A Poplar Rust Effector Protein Associates with Protein Disulfide Isomerase and Enhances Plant Susceptibility
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plants Material and Growth Conditions
2.2. Cloning Procedures and Plasmid Constructs
2.3. Protein Expression in N. benthamiana and A. thaliana
2.4. Pathogen Infections Assay
2.5. Membrane Fractionation
2.6. Sample Preparation for Mass Spectrometry
2.7. Western Blot Analysis
2.8. Confocal Microscopy
2.9. RNA Extraction and Transcriptome Analysis
2.10. Y2H Reporter Assays
2.11. Molecular Modeling of the Proteins
3. Results
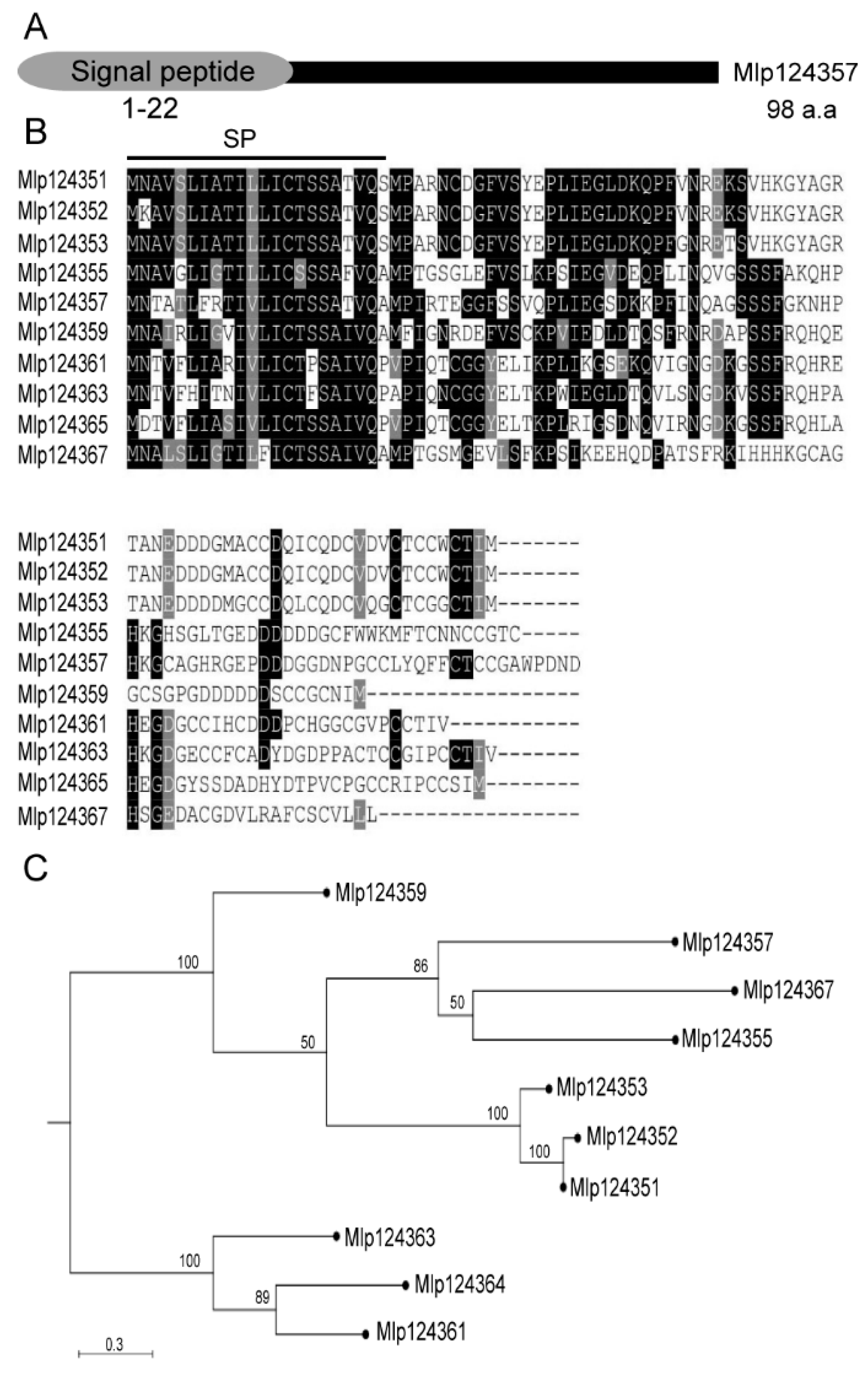
3.1. Selection and Phylogenetic Analysis of Mlp124357
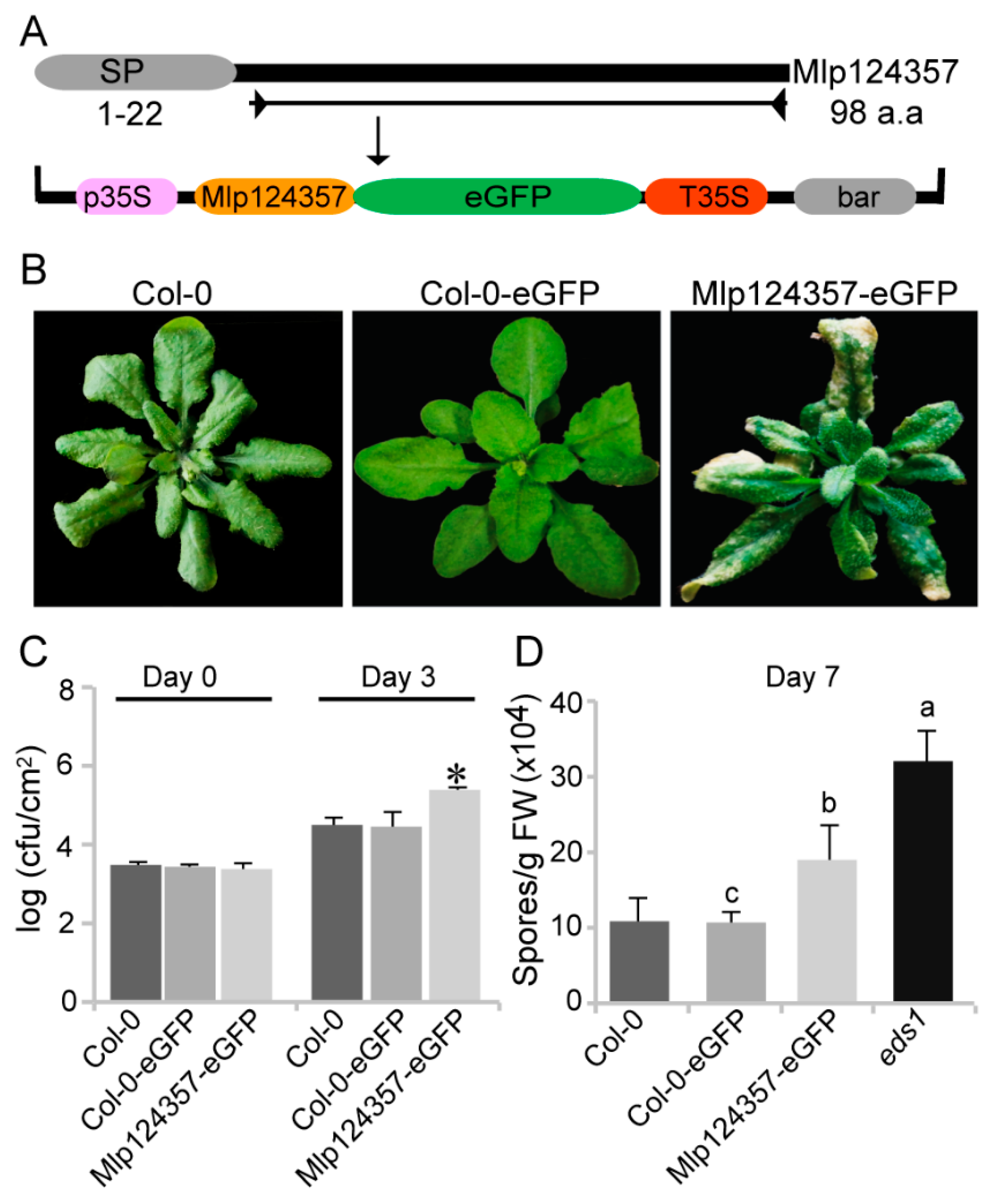
3.2. Mlp124357 Expression in Planta Affects the Plant Susceptibility to Bacterial and Oomycete Pathogens
3.3. Mlp124357 Possesses a GxxxG Motif that Is Required for the Interaction with Tonoplasts
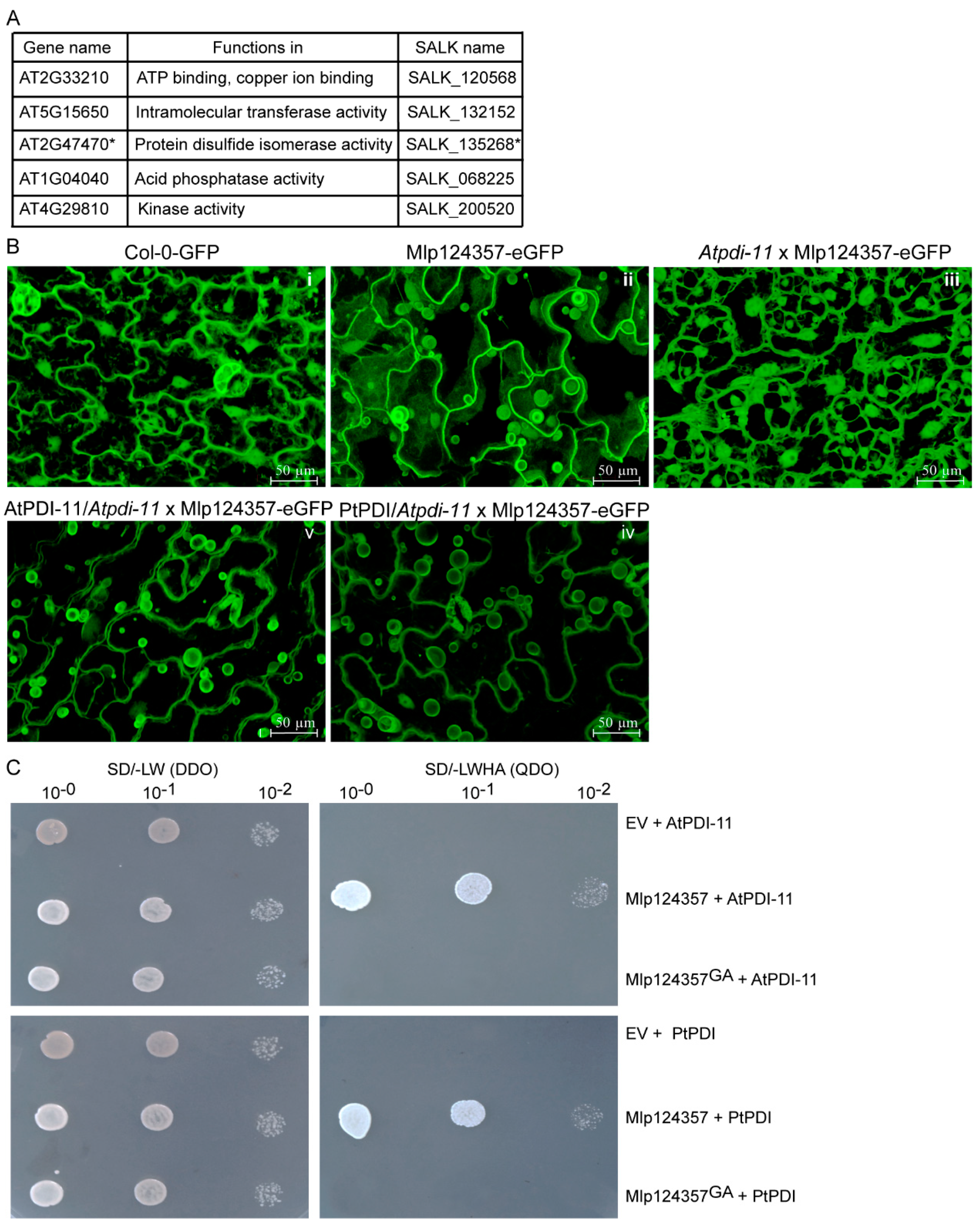
3.4. Protein Disulfide Isomerase 11 as a Potential Plant Interactor of Mlp124357
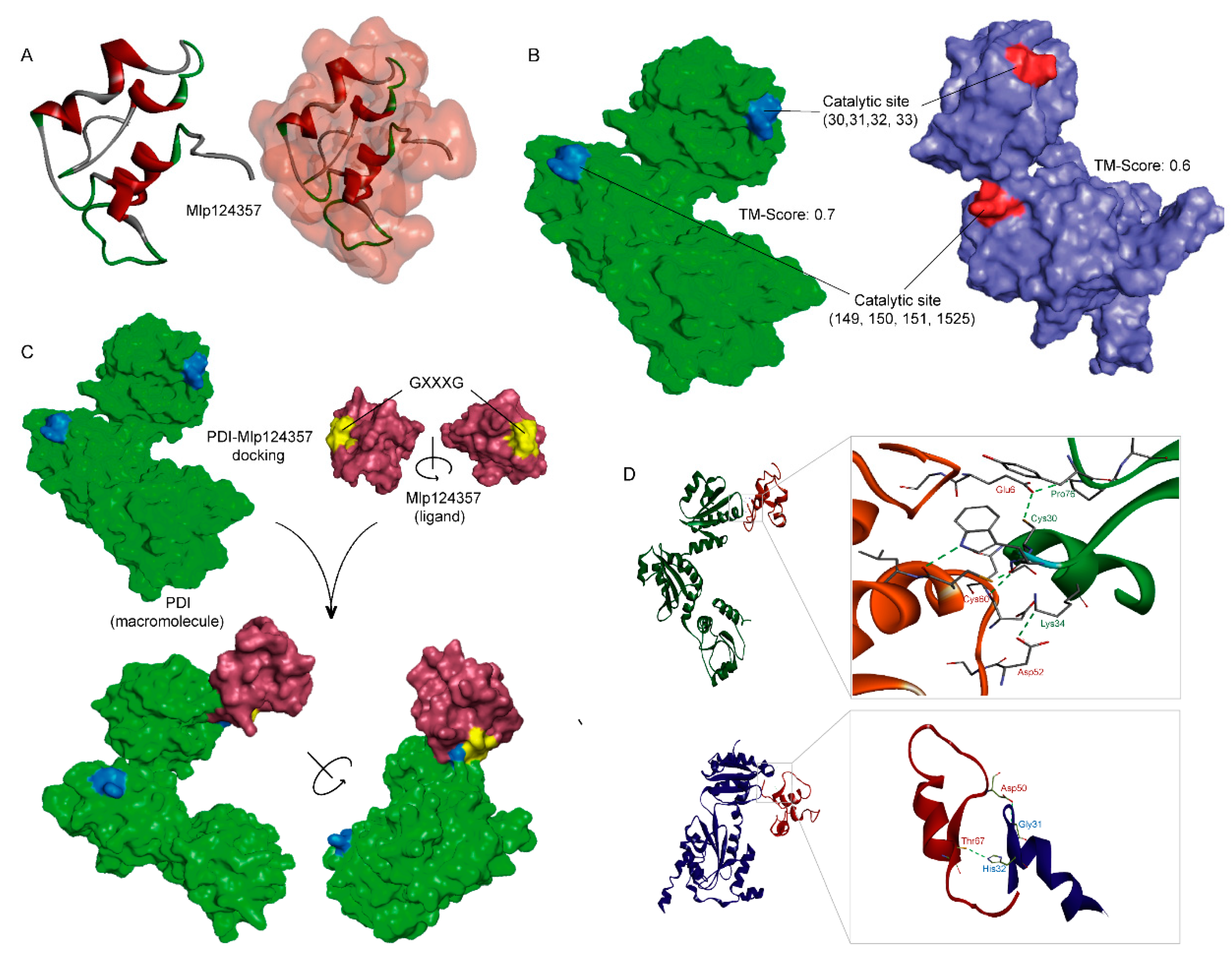
3.5. Molecular Modeling also Supports the Association of AtPDI-11 and Mlp124357 Effector
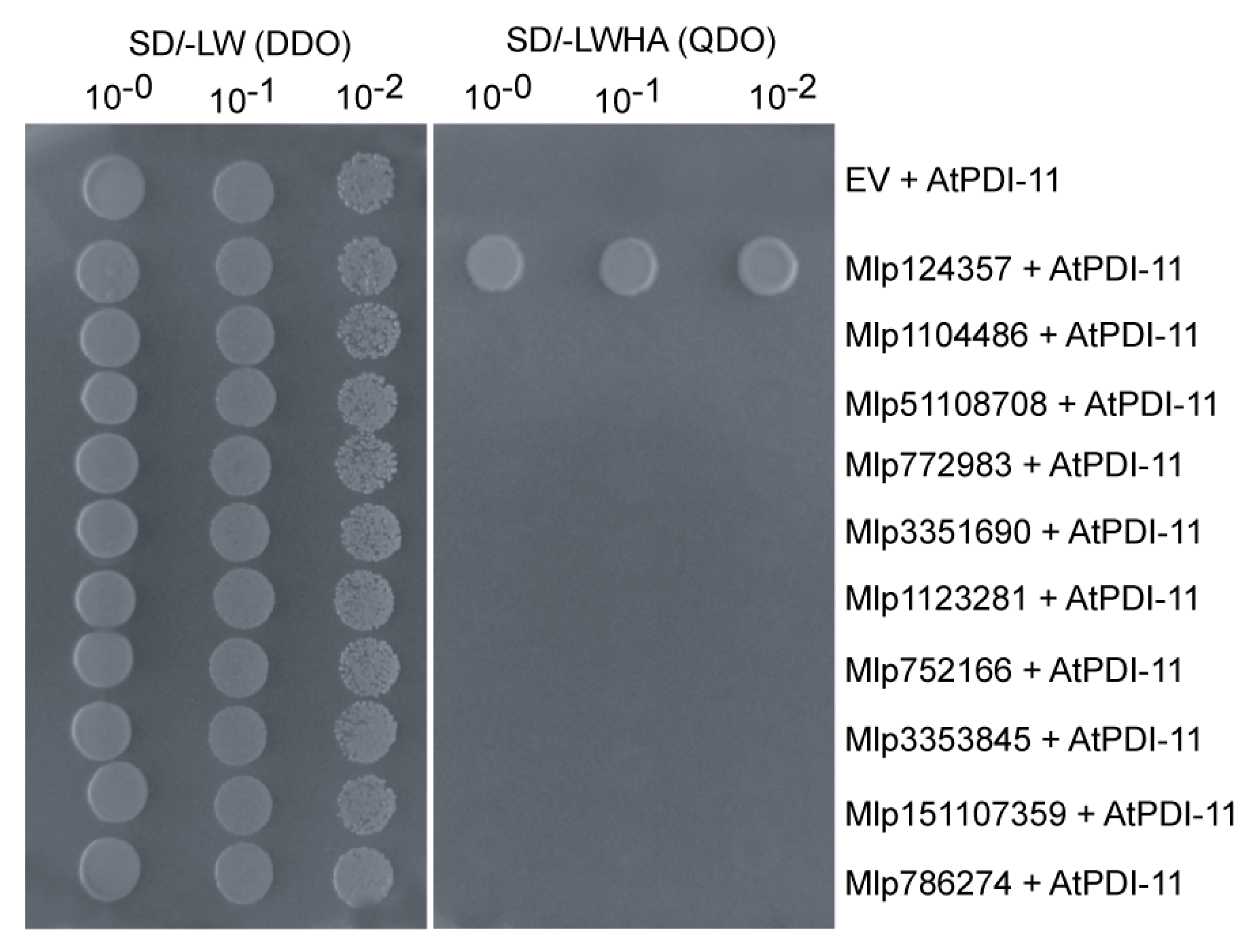
3.6. The Mlp124357-PDI Association Takes Place in an Effector-Specific Manner
3.7. Protein Disulfide-Isomerase 11 Acts as a Helper of Mlp124357
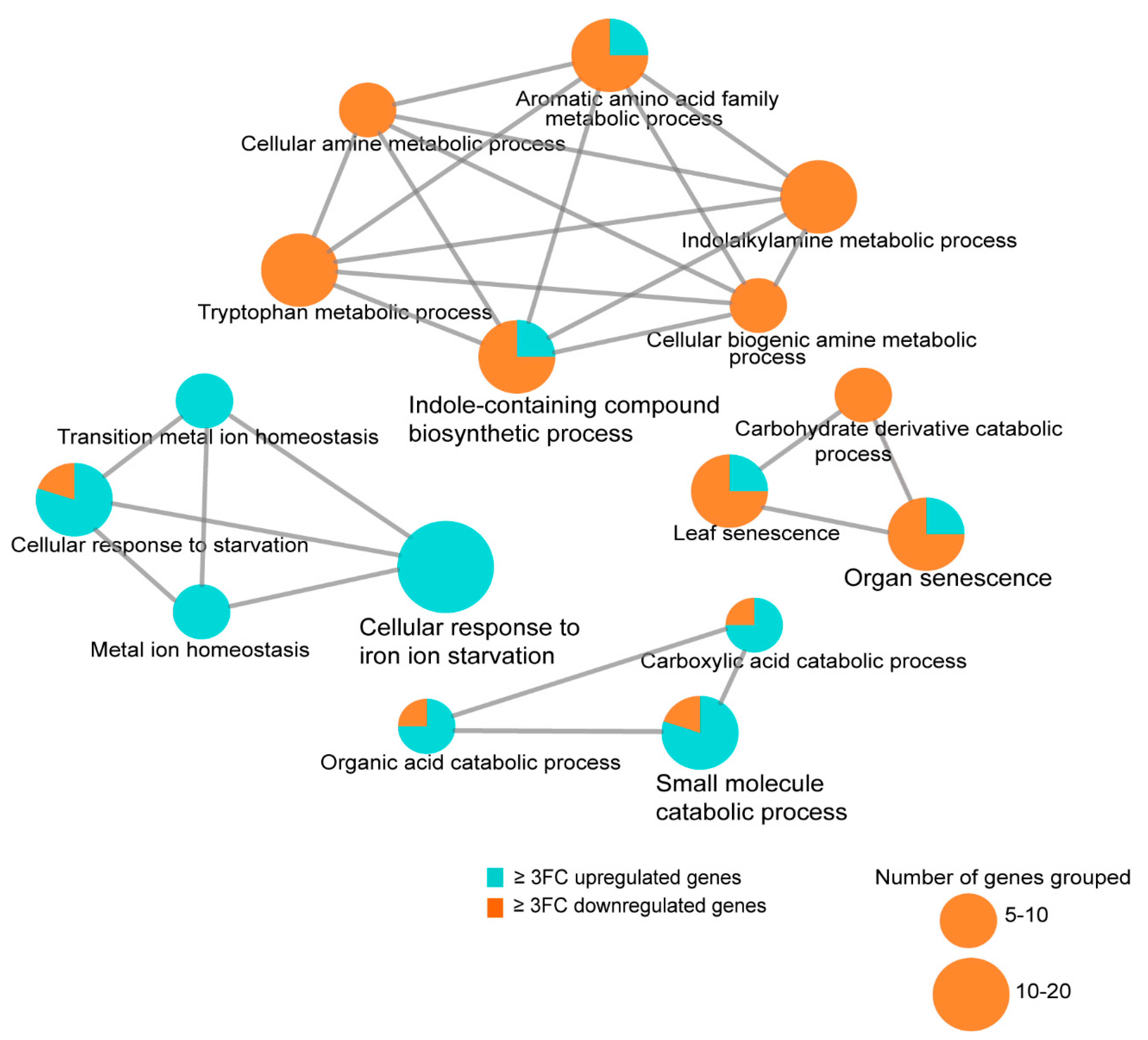
3.8. Transcriptome Analysis of the Arabidopsis Transgenics Line Expressing Mlp124357
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.B.; dos Santos, K.C.G.; Sanchez, I.B.; Petre, B.; Lorrain, C.; Plourde, M.B.; Duplessis, S.; Desgagné-Penix, I.; Germain, H. A rust fungal effector binds plant DNA and modulates transcription. Sci. Rep. 2018, 8, 14718. [Google Scholar] [CrossRef] [PubMed]
- Caillaud, M.C.; Piquerez, S.J.; Fabro, G.; Steinbrenner, J.; Ishaque, N.; Beynon, J.; Jones, J.D. Subcellular localization of the Hpa RxLR effector repertoire identifies a tonoplast-associated protein HaRxL17 that confers enhanced plant susceptibility. Plant J. 2012, 69, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Petre, B.; Lorrain, C.; Saunders, D.G.; Win, J.; Sklenar, J.; Duplessis, S.; Kamoun, S. Rust fungal effectors mimic host transit peptides to translocate into chloroplasts. Cell. Microbiol. 2016, 18, 453–465. [Google Scholar] [CrossRef] [Green Version]
