Abstract
A frequent cause of lower back pain presenting with leg pain is a herniated lumbar intervertebral disc. A herniation or a herniated lumbar disc is a change of position of disc material (nucleus pulpous or annulus fibrosis). Usually, the lower back pain goes away within days or weeks. Regular treatment techniques for lower back pain include medication, exercises, relaxation methods and surgery. Back pain and back problems regularly occur in the lumbar region. The spinal canal is made up of vertebrae; each one protects the spinal nerves. Intervertebral discs and facet joints connect the vertebrae above and below. Groups of muscles and ligaments hold the vertebrae and the discs together. Muscles support the spine and the body weight, and they allow us to move. Pressure can result in excessive wear and tear of the other structures. For example, a common problem in the lower back is disc herniation. In this case, pressure on an intervertebral disc makes its center, the nucleus pulposus, protrude backwards and push against the spinal nerves, leading to lower back pain. Detection and classification are the two most important tasks in computer aided diagnosing systems. Detection of a herniated lumbar disc from magnetic resonance imaging (MRI) is a very difficult task for radiologist. The extraction of herniated discs has been achieved by different approaches such as active contours, region growing, watershed techniques, thresholding and deep learning. In this study, to detect intervertebral disc from axial MRIs we develop a method using generative adversarial networks (GANs), especially the CycleGAN model, to automatically generate and detect intervertebral disc and to classify the type of the herniated lumbar disc such as foraminal or median. We propose to explore the importance of axial view MRI to determine the herniation type. Accurately, GANs and other generative networks have created several ways to tackle different problems well known and challenging of medical image analysis, such as segmentation, reconstruction, data simulation, medical image de-noising, and classification. Moreover, their efficiency to synthesize images and data at unprecedented levels of realism also gives hope that the chronic scarcity of labeled data in the medical field can be resolved with the help of these generative models. In our case, having a database that contains several images is a very difficult task. In this paper, we put forward a new approach based on GANs, in order to solve the problem of lumbar intervertebral disc images reduction. This method is based especially on CycleGAN. Consequently, the essential objective of our work is to generate and automatically classify the herniation type as foraminal or median using GANs. Our computer aided diagnosis (CAD) system achieved a 97.2% accuracy on our dataset. This result represents a very high-performance results by providing the state of the art and our work utilizing the GANs technique. Our CAD is very effective and efficient for classifying herniations of lumbar intervertebral discs. Therefore, the contribution of this study appears in: firstly, the use of the CycleGAN model based on convolutional layers to detect and classify the herniation type (median or foraminal) in lumbar intervertebral discs, secondly, the use of axial view MRI in order to classify the type of the herniated intervertebral disc. The main objective of this paper is to help radiologists automatically recognize and classify herniated lumbar discs.
1. Introduction
The spine is composed of 24 vertebrae including twelve dorsal, five lumbar vertebrae and seven cervical [1]. The intervertebral disc connects each two vertebrae to each other; acting like a shock absorber cushion [1]. The intervertebral discs are composed of two important components: a nucleus pulposus surrounded by a fibrous annulus [1]. When a part of the nucleus protrudes outside, this is called a herniated disc [1]. The latter represents the rupture of the fibrous of an intervertebral disc [1]. It usually occurs between 35 and 55 years old due to overweight, overwork and pregnancy [1]. Lumbar disc herniation (LDH) is a major public health problem in the world [1]. The main cause of this disease is a loss of the height of one intervertebral disc or many discs [1]. In this study, we concentrate on lumbar disc extraction from the axial view [1]. This task of features extraction represents a difficult task in computer aided diagnosis (CAD) to aid radiologist in LDH detection and classification [1]. The occurrence of LDH goes through the following steps. The first step, bulging: a bulging disc can be referred to as a protruding or slipped disc [1]. The second stage is protrusion [1]. The third step is extrusion [1]. The last one is exclusion [1]. CAD systems are based on the analyzing different types of patients (including their bodies, physical examinations or medical history) to help radiologists quickly diagnose [1]. Various medical imaging modalities can be used to extract lumbar disc herniation [1]. The focus of this work is on MRI [1]. There are two different views used to diagnose the LDH: sagittal and axial MRI [1]. The sagittal view (Figure 1) allows for defining the LDH, the normal lumbar intervertebral disc, the shape of each disc ((L1–L2), (L2–L3), (L3–L4), (L4–L5), (L5–S1)) and its situation [1]. However, the axial view MRI provides more information about the herniated disc, including the type of the LDH and its stage [1]. There are different works for intervertebral disc labeling, detection, feature extraction and segmentation [1]. Chwialkowski et al. [2] developed a novel technique to extract pathologies in lumbar intervertebral discs images by first detecting candidates’ vertebrae with an estimated model and then studying the change in gray-level intensities in healthy and damaged intervertebral discs [1]. Tsai et al. [3] presented an approach to detect the herniation from computed tomography volumes and 3D MRI of discs by using geometric features like the size, the location and the shape [1]. Rabia et al. [4] put forward models for lumbar spine intervertebral disc segmentation. They proposed a minimally supervised 3D segmentation approach of lumbar intervertebral herniated discs for MRI scans which exploits weak shape priors encoded in simplex mesh active surface models [1]. Alomari et al. [5] developed a fully automated herniation detection system using gradient vector flow snakes for an initial disc contour, and then trained a Bayesian classifier on the resulting shape features [1]. They achieved 92.5% accuracy on 65 clinical MRI cases, with the low sensitivity of 86.4%. Michopoulou et al. [6] achieved 86–88% accuracy for normal vs. degenerated disc classification [1]. The authors used fuzzy C-means to perform semi-automatic atlas-based disc detection and then used a Bayesian classifier model [1]. They also achieved 94% accuracy using texture features for 50 manually extracted discs [1]. Booth et al. [7] put forward a novel system that could automatically locate the center of the spinal canal on the axial images based on a symmetry measure [1]. They proposed an algorithm that could automatically extract the center of the spinal canal [1]. Hoad et al. [8] respectively proposed an approach to detect the spinal cord and the vertebrae on MRI [1]. For both cases, the initialization step needed user interaction to manually extract the center of the spinal cord at every spine level or