Cell Adhesion Molecules and Ubiquitination—Functions and Significance
Abstract
:1. Introduction
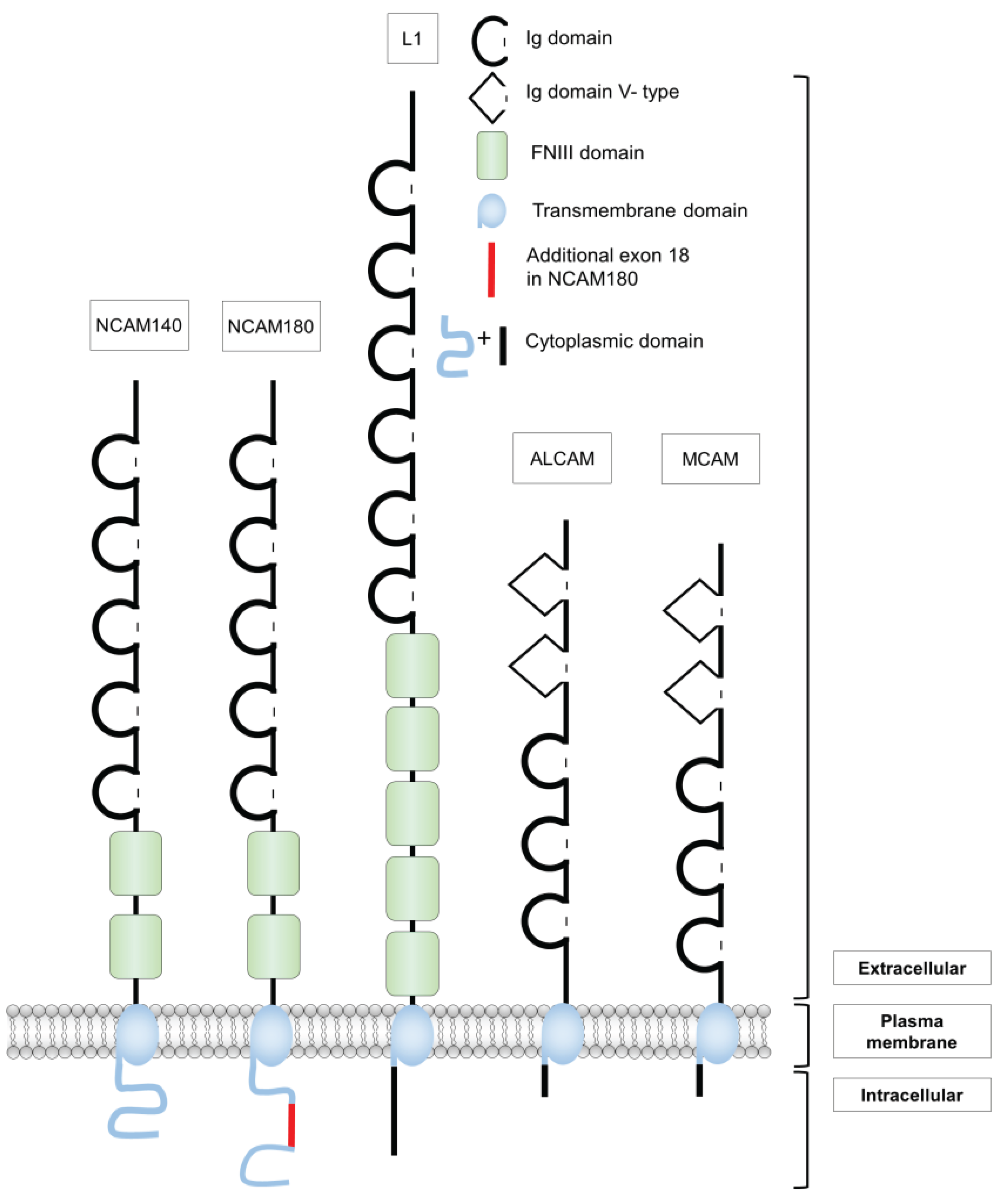
2. Structure and Functions of Different Cell Adhesion Molecules of the Ig Superfamily
2.1. The Neural Cell Adhesion Molecule NCAM
2.1.1. Expression and Functions
2.1.2. Cellular Mechanisms
2.2. The Cell Adhesion Molecule L1
2.2.1. Expression and Functions
2.2.2. Cellular Mechanisms
2.3. The Melanoma Cell Adhesion Molecule MCAM
2.3.1. Expression and Functions
2.3.2. Cellular Mechanisms
2.4. The Activated Leukocyte Cell Adhesion Molecule ALCAM
2.4.1. Expression and Functions
2.4.2. Cellular Mechanisms
| Cell Adhesion Molecule | Modification | References |
|---|---|---|
| NCAM | Polysialylation * (EC) | [306,307] |
| N-glycosylation (EC) | [97] | |
| Palmitoylation (IC) | [90] | |
| Phosphorylation (IC) | [91,92,93,94,95] | |
| Ufmylation (IC) | [96] | |
| L1 | N-glycosylation (EC) | [308,309,310,311] |
| Phosphorylation (IC) | [144,145,146,147,148,161,162,163,308,312] | |
| Sumoylation (IC) | [168] | |
| MCAM | N-glycosylation (EC) | [198] |
| Phosphorylation (IC) | [200] | |
| ALCAM | N-glycosylation (EC) | [243,244] |
| Cell Adhesion Molecule | Interaction Partners | References |