- Petre, B.; Saunders, D.G.; Sklenar, J.; Lorrain, C.; Win, J.; Duplessis, S.; Kamoun, S. Candidate effector proteins of the rust pathogen Melampsora larici-populina target diverse plant cell compartments. Mol. Plant-Microbe Interact. 2015, 28, 689–700. [Google Scholar] [CrossRef] [Green Version]
- Vargas, W.A.; Sanz-Martín, J.M.; Rech, G.E.; Armijos-Jaramillo, V.D.; Rivera, L.P.; Echeverria, M.M.; Díaz-Mínguez, J.M.; Thon, M.R.; Sukno, S.A. A fungal effector with host nuclear localization and DNA-binding properties is required for maize anthracnose development. Mol. Plant-Microbe Interact. 2016, 29, 83–95. [Google Scholar] [CrossRef]
- Schornack, S.; van Damme, M.; Bozkurt, T.O.; Cano, L.M.; Smoker, M.; Thines, M.; Gaulin, E.; Kamoun, S.; Huitema, E. Ancient class of translocated oomycete effectors targets the host nucleus. Proc. Natl. Acad. Sci. USA 2010, 107, 17421–17426. [Google Scholar] [CrossRef] [Green Version]
- Wirthmueller, L.; Roth, C.; Fabro, G.; Caillaud, M.C.; Rallapalli, G.; Asai, S.; Sklenar, J.; Jones, A.M.; Wiermer, M.; Jones, J.D. Probing formation of cargo/importin-α transport complexes in plant cells using a pathogen effector. Plant J. 2015, 81, 40–52. [Google Scholar] [CrossRef] [Green Version]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [Green Version]
- Bombarely, A.; Rosli, H.G.; Vrebalov, J.; Moffett, P.; Mueller, L.A.; Martin, G.B. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant-Microbe Interact. 2012, 25, 1523–1530. [Google Scholar] [CrossRef] [Green Version]
- Germain, H.; Joly, D.L.; Mireault, C.; Plourde, M.B.; Letanneur, C.; Stewart, D.; Morency, M.J.; Petre, B.; Duplessis, S.; Séguin, A. Infection assays in Arabidopsis reveal candidate effectors from the poplar rust fungus that promote susceptibility to bacteria and oomycete pathogens. Mol. Plant Pathol. 2018, 19, 191–200. [Google Scholar] [CrossRef] [Green Version]