to manually locate four points on each vertebral body [1]. On the other hand, they put forward a system that could automatically detect the center of the spinal canal on the axial MRI based on a symmetry measure [1]. The authors applied an active contour model to detect the spinal canal [1]. Ghosh et al. [9] developed a majority voting algorithm for LDH that used planar shape features texture features and intensities, extracted by the gray-level co-occurrence matrix [1]. Their system was tested on a dataset containing 35 subjects and the accuracy achieved was 94.86%. They put forward a robust and fully automated LDH diagnosis system [1]. They reduced the time to analyze each case [1]. They developed five several classifiers (SVM, PCA + LDA, Bayes, PCA + QDA, PCA + SVM) and combined them to achieve the best results; they got 94.86% accuracy and 95.90% specificity. Shi et al. [10] put forward an unsupervised segmentation method which did not require initialization and admitted combinations of different features such as the position and the brightness. Peng et al. [11] achieved the best sagittal slice to extract the lumbar intervertebral disc based on a canny edge operator creating open contours. Elias et al. [12] suggested a novel system in order to diagnose LDH disease by MRI. The authors used the Otsu thresholding method and detected features by calculating the shape feature, and they finally found the classification by using the k-nearest neighbor (KNN), support vector machine and multi-layer perceptron (MLP). The KNN and MLP classifiers showed around 92.38% and 91.90% of accuracy, respectively. Chevrefils et al. [13] put forward a novel technique by combining two approaches, the morphological operations and the watershed method, to extract the intervertebral discs from MRI. An automated approach was proposed to regulate the initial values of the centroid of the cluster using intuitionistic fuzzy clustering. The writers suggested a new function complement, which was introduced for intuitionistic fuzzy clustering using axial MRI. The evaluation was done using recall, precision, Jaccard coefficient and Dice coefficient. Alawneh et al. [14] built a simple CAD that would diagnose lumbar disc herniation using MRI as a normal or herniated disc. In Alwaneh et al. [14], the herniated lumbar disc was detected over two passes: horizontal and vertical extraction. Each image was read and converted to a gray scale if the image was in RGB (Red Green Blue). Then, they sliced an initial strip around the center column of the image. The initial vertical strip was converted to a binary one using adaptive thresholding. Added to that, a random threshold was chosen and the image histogram was divided into two parts around this threshold. After that, a threshold value was calculated as the average of the two means of the two histograms. They built a CAD that diagnoses lumbar disc cases using axial view MRI as normal or abnormal. Seifert et al. [15] put forward a novel system for cervical intervertebral disc detection using the statistical shapeware deformable models and the Hough transform.
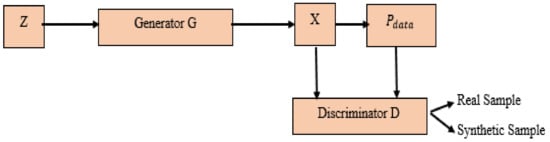
Figure 1.
GAN (Generative Adversarial Networks).
To overcome the shortfalls of the existing techniques, we have suggested in the present work to use the cycle generative adversarial network (CycleGAN) model on axial view MRI and have improved the lumbar intervertebral disc generation, detection and classification by implementing a powerful classifier based on GANs. Lumbar discs are classified using CycleGAN to have the probabilities of the presence of a foraminal or median herniation in the studied dataset. The CycleGAN uses a combination of two generators and a combination of two discriminators in order to select real and fake samples. Furthermore, we propose to use axial view MRI. Thus, the suggested technique allows us to generate, localize and classify the type of lumbar intervertebral discs, contrarily to the above-mentioned existing approaches, which can only detect lumbar intervertebral discs.
We develop a novel approach based on CycleGAN for generating, detecting and classifying lumbar intervertebral discs using axial view MRI. This system is used for diagnosing the LDH; it is based on GANs, which have achieved great success in artificial intelligence and in several applications in recent years. Therefore, GANs approaches have been extensively used to analyze and synthesize biomedical images. Indeed, CycleGAN is a model used to solve MRI-to-MRI translation tasks, the objective being to learn the mapping between an input image (class A) and an output image (class B) using training based on a cycle consistent adversarial network.
The aim of this paper is to automatically generate lumbar intervertebral discs in MRI and classify the type of herniation as foraminal (classA) or median (classB), which can assist radiologists and clinicians in diagnosing based on a large dataset of axial view MRI. A novel algorithm is proposed using the CycleGAN based on convolutional layers. The results of our study are evaluated with regards the accuracy of real and fake samples as well as the generator accuracy. We provide for GANs general introduction and a state of the art. Next, we introduce results obtained of generated data and classified herniations using CycleGANs with a discussion of the limitations of GANs methods. Herniated intervertebral discs represent the modification of their height (one disc or more discs). This herniation is the main cause of lower back pain or sciatica. Little LDH MRI is a challenging problem in the various research works. Furthermore, the herniation classification as foraminal (class A) or median (class B) represents the most difficult problem. To help radiologists recognize the type of the herniated lumbar intervertebral disc and to increase the number of MRI cases, we suggest exploiting the CycleGANs to generate the dataset. Moreover, to automatically classify the herniation type. GAN models must learn to capture the statistical distribution of training data [16], allowing us to synthesize samples from the learned distribution [16]. GANs provide a way to learn deep representations without any extensively annotated training data [17]. GANs are made up of two essential neural networks, the discriminative models in which the convolutional neural networks try to discriminate between image pixels that correspond to different classes [18]. The optimization of these convolutional network is supervised by a loss function that quantifies the agreement between reference labels and model predictions [18]. Second, the generative models aim to learn the underlying distribution of the data and the generative process that creates them [18]. We summarize various existing techniques for GANs, which are highlighted in Table 1.

Table 1.
Different techniques in biomedical images based on Generative Adversarial Networks (GANs).
The present paper is organized as follows: we provide an introduction to GAN and CycleGAN methods with a focus on applications in biomedical image analysis.
In Section 3, we describe state of the art of GANs networks in biomedical images. In Section 4, we describe the use of CycleGAN network to map images from one class (foraminal herniation) to another class (median herniation). Section 5 explains results found from CycleGAN on our dataset collected from the university hospital of Sousse. Section 6 presents the conclusion of our work.
