.9242 | 0.9133 | 0.4768 | 0.5232 | 0.6451 |
| Sensitivity | Specificity | IoU | VOE | F1-Score/Dice | |
|---|---|---|---|---|---|
| Retina | 0.6244 | 0.9441 | 0.3648 | 0.6352 | 0.5348 |
| Coronary artery | 0.9551 | 0.9144 | 0.4677 | 0.5323 | 0.6377 |
| Brain | 0.9139 | 0.9166 | 0.4804 | 0.5196 | 0.6487 |
| Sensitivity | Specificity | IoU | VOE | F1-Score/Dice | |
|---|---|---|---|---|---|
| Retina | 0.6506 | 0.9152 | 0.3130 | 0.6870 | 0.4767 |
| Coronary artery | 0.8088 | 0.8242 | 0.2577 | 0.7423 | 0.4095 |
| Brain | 0.6457 | 0.6751 | 0.1430 | 0.8570 | 0.2506 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hachaj, T.; Piekarczyk, M. High-Level Hessian-Based Image Processing with the Frangi Neuron. Electronics 2023, 12, 4159. https://doi.org/10.3390/electronics12194159
Hachaj T, Piekarczyk M. High-Level Hessian-Based Image Processing with the Frangi Neuron. Electronics. 2023; 12(19):4159. https://doi.org/10.3390/electronics12194159
Chicago/Turabian StyleHachaj, Tomasz, and Marcin Piekarczyk. 2023. "High-Level Hessian-Based Image Processing with the Frangi Neuron" Electronics 12, no. 19: 4159. https://doi.org/10.3390/electronics12194159
APA StyleHachaj, T., & Piekarczyk, M. (2023). High-Level Hessian-Based Image Processing with the Frangi Neuron. Electronics, 12(19), 4159. https://doi.org/10.3390/electronics12194159