- Goodin, M.M.; Zaitlin, D.; Naidu, R.A.; Lommel, S.A. Nicotiana benthamiana: Its history and future as a model for plant–pathogen interactions. Mol. Plant-Microbe Interact. 2008, 21, 1015–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Win, J.; Chaparro-Garcia, A.; Belhaj, K.; Saunders, D.; Yoshida, K.; Dong, S.; Schornack, S.; Zipfel, C.; Robatzek, S.; Hogenhout, S. Effector biology of plant-associated organisms: Concepts and perspectives. Proc. Cold Spring Harb. Symp. Quant. Biol. 2012, 77, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Macho, A.P.; Guevara, C.M.; Tornero, P.; Ruiz-Albert, J.; Beuzon, C.R. The Pseudomonas syringae effector protein HopZ1a suppresses effector-triggered immunity. New Phytol. 2010, 187, 1018–1033. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Wang, Y.; Xue, L.; Chu, J.; Yan, C.; Fu, J.; Chen, M.; Innes, R.W.; Zhou, J.-M. Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe 2010, 7, 164–175. [Google Scholar] [CrossRef] [Green Version]
- Anderson, J.C.; Pascuzzi, P.E.; Xiao, F.; Sessa, G.; Martin, G.B. Host-mediated phosphorylation of type III effector AvrPto promotes Pseudomonas virulence and avirulence in tomato. Plant Cell 2006, 18, 502–514. [Google Scholar] [CrossRef] [Green Version]
- Nimchuk, Z.; Marois, E.; Kjemtrup, S.; Leister, R.T.; Katagiri, F.; Dangl, J.L. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell 2000, 101, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Bai, X.; Correa, V.R.; Toruño, T.Y.; Ammar, E.-D.; Kamoun, S.; Hogenhout, S.A. AY-WB phytoplasma secretes a protein that targets plant cell nuclei. Mol. Plant-Microbe Interact. 2009, 22, 18–30. [Google Scholar] [CrossRef] [Green Version]
- Szurek, B.; Marois, E.; Bonas, U.; Van den Ackerveken, G. Eukaryotic features of the Xanthomonas type III effector AvrBs3: Protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J. 2001, 26, 523–534. [Google Scholar] [CrossRef]
- Duplessis, S.; Joly, D.L.; Dodds, P.N. Rust effectors. In Effectors in Plant-Microbe Interactions; John Wiley & Sons: Hoboken, NJ, USA, 2012; pp. 155–193. [Google Scholar]
- Pennisi, E. Armed and dangerous. Science 2010, 327, 804–805. [Google Scholar]