2. Overview of GANs in Medical Images
From the early days of medical image analysis, machine learning and artificial intelligence driven systems have been a key component for complex decision-making. There are different networks based on CNN, spiking neural networks (SNN), GANs and other networks applied on medical images. The reconstruction of many research-based magnetic resonance images focuses on data augmentation generative adversarial network (DAGAN) architecture [19]. In this approach, a perceptual loss was added to pixel-wise and adversarial (Adv) losses to compare generated information and deep extracted features in real time, which would enhance the stability of GANs. Different work has refined the DAGAN architecture over time [20]. The next update of DAGAN was presented in [21], which proposed adding a refinement model in order to separate perceptual information and a similar architecture was proposed in [22], where the authors proposed a chain of generators to address the ambiguities made in other ones. The authors in [23] adapt the super-resolution using a generative adversarial network (SRGAN) with 3D convolutional layers to enhance the stability of the proposed GAN model. The writers in [24] developed a novel approach based on 3DSRGAN, along with two loss of functions to prevent over-fitting and to control data interpolation. The authors in [10] put forward a Mdcsrn, multilevel densely connected super-resolution network, representing a combination of a modified version of dense net and the Wasserstein generative adversarial network (WGAN) model. The writers in [25] developed a novel technique based on generative adversarial networks, especially based on least squares generative adversarial networks (LSGAN) as a solution to address training instability. The authors in [26] proposed a multi-channel deep GAN for MRI reconstruction. The writers in [27] suggested an image super resolution technique using generative adversarial networks which would take low-resolution input fundus images and generate high-resolution ones for retinal image analysis. The authors in [28] suggested a novel algorithm based on GANs for phase contrast microscopy image super-resolution. Daniele et al. [29] have proposed an unsupervised super resolution framework based on adversarial deep neural network with a physically inspired cycle consistency, designed to impose some acquisition properties on the super-resolved images [30].
Thomas et al. performed unsupervised learning based on GANs to identify anomalies in imaging data as candidates for markers [30]. Chen et al. [31] have studied the detection of lesions in brain MRI using constrained adversarial auto-encoders by learning the data distribution of brain MRI of healthy subjects using auto-encoder based methods [31]. The authors in [32] have suggested the visual attribution GAN (VA-GAN) for Alzheimer’s disease detection. For aggressive prostate cancer detection [33] and skin lesion detection, the authors in [34] have put forward the use of U-Net architecture as the generator of a GAN and a cGAN, respectively. In prostate US, the low contrast of tumor boundaries was challenged contour detection. The writers in [35] address this problem using GANs. The authors in [36] have suggested learning tissue texture information from a small amount of paired data and address the blurring effect using GANs [36]. Correspondingly, [21] have developed a novel technique based on conditional GAN (Cgan) in order to remove metal artifacts from CT images. The authors in [37] have proposed the sharpness aware generative adversarial network (SAGAN) to work on the sharpness of the denoised images. The authors in [38] presented a biologically-inspired cognitive supercomputing system (BiCoSS) that integrates multiple granules (GRs) of SNNs to realize a hybrid compatible neuromorphic platform. The writers in [39] developed a large-scale cerebellar network model for supervised learning, as well as a cerebellum-inspired neuromorphic architecture to map the cerebellar anatomical structure onto the large-scale model. The writers in [40] presented a novel technique; one of the challenging problems in real-time control of movement disorders is the effective handling of time-variant brain activities that involve stochastic functional networks with nonlinear dynamics. For such challenges in neuromodulator tasks, fuzzy logic control (FLC) has shown significant potential. Their objective was to present a FLC based strategy to treat pathological symptoms of movement disorders with higher performance. The authors in [41] presented a real-time digital neuromorphic system for the simulation of large-scale conductance-based spiking neural networks which has been the advantages of both high biological realism and large network scale. Using this system, a detailed large-scale cortico-basal ganglia-thalamocortical loop is simulated using a scalable 3-D network-on-chip topology with six Altera Stratix III field-programmable gate arrays simulate one million neurons. The writers in [42] proposed a novel multilayer extreme learning machine (ELM) classification model combined with dynamic generative adversarial net (GAN) to tackle limited and imbalanced biomedical data. In [43], hyaluronic acid (HA) is represented; a natural linear polysaccharide that has been used extensively in the biomedical field, as it is a biocompatible, biodegradable, nontoxic and non-immunogenic polymer with high water affinity. Besides, the presence of multiple acid and hydroxyl groups in the HA molecule makes it an ideal candidate for chemical modification. They described the synthesis and characterization of HA-based hydrogels. For this purpose, an aqueous mixtures containing 5% (w/w) of HA and different concentrations of Gantrez S97 (GAN) (1%, 3% and 5% w/w) were used to prepare HA-based hydrogels. There are several studies based on GANs, Yu et al. [44] developed a novel system based on generative networks for fast compressive sensing MRI. This model can lessen parameter tuning and stabilize training with fast convergence. They used k-space information to reconstruct images. The authors in [40] developed a FLC-based strategy to treat pathological symptoms of movement disorders with higher performance. Multicompetent emulation is an essential step to enhance the biological realism of neuromorphic systems and to further understand the computational power of neurons. In [41], the writers present a hardware scalable, efficient and real-time computing strategy for the implementation of large-scale biologically meaningful neural networks with one million multi-compartment neurons (CMNs). The writers in [45] developed a model named dendritic event-based processing (DEP) that effectively solves the credit assignment challenging. The results show that the suggested DEP achieved high performance and spiking representations can rapidly learn. The authors in [46,47] provide an overview of the developments in neuromorphic computing for both algorithms and hardware and highlight the fundamentals of learning and hardware frameworks. They discussed the main challenges and the future prospects of neuromorphic computing, with emphasis on algorithm hardware code sign.
3. Generative Adversarial Networks
GANs consist of a training dataset X (in our case there are two datasets X1: foraminal cases and X2: median cases) and a pair of competing models: Discriminator (D) with weights ( and generator (G) with parameters () (see Figure 1). G focuses on mapping which draws latent random variables z ~ (z) mapping from a prior distribution to generated data x ∈ X^, which is expected to follow the distribution (x|z). The primary objective is to optimize this drawn such that the generated distribution data X resembles the distribution of the training data X, i.e., (x|z) ∼. In other words, G is supposed to generate fake data, which must not be distinguishable from real data. This is achieved with the help of the discriminator network D, whose task is to classify between fake and real samples. Essentially, D is a binary classifier which yields D(x) = 1 for real samples and D(x) = 0 for fake data. Both networks are adversaries, as G attempts to gradually synthesize more and more realistic samples that D would misclassify as real, while D constantly learns to differentiate between real and synthesized samples. Mathematically speaking, D and G play a two-player minimax game with the following value function V(G, D) (see Equation (1)). In order to optimize Equation (1), the discriminator D is trained to maximize the probability of correct label assignment for real and fake samples, however, the generator G is trained to make D think that a generated data is real by minimizing log(1 − D(G (z))) [48]. These two networks are regularly implemented as convolutional neural networks or multi-layer perceptron and trained with minibatch stochastic gradient descend [48]. Once learned, it is sufficient to sample a random z and feed it through the generator to synthesize data [48]. This generative adversarial network shows a few interesting properties: (1) the generator G is updated only through gradients back propagated from the discriminator; and (2) no explicit correspondences between z, x and x are required, such that input is not explicitly memorized by G [48]. Finally, (3) as proven by the authors, optimization for Equation (1) minimizes the Jensen–Shannon (JS) divergence between the distributions of real and synthetic data [48]. Although theoretically well grounded, the vanilla GAN has proven to be quite hard to train [48]. General convergence is heavily dependent on hyper parameter tuning to avoid vanishing or exploding gradients, and they are prone to mode collapse [48]. This term describes a phenomenon where GANs map all z to very similar synthetic samples covering only a single mode of the data distribution [48]. During optimization, these modes might also change (so-called mode hopping) [48]. A plethora of extensions and subclasses has been proposed to cope with these problems, a selection which is introduced in the following subsections [48].