|---|---|---|
| NCAM | β-actin (IC) | [80] |
| α-actinin (IC) | [80] | |
| ADAM (EC) | [19,102,103] | |
| ATP (EC) | [73] | |
| Axonin-1/TAG-1 (EC) | [59] | |
| CKI (IC) | [93] | |
| EphA3 (EC) | [65,66] | |
| FGFR (EC) | [61,63,64] | |
| GAP-43 (IC) | [78] | |
| GFR-α1, GDNF (EC) | [67] | |
| GSK-3 (IC) | [93] | |
| Heparin, HSPGs, CSPGs (EC) | [68,69,70] | |
| Kinesin-1 (IC) | [81] | |
| L1 (EC) | [58] | |
| LANP (IC) | [79] | |
| NCAM (EC) | [313] | |
| MAP1A (IC) | [80] | |
| p59fyn (IC) | [86] | |
| PLC-γ (IC) | [79] | |
| PP1, PP2A (IC) | [79] | |
| PrP (EC) | [71,72] | |
| RhoA-binding kinase-α (IC) | [80] | |
| RPTPα (IC) | [87] | |
| Spectrin (IC) | [74] | |
| ST8SiaII, ST8SiaIV | [314] | |
| Syndapin (IC) | [79] | |
| Tropomyosin (IC) | [80] | |
| TOAD-64 (IC) | [79] | |
| TRKB (IC) | [95] | |
| α- and β-tubulin (IC) | [80] | |
| Ufc-1 (IC) | [96] | |
| L1 | ADAM (EC) | [134,179,181,183] |
| AP-2 (μ-subunit) (IC) | [178] | |
| ALCAM/DM-GRASP (EC) | [129] | |
| Ankyrin (IC) | [156] | |
| Axonin-1/TAG-1 (EC) | [130,131] | |
| CD24 (EC) | [128] | |
| CK II (IC) | [177] | |
| Doublecortin | [165] | |
| Erk2 | [146] | |
| ERM proteins (IC) | [141,142] | |
| F3/F11/contactin (EC) | [124] | |
| FGFR (EC) | [138] | |