- Duplessis, S.; Major, I.; Martin, F.; Séguin, A. Poplar and pathogen interactions: Insights from Populus genome-wide analyses of resistance and defense gene families and gene expression profiling. Crit. Rev.Plant Sci. 2009, 28, 309–334. [Google Scholar] [CrossRef]
- Hacquard, S.; Petre, B.; Frey, P.; Hecker, A.; Rouhier, N.; Duplessis, S. The poplar-poplar rust interaction: Insights from genomics and transcriptomics. J. Pathog. 2011, 2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Major, I.T.; Nicole, M.-C.; Duplessis, S.; Séguin, A. Photosynthetic and respiratory changes in leaves of poplar elicited by rust infection. Photosynth. Res. 2010, 104, 41–48. [Google Scholar] [CrossRef]
- Duplessis, S.; Cuomo, C.A.; Lin, Y.-C.; Aerts, A.; Tisserant, E.; Veneault-Fourrey, C.; Joly, D.L.; Hacquard, S.; Amselem, J.; Cantarel, B.L. Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc. Natl. Acad. Sci. USA 2011, 108, 9166–9171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hacquard, S.; Delaruelle, C.; Legué, V.; Tisserant, E.; Kohler, A.; Frey, P.; Martin, F.; Duplessis, S. Laser capture microdissection of uredinia formed by Melampsora larici-populina revealed a transcriptional switch between biotrophy and sporulation. Mol. Plant-Microbe Interact. 2010, 23, 1275–1286. [Google Scholar] [CrossRef]
- Hacquard, S.; Joly, D.L.; Lin, Y.-C.; Tisserant, E.; Feau, N.; Delaruelle, C.; Legué, V.; Kohler, A.; Tanguay, P.; Petre, B. A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici-populina (poplar leaf rust). Mol. Plant-Microbe Interact. 2012, 25, 279–293. [Google Scholar] [CrossRef] [Green Version]
- Saunders, D.G.; Win, J.; Cano, L.M.; Szabo, L.J.; Kamoun, S.; Raffaele, S. Using hierarchical clustering of secreted protein families to classify and rank candidate effectors of rust fungi. PLoS ONE 2012, 7, e29847. [Google Scholar] [CrossRef]
- Madina, M.H.; Zheng, H.; Germain, H. New insight into bulb dynamics in the vacuolar lumen of Arabidopsis cells. Botany 2018, 96, 511–520. [Google Scholar] [CrossRef]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019. [Google Scholar] [CrossRef]
- Mireault, C.; Paris, L.-E.; Germain, H. Enhancement of the Arabidopsis floral dip method with XIAMETER OFX-0309 as alternative to Silwet L-77 surfactant. Botany 2014, 92, 523–525. [Google Scholar] [CrossRef] [Green Version]