Each GAN consists of two essential neural networks. Firstly, the generative model aims to generate synthetic samples x from a real data . A noise vector z is transformed from (distribution) into new samples: X = G(z) (as shown in Figure 1).
Secondly, generator G takes a noise vector z sampled from distribution as an input and uses fully connected or convolutional layers to transform this vector into a sample x. The discriminator D tries to distinguish these samples from samples drawn from the real data distribution . The Figure 1 illustrates GAN model architecture, which involves two sub-models: a generator model for generating new examples and a discriminator model for classifying whether generated examples are real, from the domain, or fake, generated by the generator model, and also classifies lumbar intervertebral discs as foraminal or median.
4. Proposed Approach
We have put forward a novel approach based on CycleGAN as illustrated in Figure 2 to solve the problem of the lack of background information [48]. The generative model consists of two mapping functions (i.e., F: X → Y and G: Y → X) and adversarial discriminators (i.e., DX and DY). Actually, DX makes the outputs of G(y) distinguishable from domain X. Similarly, DY makes the outputs of F(x) distinguishable from domain Y. To further regularize the mappings, we introduce a kind of cycle consistency loss where we can arrive at our starting point if we translate from one domain to another and back again. In Figure 3, the image translation cycle brings X and Y back to themselves through functions F(x) and G(y). This cycle consistency loss can be expressed as:
where (x) and (y) are the distributions of X and Y, respectively. Minimizing the loss between G (F(x) and x and between F (G(y) and y can make the generated images close to real images [48]. Our objective is to learn a mapping from a source domain A with foraminal lumbar intervertebral discs herniations images to a target domain B, which is a median one. The inputs of the network are unpaired training image samples a in A and b in B. As shown in Figure 4, the CycleGAN that consists of two generators GA and GB and two discriminators DA and DB is an improvement on the convolution level from the original CycleGAN. The images taken from two datasets or two different classes, converting an image from the representation of one class to another (e.g., foraminal intervertebral discs to median ones or vice versa) is suggested as an image-to-image translation [23]. We can summarize the principle of CycleGAN as it converts one image classified into class X to another image classified into class Y, with the aim to learn the mapping (F) from X to Y, the F mapping corresponds to the GAN builder and F can convert the image x in X to image F(x) in Y. The mapping G can transform the image y into G(y) in X.
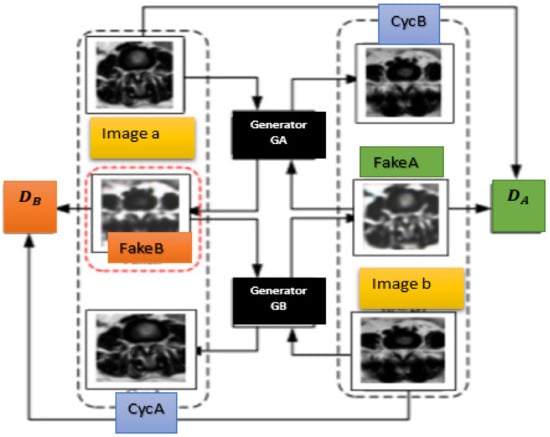
Figure 2.
CycleGAN architecture.

Figure 3.
Some examples of lumbar intervertebral disc detection results using the CycleGAN model: (a) input images; (b) detected masks.
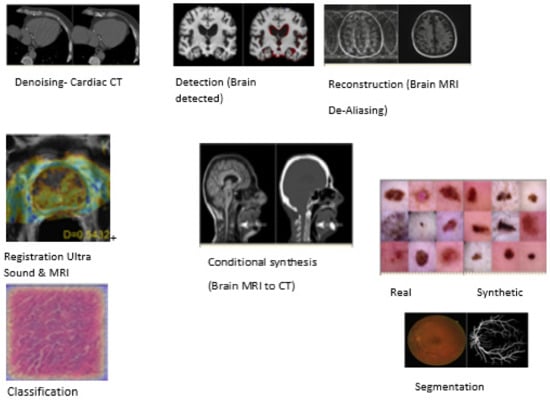
Figure 4.
A survey on generative adversarial networks for medical image analysis: The pie chart represents the visual examples of GANs applications. Examples are taken from papers as follows: denoising, reprinted with permission from ref. [36]. Copyright 2017 IEEE. conditional synthesis, reprinted from ref. [49]; registration, reprinted from ref. [50], reconstruction reprinted with permission from ref. [44]. Copyright 2018 Springer. detection reprinted from ref. [51], classification reprinted from ref. [52], unconditional synthesis reprinted from ref. [17], and segmentation reprinted from ref. [53] Source: reprinted from ref. [46].
4.1. System Based on CycleGAN
We evaluate our developed CycleGAN classification method on our MRI dataset collected from the University Hospital of Sahloul, Sousse using axial view MRI. There are two classes: class A, which contains the median herniations, and class B which contains the foraminal herniations. These experimentations were conducted in the Informatics and systems Laboratory, France. All data sets and results were staged, analyzed and evaluated by an expert radiologist. The dataset A is divided into two folders: train A and test A. The dataset B is segmented into train B and test B.
4.2. Evaluation Metrics
The best metric to evaluate our detection approach on our dataset is the intersection over union (IoU). The IoU is used usually for evaluation segmentation (show results in Figure 3) and it calculates the ratio of the area intersection to the area of unions. We need two parameters:
- (1)
- Predicted bounding boxes from our model.
- (2)
- Ground truth bounding boxes.