| Integrins (EC) | [315] | |
| L1 (EC) | [123] | |
| Laminin (EC) | [125] | |
| NCAM (EC) | [58] | |
| Neurocan (EC) | [126] | |
| Neuropilin-1 (EC) | [136] | |
| P90rsk (IC) | [145] | |
| Phosphocan (EC) | [127] | |
| Rabex-5 (IC) | [154] | |
| (RanBPM) | [175] | |
| MCAM | Actin (IC) | [200] |
| ERM family | [232] | |
| Galectin-1 (EC) | [316] | |
| Laminin-411 (EC) | [317] | |
| MCAM (EC) | [224] | |
| Neurite outgrowth factor (EC) | [318] | |
| VEGFR2 (EC) | [319] | |
| Wnt5a (EC) | [320] | |
| ALCAM | ADAM17 (EC) | [247] |
| ALCAM (EC) | [276,321] | |
| CD6 (EC) | [292] | |
| L1 (EC) | [129] |
3. Structure and Function of Ubiquitin
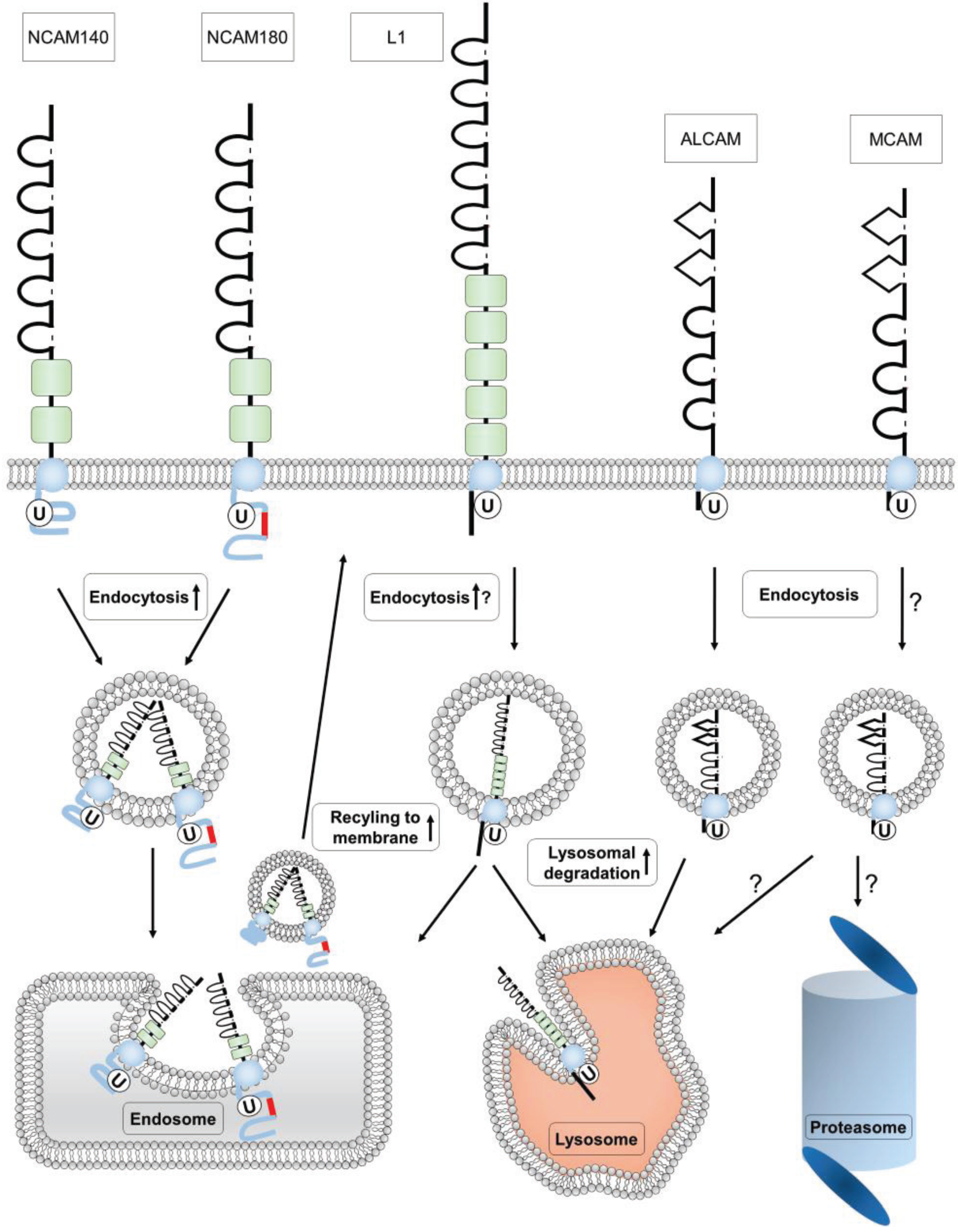
4. Ubiquitination of Ig Superfamily Members
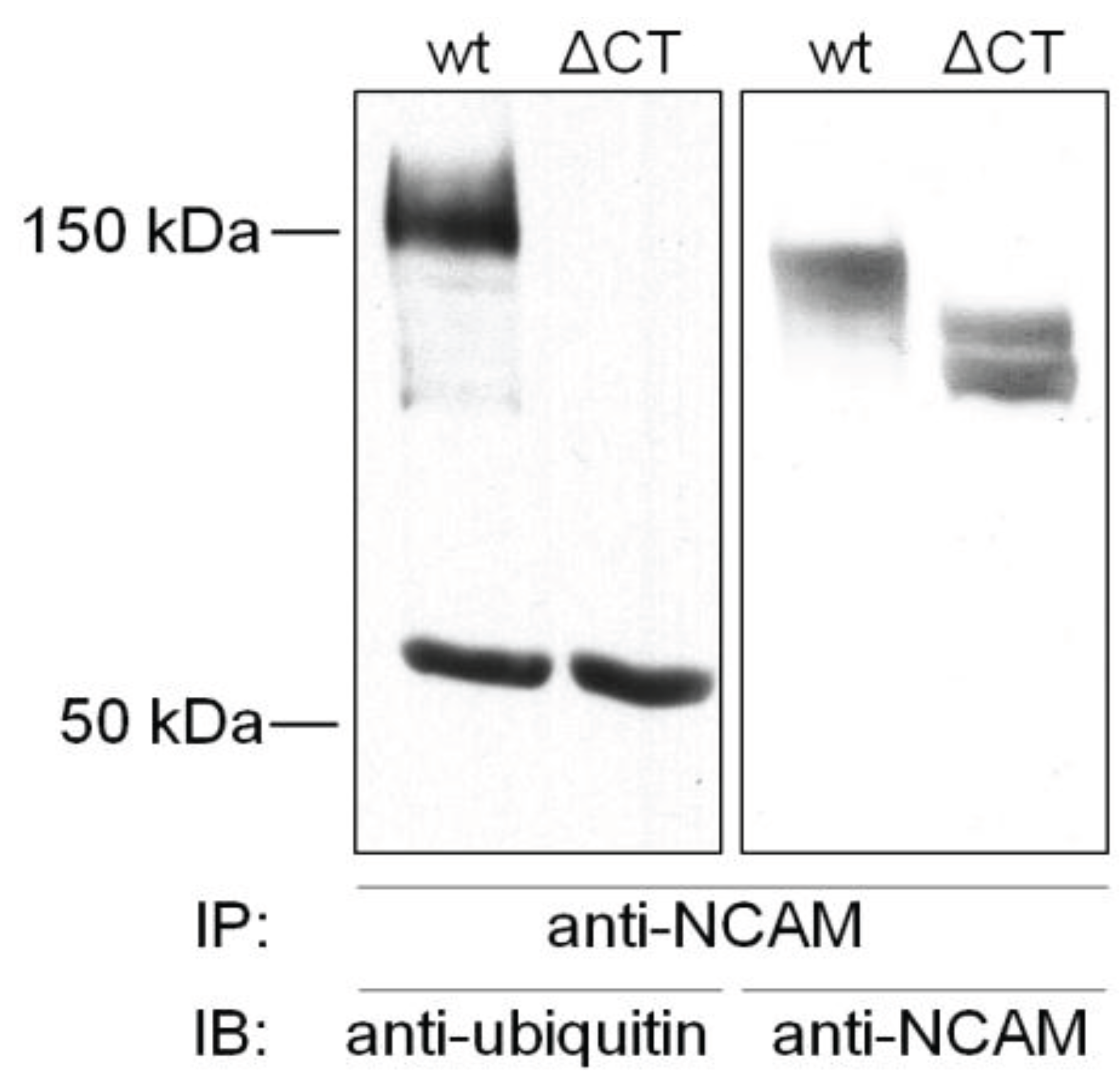
4.1. Ubiquitination of NCAM
4.2. Ubiquitination of L1
4.3. Ubiquitination of MCAM
4.4. Ubiquitination of ALCAM
5. Conclusions and Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Edelman, G.M. Cell adhesion molecules. Science 1983, 219, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Edelman, G.M.; Crossin, K.L. Cell adhesion molecules: Implications for a molecular histology. Annu. Rev. Biochem. 1991, 60, 155–190. [Google Scholar] [CrossRef] [PubMed]