- Dong, O.X.; Meteignier, L.V.; Plourde, M.B.; Ahmed, B.; Wang, M.; Jensen, C.; Jin, H.; Moffett, P.; Germain, H. Arabidopsis TAF15b Localizes to RNA Processing Bodies and Contributes to snc1-Mediated Autoimmunity. Mol. Plant Microbe Interact. 2016, 29, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Paleontol. Electron. 2001, 4, 9. [Google Scholar]
- Germain, H.; Gray-Mitsumune, M.; Houde, J.; Benhamman, R.; Sawasaki, T.; Endo, Y.; Matton, D.P. The Solanum chacoense ovary receptor kinase 11 (ScORK11) undergoes tissue-dependent transcriptional, translational and post-translational regulation. Plant Physiol. Biochem. 2013, 70, 261–268. [Google Scholar] [CrossRef]
- Widell, S.; Larsson, C. Separation of presumptive plasma membranes from mitochondria by partition in an aqueous polymer two-phase system. Physiol. Plant. 1981, 51, 368–374. [Google Scholar] [CrossRef]
- Serino, G.; Deng, X.W. Protein coimmunoprecipitation in Arabidopsis. CSH Prot. 2007, 2007, pdb prot 4683. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef]
- Germain, H.; Gray-Mitsumune, M.; Lafleur, E.; Matton, D.P. ScORK17, a transmembrane receptor-like kinase predominantly expressed in ovules is involved in seed development. Planta 2008, 228, 851–862. [Google Scholar] [CrossRef]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins Struct. Funct. Bioinform. 2012, 80, 1715–1735. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.G.; Hourai, Y.; Weng, Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS ONE 2011, 6, e24657. [Google Scholar] [CrossRef]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef] [Green Version]
- Tovchigrechko, A.; Vakser, I.A. GRAMM-X public web server for protein–protein docking. Nucleic Acids Res. 2006, 34, W310–W314. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System. 2002. Available online: http://www.pymol.org (accessed on 7 September 2020).
- Yuan, S.; Chan, H.S.; Hu, Z. Using PyMOL as a platform for computational drug design. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2017, 7, e1298. [Google Scholar] [CrossRef]