Table 2 summarizes the IoU results, comparing our performed result and other results for other work. We can conclude that the CycleGAN is the best one for detecting lumbar discs in order to recognize type of herniation (foraminal or median), as shown in Figure 4. Our system CAD is made up of two essential steps, the first one is generation and localization. The second one represents classification (automatically classifying herniation type, as given in Table 3). Table 3 summarizes results found using the CycleGAN model based on three essential metrics, namely precision, recall and average precision (see Table 3). GANs built a two-player game between the generator model and the discriminator one. The first generates fake or synthetic samples and the second one decides whether these images are real or fake samples. This game takes up as the generator network learns well how to produce more realistic samples and the discriminator network improves its capacity to classify samples as real or fake images. Although our model based on the CycleGAN model is utilized to generate synthetic data, we use the process to better improve the segmentation results and effectively ameliorate classification performance. Our datasets were divided into two classes (class A: foraminal herniations and class B: Median herniations), and were randomly divided into training and test sets (as depicted in Figure 4). Approximately 2/3 of lumbar intervertebral discs images are utilized for training and 1/3 for testing. The CycleGAN model was tested with the remaining 1/3 of our dataset. Our system consists of three stages: the generation of lumbar intervertebral discs, the detection of intervertebral discs and finally the classification of herniated lumbar discs (as foraminal or median). We evaluated the detection results using intersection over union (IoU) and classification ones utilizing accuracy (see Figure 4). The latter are the precision of our system; we propose using precision, average precision and recall (see Table 3).

Table 2.
IoU results using different architectures.

Table 3.
CycleGAN model results (%).
The generator model (as illustrated in Figure 3) is fixed with ten hidden nodes, one random value input attribute, and three output values. We performed the generative adversarial network experiments multiple times using several discriminator configurations and note the performance of our system. The discriminator model includes either one or two hidden layers. The number of nodes per hidden layer changes between 2 and 100. For coherence, two hidden layer experiments included the same number of nodes for both hidden layers. For each trial, training is stopped when the mapping error on the test set increases. The mapping error is defined as the distance between the inferred output array and the known labels, using one-hot encoding. The mapping error is also known as the validation loss. The discriminator model takes as an input one 28 × 28 grayscale image and it outputs a binary prediction as to whether the image is real (class = 1) or fake (class = 0). The last one is implemented as a modest convolutional neural network (CNN) using the LeakyRelu activation function, Adam version of the stochastic gradient descent, a momentum of 0.5, learning rate of 0.0002 and a 2 × 2 stride downsampling. The generator network takes as input a point in the latent space and it outputs a single 28 × 28 grayscale image. This was performed by utilizing a fully connected layer to clarify the point in the latent space and to achieve sufficient activations that can be reshaped into many copies of a low-resolution version of the output image. The results associating to the training of this network are illustrated in Table 2, and the metrics that are selected for evaluating the performance of our system are: recall (R), precision (P), accuracy and average precision (AP) (see Equations (4)–(7)).
4.3. Dataset
We collected our dataset MRI from the University hospital of Sousse. It includes axial view MRIs for each lumbar intervertebral disc. Our images were divided into 1500 cases for training and 500 cases for testing. The CycleGAN model had two generators (generators A and B) and two discriminators (discriminators A and B).
4.4. Results
The results are described in Table 3. In fact, we performed with higher precision of our CycleGAN model on our dataset. The generated and reconstructed images are presented in Figure 5. The architecture of generator network and discriminator network are described in Figure 6 and Figure 7. Therefore, the accuracy of real and fake samples and of the generator are shown in Figure 8.
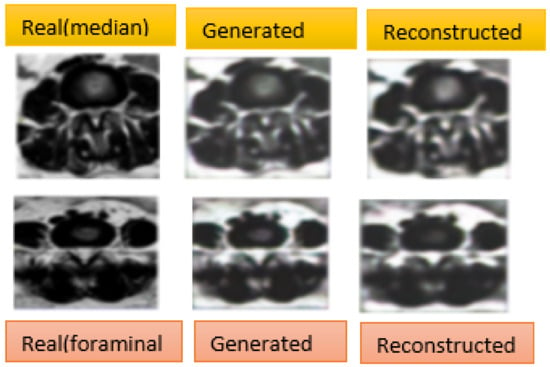
Figure 5.
Generated and reconstructed images results using CycleGAN. On the left, we have the original images, in the center, we have the first generated images, and on the right, we have the second generated images which should look like the original images.
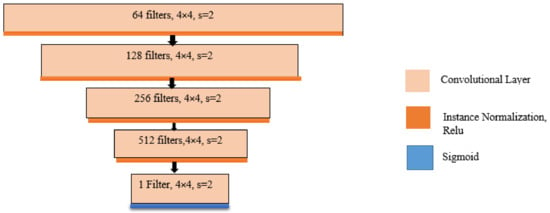
Figure 6.
Generator architecture.
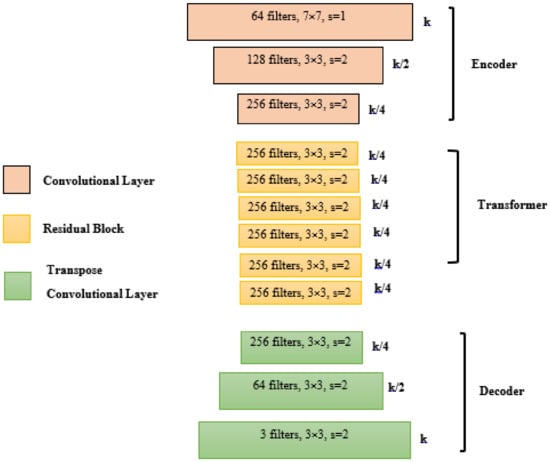
Figure 7.
Discriminator architecture.
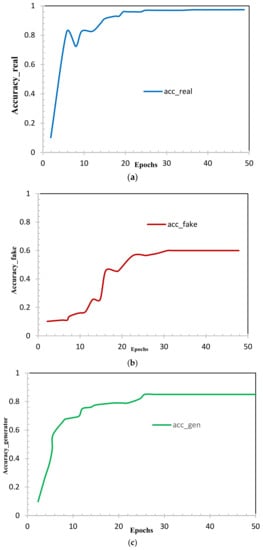
Figure 8.
Accuracy of (a) real samples, (b) fake samples and (c) generator.
Figure 8 shows us the accuracy curves of real samples (Figure 8a), fake samples (Figure 8b), and generator (Figure 8c). This also shows that the generalized dataset through the CycleGAN model performed appropriately to target the lumbar discs images and preserved the most important features; we can conclude through our experiments that the intensity generalization of CycleGAN are very efficient. The loss of the discriminator (as illustrated in Figure 9a,b) is expected to rapidly decrease to a value close to zero, where it remains during training. The loss of the generator (as illustrated in Figure 9c) is expected to either decrease to zero or continually decrease during training. The generator is expected to produce extremely low-quality images that are easily identified as fake by the discriminator (as illustrated in Figure 9). We remark on the performance of all the experiments using the CycleGAN model.
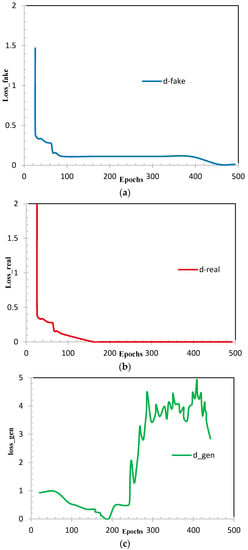
Figure 9.
Loss of (a) fake samples; (b) real samples and (c) generator.
5. Discussion
We have conducted different experiments using the CycleGAN model to validate our own dataset and to open perspectives for future works. We have performed the best results in generating and classifying lumbar intervertebral discs on axial view MRI. From the analysis and discussion results introduced in Table 2 and Table 3, we remark that a distinct performance has been achieved in terms of IoU and accuracy. Our system has a higher and clearer performance compared to other CAD systems. Additionally, using axial view MRI aids radiologists in the recognition of herniation types (median, foraminal). Moreover, locating intervertebral discs on axial view MRI is more significant than on sagittal views. The performed results for lumbar herniated disc detection and classification of herniated discs are promising, and highlight the importance of CycleGAN for dataset generation, classification and detection. To the best of our knowledge, this is the first use of CycleGANs on our dataset including axial view MRIs and collected from university hospital of Sousse, which presents the contribution and originality of our work. For this reason, we will further continue research on putting forward methods and techniques based on GANs, which are very interesting models applied in many fields.
6. Conclusions
We have developed a system based on GANs using the CycleGAN model. We have utilized intervertebral discs on axial view MRI in order to help radiologists in performing lumbar disc stenosis. Our dataset includes a clinical lumbar intervertebral disc MRI study of 1500 patients with symptomatic lower back pain. We have proposed an image synthesis approach using the CycleGAN model. We have presented data including a lumbar intervertebral disc generation technique using an adversarial network and random sampling, and have demonstrated it on lumbar disc localization and classification. We began with an incompletely labeled dataset and generated newly labeled images by randomly sampling labeled lumbar intervertebral discs and generating lumbar intervertebral disc images with backgrounds. We used these generated examples to train a supervised intervertebral discs detection system. This model outperformed existing state-of-the-art detection detectors when doing testing on our test set. The performance of the developed system is close to that of a model on fully labeled samples. Even though the implementation of the new loss function can effectively reduce the occurrence of unknown cells, given the result of the CycleGAN model, few new intervertebral discs that can affect the accuracy of the results can still be generated.
The advantages of the proposed technique with respect to other lumbar herniated detection and classification approaches can be summarized in two points. The first one is the parallelism of the generation and detection of lumbar intervertebral discs using the CycleGAN model performing at 97.2% IoU, which represents the highest result compared to previous methods. The lumbar intervertebral discs of generation and detection using the proposed method can easily and rapidly generate and localize intervertebral discs from axial view MRI compared to conventional lumbar disc detection methods and U-net convolutional networks, which require a lot of computation time as well as a large amount of data, which is sometimes not easily available. Additionally, we have worked on a challenging database by adding normal and herniated intervertebral discs. Compared to most intervertebral disc detection and classification methods using deep convolutional neural networks, the proposed method has the highest detection rate values, which can reach 97.2%, even in a noisy context. We conclude that our method is more reliable than many other methods and can provide high detection and classification rates. Moreover, the capacity of treating a huge number of axial view MRIs is a real advantage of this approach. In future works, we will propose the use of a pix2pix that represents a type of conditional GAN to improve the detection and classification rates and lumbar intervertebral disc localization for each case. For future work, we will use an optimizer that merges the speediness of the capacity of generalization and Adam on new data. A possible improvement to the suggested system is the incorporation of AdaBound to significantly improve the detection rate, and more magnetic resonance images to train the network for higher accuracy.
Author Contributions
W.M. conceived of the presented idea. W.M. developed the theory and performed the computations. M.B. and M.S. verified the analytical methods, were involved in planning, and supervised the work. M.B., M.S. and E.M. encouraged W.M. designed and they supervised the findings of this work. F.T.T. verified all the found results. All authors discussed the results and contributed to the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mbarki, W.; Bouchouicha, M.; Frizzi, S.; Tshibasu, F.; Ben Farhat, L.; Sayadi, M. A novel method based on deep learning for herniated lumbar disc segmentation. In Proceedings of the 2020 4th International Conference on Advanced Systems and Emergent Technologies (IC_ASET), Hammamet, Tunisia, 15–18 December 2020; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2020; pp. 394–399. [Google Scholar]
- Chwialkowski, M.P.; Shile, P.E.; Peshock, R.M.; Pfeifer, D.; Parkey, R.W. Automated detection and evaluation of lumbar discs in MR images. In Proceedings of the Images of the Twenty-First Century. Proceedings of the Annual International Engineering in Medicine and Biology Society, Seattle, WA, USA, 9–12 November 1989; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2003; pp. 571–572. [Google Scholar]