- Bronnimann, M.P.; Chapman, J.A.; Park, C.K.; Campos, S.K. A transmembrane domain and GxxxG motifs within L2 are essential for papillomavirus infection. J. Virol. 2013, 87, 464–473. [Google Scholar] [CrossRef] [Green Version]
- Cabanillas, D.G.; Jiang, J.; Movahed, N.; Germain, H.; Yamaji, Y.; Zheng, H.; Laliberte, J.-F. Turnip mosaic virus uses the SNARE protein VTI11 in an unconventional route for replication vesicle trafficking. Plant Cell 2018, 30, 2594–2615. [Google Scholar] [CrossRef] [Green Version]
- Teese, M.G.; Langosch, D. Role of GxxxG motifs in transmembrane domain interactions. Biochemistry 2015, 54, 5125–5135. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. Protein Structure and Function Prediction Using I-TASSER. Curr. Protoc. Bioinform. 2015, 52, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Speth, E.B.; Lee, Y.N.; He, S.Y. Pathogen virulence factors as molecular probes of basic plant cellular functions. Curr. Opin. Plant Biol. 2007, 10, 580–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunnac, S.; Lindeberg, M.; Collmer, A. Pseudomonas syringae type III secretion system effectors: Repertoires in search of functions. Curr. Opin. Microbiol. 2009, 12, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Torto, T.A.; Li, S.; Styer, A.; Huitema, E.; Testa, A.; Gow, N.A.; Van West, P.; Kamoun, S. EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res. 2003, 13, 1675–1685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faris, J.D.; Zhang, Z.; Lu, H.; Lu, S.; Reddy, L.; Cloutier, S.; Fellers, J.P.; Meinhardt, S.W.; Rasmussen, J.B.; Xu, S.S. A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. USA 2010, 107, 13544–13549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Zhang, Z.; Faris, J.D.; Oliver, R.P.; Syme, R.; McDonald, M.C.; McDonald, B.A.; Solomon, P.S.; Lu, S.; Shelver, W.L. The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PLoS Pathog. 2012, 8, e1002467. [Google Scholar] [CrossRef] [Green Version]
- Brar, H.K.; Swaminathan, S.; Bhattacharyya, M.K. The Fusarium virguliforme toxin FvTox1 causes foliar sudden death syndrome-like symptoms in soybean. Mol. Plant-Microbe Interact. 2011, 24, 1179–1188. [Google Scholar] [CrossRef] [Green Version]