- Tsai, M.-D.; Yeh, Y.-D.; Hsieh, M.-S.; Tsai, C.-H. Automatic spinal disease diagnoses assisted by 3D unaligned transverse CT slices. Comput. Med. Imaging Graph. 2004, 28, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Haq, R.; Aras, R.; Besachio, D.A.; Borgie, R.C.; Audette, M.A. 3D lumbar spine intervertebral disc segmentation and compression simulation from MRI using shape-aware models. Int. J. Comput. Assist. Radiol. Surg. 2014, 10, 45–54. [Google Scholar] [CrossRef] [PubMed]
- AlOmari, R.S.; Corso, J.J.; Chaudhary, V.; Dhillon, G. Automatic diagnosis of lumbar disc herniation with shape and appearance features from MRI. In Medical Imaging 2010: Computer-Aided Diagnosis; International Society for Optics and Photonics: Bellingham, WA, USA, 2010; Volume 7624, p. 76241A. [Google Scholar]
- Michopoulou, S. Image Analysis for the Diagnosis of MR Images of the Lumbar Spine. Ph.D. Thesis, UCL (University College London), London, UK, 2011. [Google Scholar]
- Cuckler, J.M.; Bernini, P.; Wiesel, S.W.; Booth, R.E., Jr.; Rothman, R.H.; Pickens, G.T. The use of epidural steroids in the treat-ment of lumbar radicular pain. A prospective, randomized, double-blind study. J. Bone Jt. Surg. 1985, 67, 63–66. [Google Scholar] [CrossRef]
- Hoad, C.L.; Martel, A.L. Segmentation of MR images for computer-assisted surgery of the lumbar spine. Phys. Med. Biol. 2002, 47, 3503–3517. [Google Scholar] [CrossRef]
- Ghosh, S.; Chaudhary, V. Supervised methods for detection and segmentation of tissues in clinical lumbar MRI. Comput. Med. Imaging Graph. 2014, 38, 639–649. [Google Scholar] [CrossRef]
- Chen, Y.; Shi, F.; Christodoulou, A.G.; Xie, Y.; Zhou, Z.; Li, D. Efficient and Accurate MRI Super-Resolution Using a Generative Adversarial Network and 3D Multi-level Densely Connected Network. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Metzler, J.B., Ed.; Springer: Cham, Switzerland, 2018; pp. 91–99. [Google Scholar]
- Jiang, J.; Hu, Y.-C.; Tyagi, N.; Zhang, P.; Rimner, A.; Mageras, G.S.; Deasy, J.O.; Veeraraghavan, H. Tumor-Aware, Adversarial Domain Adaptation from CT to MRI for Lung Cancer Segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Metzler, J.B., Ed.; Springer: Cham, Switzerland, 2018; Volume 11071, pp. 777–785. [Google Scholar]
- Ebrahimzadeh, E.; Fayaz, F.; Ahmadi, F.; Nikravan, M. A machine learning-based method in order to diagnose lumbar disc herniation disease by MR image processing. MedLife Open Access 2018, 1, 1. [Google Scholar]
- Chevrefils, C.; Chériet, F.; Grimard, G.; Aubin, C.-E. Watershed Segmentation of Intervertebral Disk and Spinal Canal from MRI Images. In International Conference Image Analysis and Recognition; Springer: Berlin/Heidelberg, Germany, 2007; pp. 1017–1027. [Google Scholar]
- Alawneh, K.; Al-Dwiekat, M.; Alsmirat, M.; Al-Ayyoub, M. Computer-aided diagnosis of lumbar disc herniation. In Proceedings of the 2015 6th International Conference on Information and Communication Systems (ICICS), Amman, Jordan, 7–9 April 2015; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2015; pp. 286–291. [Google Scholar]
- Marquardt, G.; Bruder, M.; Theuss, S.; Setzer, M.; Seifert, V. Ultra-long-term outcome of surgically treated far-lateral, extraforaminal lumbar disc herniations: A single-center series. Eur. Spine J. 2011, 21, 660–665. [Google Scholar] [CrossRef]
- Creswell, A.; White, T.; Dumoulin, V.; Arulkumaran, K.; Sengupta, B.; Bharath, A.A. Generative ad-versarial networks: An overview. IEEE Signal Process. Mag. 2018, 35, 53–65. [Google Scholar] [CrossRef]
- Baur, C.; Albarqouni, S.; Navab, N. Generating Highly Realistic Images of Skin Lesions with GANs. In Lecture Notes in Computer Science; Metzler, J.B., Ed.; Springer: Cham, Switzerland, 2018; pp. 260–267. [Google Scholar]
- Wolterink, J.M.; Kamnitsas, K.; Ledig, C.; Išgum, I. Generative adversarial networks and ad-versarial methods in biomedical image analysis. arXiv 2018, arXiv:1810.10352. [Google Scholar]
- Yu, S.; Dong, H.; Yang, G.; Slabaugh, G.; Dragotti, P.L.; Ye, X.; Guo, Y. Deep de-aliasing for fast compressive sensing mri. arXiv 2017, arXiv:1705.07137. [Google Scholar]
- Yang, G.; Yu, S.; Dong, H.; Slabaugh, G.; Dragotti, P.L.; Ye, X.; Firmin, D. Dagan: Deep de-aliasing generative adversarial networks for fast compressed sensing mri recon-struction. IEEE Trans. Med. Imaging 2017, 37, 1310–1321. [Google Scholar] [CrossRef]
- Seitzer, M.; Yang, G.; Schlemper, J.; Oktay, O.; Würfl, T.; Christlein, V.; Wong, T.; Mohiaddin, R.; Firmin, D.; Keegan, J.; et al. Adversarial and perceptual refinement for compressed sensing mri reconstruction. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2018; pp. 232–240. [Google Scholar]
- Quan, T.M.; Nguyen-Duc, T.; Jeong, W.K. Compressed sensing mri reconstruction with cyclic loss in genera-tive adversarial networks. arXiv 2017, arXiv:1709.00753. [Google Scholar]
- Sánchez, I.; Vilaplana, V. Brain mri super-resolution using 3d generative adversarial networks. arXiv 2018, arXiv:1812.11440. [Google Scholar]
- Li, Z.; Wang, Y.; Yu, J. Reconstruction of Thin-Slice Medical Images Using Generative Adversarial Network. In Tools and Algorithms for the Construction and Analysis of Systems; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2017; Volume 10541, pp. 325–333. [Google Scholar]
- Mardani, M.; Gong, E.; Cheng, J.Y.; Vasanawala, S.S.; Zaharchuk, G.; Xing, L.; Pauly, J.M. Deep Generative Adversarial Neural Networks for Compressive Sensing MRI. IEEE Trans. Med. Imaging 2019, 38, 167–179. [Google Scholar] [CrossRef]
- Shitrit, O.; Raviv, T.R. Accelerated Magnetic Resonance Imaging by Adversarial Neural Network. In Lecture Notes in Computer Science; Metzler, J.B., Ed.; Springer: Cham, Switzerland, 2017; pp. 30–38. [Google Scholar]
- Mahapatra, D.; Bozorgtabar, B.; Hewavitharanage, S.; Garnavi, R. Image Super Resolution Using Generative Adversarial Networks and Local Saliency Maps for Retinal Image Analysis. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Metzler, J.B., Ed.; Springer: Cham, Switzerland, 2017; pp. 382–390. [Google Scholar]
- Han, L.; Yin, Z. A Cascaded Refinement GAN for Phase Contrast Microscopy Image Super Resolution. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Metzler, J.B., Ed.; Springer: Cham, Switzerland, 2018; pp. 347–355. [Google Scholar]