- Shang, Y.; Li, X.; Cui, H.; He, P.; Thilmony, R.; Chintamanani, S.; Zwiesler-Vollick, J.; Gopalan, S.; Tang, X.; Zhou, J.-M. RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. Proc. Natl. Acad. Sci. USA 2006, 103, 19200–19205. [Google Scholar] [CrossRef] [Green Version]
- Kanneganti, T.-D.; Huitema, E.; Cakir, C.; Kamoun, S. Synergistic interactions of the plant cell death pathways induced by Phytophthora infestans Nep1-like protein PiNPP1.1 and INF1 elicitin. Mol. Plant-Microbe Interact. 2006, 19, 854–863. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.-X.; Domier, L.L.; Radwan, O.; Yendrek, C.R.; Hudson, M.E.; Hartman, G.L. Identification of multiple phytotoxins produced by Fusarium virguliforme including a phytotoxic effector (FvNIS1) associated with sudden death syndrome foliar symptoms. Mol. Plant-Microbe Interact. 2016, 29, 96–108. [Google Scholar] [CrossRef] [Green Version]
- Madina, M.H.; Rahman, M.S.; Zheng, H.; Germain, H. Vacuolar membrane structures and their roles in plant-pathogen interactions. Plant Mol. Biol. 2019, 101, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Verchot, J. Cellular chaperones and folding enzymes are vital contributors to membrane bound replication and movement complexes during plant RNA virus infection. Front. Plant Sci. 2012, 3, 275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuen, C.; Matsumoto, K.; Christopher, D. Variation in the subcellular localization and protein folding activity among Arabidopsis thaliana homologs of protein disulfide isomerase. Biomolecules 2013, 3, 848–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, E.J.; Yuen, C.Y.; Kang, B.-H.; Ondzighi, C.A.; Staehelin, L.A.; Christopher, D.A. Protein disulfide isomerase-2 of Arabidopsis mediates protein folding and localizes to both the secretory pathway and nucleus, where it interacts with maternal effect embryo arrest factor. Mol. Cells 2011, 32, 459–475. [Google Scholar] [CrossRef] [Green Version]
- Ondzighi, C.A.; Christopher, D.A.; Cho, E.J.; Chang, S.-C.; Staehelin, L.A. Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell 2008, 20, 2205–2220. [Google Scholar] [CrossRef] [Green Version]
- Wittenberg, G.; Levitan, A.; Klein, T.; Dangoor, I.; Keren, N.; Danon, A. Knockdown of the A rabidopsis thaliana chloroplast protein disulfide isomerase 6 results in reduced levels of photoinhibition and increased D 1 synthesis in high light. Plant J. 2014, 78, 1003–1013. [Google Scholar] [CrossRef]
- Turano, C.; Coppari, S.; Altieri, F.; Ferraro, A. Proteins of the PDI family: Unpredicted non-ER locations and functions. J. Cell Physiol. 2002, 193, 154–163. [Google Scholar] [CrossRef]
- Rovenich, H.; Boshoven, J.C.; Thomma, B.P. Filamentous pathogen effector functions: Of pathogens, hosts and microbiomes. Curr. Opin. Plant Biol. 2014, 20, 96–103. [Google Scholar] [CrossRef] [Green Version]
- Coaker, G.; Falick, A.; Staskawicz, B. Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 2005, 308, 548–550. [Google Scholar] [CrossRef] [Green Version]
- Coaker, G.; Zhu, G.; Ding, Z.; Van Doren, S.R.; Staskawicz, B. Eukaryotic cyclophilin as a molecular switch for effector activation. Mol. Microbiol. 2006, 61, 1485–1496. [Google Scholar] [CrossRef]
- Kong, G.; Zhao, Y.; Jing, M.; Huang, J.; Yang, J.; Xia, Y.; Kong, L.; Ye, W.; Xiong, Q.; Qiao, Y. The activation of Phytophthora effector Avr3b by plant cyclophilin is required for the nudix hydrolase activity of Avr3b. PLoS Pathog. 2015, 11, e1005139. [Google Scholar] [CrossRef] [PubMed] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madina, M.H.; Rahman, M.S.; Huang, X.; Zhang, Y.; Zheng, H.; Germain, H. A Poplar Rust Effector Protein Associates with Protein Disulfide Isomerase and Enhances Plant Susceptibility. Biology 2020, 9, 294. https://doi.org/10.3390/biology9090294
Madina MH, Rahman MS, Huang X, Zhang Y, Zheng H, Germain H. A Poplar Rust Effector Protein Associates with Protein Disulfide Isomerase and Enhances Plant Susceptibility. Biology. 2020; 9(9):294. https://doi.org/10.3390/biology9090294
Chicago/Turabian StyleMadina, Mst Hur, Md Saifur Rahman, Xiaoqiang Huang, Yang Zhang, Huanquan Zheng, and Hugo Germain. 2020. "A Poplar Rust Effector Protein Associates with Protein Disulfide Isomerase and Enhances Plant Susceptibility" Biology 9, no. 9: 294. https://doi.org/10.3390/biology9090294