- Ravì, D.; Szczotka, A.B.; Pereira, S.P.; Vercauteren, T. Adversarial training with cycle con-sistency for unsupervised super-resolution in endomicroscopy. Med. Image Anal. 2019, 53, 123–131. [Google Scholar] [CrossRef]
- Schlegl, T.; Seeböck, P.; Waldstein, S.M.; Schmidt-Erfurth, U.; Langs, G. Unsupervised Anomaly Detection with Generative Adversarial Networks to Guide Marker Discovery. In International Conference on Information Processing in Medical Imaging; Springer: Cham, Switzerland, 2017; pp. 146–157. [Google Scholar]
- Chen, X.; Konukoglu, E. Unsupervised detection of lesions in brain mri using constrained adversarial au-to-encoders. arXiv 2018, arXiv:1806.04972. [Google Scholar]
- Kohl, S.; Bonekamp, D.; Schlemmer, H.P.; Yaqubi, K.; Hohenfellner, M.; Hadaschik, B.; Radtke, J.P.; Maier-Hein, K. Adversarial networks for the detection of aggressive prostate cancer. arXiv 2017, arXiv:1702.08014. [Google Scholar]
- Udrea, A.; Mitra, G.D. Generative Adversarial Neural Networks for Pigmented and Non-Pigmented Skin Lesions Detection in Clinical Images. In Proceedings of the 2017 21st International Conference on Control Systems and Computer Science (CSCS), Bucharest, Romania, 29–31 May 2017; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2017; pp. 364–368. [Google Scholar]
- Tuysuzoglu, A.; Tan, J.; Eissa, K.; Kiraly, A.P.; Diallo, M.; Kamen, A. Deep adversarial con-text-aware landmark detection for ultrasound imaging. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2018; pp. 151–158. [Google Scholar]
- Yi, X.; Babyn, P. Sharpness-Aware Low-Dose CT Denoising Using Conditional Generative Adversarial Network. J. Digit. Imaging 2018, 31, 655–669. [Google Scholar] [CrossRef]
- Wolterink, J.M.; Leiner, T.; Viergever, M.A.; Isgum, I. Generative Adversarial Networks for Noise Reduction in Low-Dose CT. IEEE Trans. Med. Imaging 2017, 36, 2536–2545. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, Y.; Noble, J.H.; Dawant, B.M. Conditional Generative Adversarial Networks for Metal Artifact Reduction in CT Images of the Ear. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Metzler, J.B., Ed.; Springer: Cham, Switzerland, 2018; pp. 3–11. [Google Scholar]
- Yang, S.; Wang, J.; Hao, X.; Li, H.; Wei, X.; Deng, B.; Loparo, K.A. BiCoSS: Toward Large-Scale Cognition Brain with Multigranular Neuromorphic Architecture. IEEE Trans. Neural Netw. Learn. Syst. 2021, 1–15. [Google Scholar] [CrossRef]
- Yang, S.; Wang, J.; Zhang, N.; Deng, B.; Pang, Y.; Azghadi, M.R. CerebelluMorphic: Large-Scale Neuromorphic Model and Architecture for Supervised Motor Learning. IEEE Trans. Neural Netw. Learn. Syst. 2021, 1–15. [Google Scholar] [CrossRef]
- Yang, S.; Deng, B.; Wang, J.; Liu, C.; Li, H.; Lin, Q.; Fietkiewicz, C.; Loparo, K.A. Design of Hidden-Property-Based Variable Universe Fuzzy Control for Movement Disorders and Its Efficient Reconfigurable Implementation. IEEE Trans. Fuzzy Syst. 2019, 27, 304–318. [Google Scholar] [CrossRef]
- Yang, S.; Deng, B.; Wang, J.; Li, H.; Lu, M.; Che, Y.; Wei, X.; Loparo, K.A. Scalable Digital Neuromorphic Architecture for Large-Scale Biophysically Meaningful Neural Network With Multi-Compartment Neurons. IEEE Trans. Neural Netw. Learn. Syst. 2020, 31, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, H.; Jiang, Z. Imbalanced biomedical data classification using self-adaptive multilayer ELM com-bined with dynamic GAN. Biomed. Eng. Online 2018, 17, 181. [Google Scholar] [CrossRef] [PubMed]
- Larrañeta, E.; Henry, M.; Irwin, N.J.; Trotter, J.; Perminova, A.A.; Donnelly, R.F. Synthesis and characterization of hyaluronic acid hydrogels crosslinked using a solvent-free process for potential biomedical applications. Carbohydr. Polym. 2018, 181, 1194–1205. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, F.; Xu, W.; Li, Y. Multi-channel Generative Adversarial Network for Parallel Magnetic Resonance Image Reconstruction in K-space. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Metzler, J.B., Ed.; Springer: Cham, Switzerland, 2018; pp. 180–188. [Google Scholar]
- Yang, S.; Gao, T.; Wang, J.; Deng, B.; Lansdell, B.; Linares-Barranco, B. Efficient Spike-Driven Learning with Den-dritic Event-Based Processing. Front. Neurosci. 2021, 15, 97. [Google Scholar] [CrossRef] [PubMed]
- Roy, K.; Jaiswal, A.; Panda, P. Towards spike-based machine intelligence with neuromorphic computing. Nature 2019, 575, 607–617. [Google Scholar] [CrossRef]
- Yang, S.; Wang, J.; Deng, B.; Liu, C.; Li, H.; Fietkiewicz, C.; Loparo, K.A. Real-Time Neuromorphic System for Large-Scale Conductance-Based Spiking Neural Networks. IEEE Trans. Cybern. 2019, 49, 2490–2503. [Google Scholar] [CrossRef]
- Kazeminia, S.; Baur, C.; Kuijper, A.; van Ginneken, B.; Navab, N.; Albarqouni, S.; Mukhopadhyay, A. GANs for medical image analysis. Artif. Intell. Med. 2020, 109, 101938. [Google Scholar] [CrossRef]
- Wolterink, J.M.; Dinkla, A.M.; Savenije, M.H.; Seevinck, P.R.; van den Berg, C.A.; Isgum, I. Deep MR to CT synthesis using unpaired data. In International Workshop on Simulation and Synthesis in Medical Imaging; Springer: Cham, Switzerland, 2017; pp. 14–23. [Google Scholar]
- Yan, P.; Xu, S.; Rastinehad, A.R.; Wood, B.J. Adversarial Image Registration with Application for MR and TRUS Image Fusion. In Lecture Notes in Computer Science; Metzler, J.B., Ed.; Springer: Cham, Switzerland, 2018; pp. 197–204. [Google Scholar]
- Baumgartner, C.F.; Koch, L.M.; Tezcan, K.C.; Ang, J.X.; Konukoglu, E. Visual Feature Attribution Using Wasserstein GANs. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2018; pp. 8309–8319. [Google Scholar]
- Ren, J.; Hacihaliloglu, I.; Singer, E.A.; Foran, D.J.; Qi, X. Adversarial Domain Adaptation for Classification of Prostate Histopathology Whole-Slide Images. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Metzler, J.B., Ed.; Springer: Cham, Switzerland, 2018; Volume 11071, pp. 201–209. [Google Scholar]
- Son, J.; Park, S.J.; Jung, K.-H. Retinal vessel segmentation in fundoscopic images with generative adversarial networks. arXiv 2017, arXiv:1706.09318. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).