- Takeichi, M. Cadherins: A molecular family important in selective cell-cell adhesion. Annu. Rev. Biochem. 1990, 59, 237–252. [Google Scholar] [CrossRef] [PubMed]
- Reichardt, L.F.; Tomaselli, K.J. Extracellular matrix molecules and their receptors: Functions in neural development. Annu. Rev. Neurosci. 1991, 14, 531–570. [Google Scholar] [CrossRef] [PubMed]
- Lasky, L.A. Selectin-carbohydrate interactions and the initiation of the inflammatory response. Annu. Rev. Biochem. 1995, 64, 113–139. [Google Scholar] [CrossRef] [PubMed]
- Brümmendorf, T.; Rathjen, F.G. Structure/function relationships of axon-associated adhesion receptors of the immunoglobulin superfamily. Curr. Opin. Neurobiol. 1996, 6, 584–593. [Google Scholar] [CrossRef]
- Gumbiner, B.M. Cell adhesion: The molecular basis of tissue architecture and morphogenesis. Cell 1996, 84, 345–357. [Google Scholar] [CrossRef]
- Murai-Takebe, R.; Noguchi, T.; Ogura, T.; Mikami, T.; Yanagi, K.; Inagaki, K.; Ohnishi, H.; Matozaki, T.; Kasuga, M. Ubiquitination-mediated regulation of biosynthesis of the adhesion receptor SHPS-1 in response to endoplasmic reticulum stress. J. Biol. Chem. 2004, 279, 11616–11625. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, O.S.; Bock, E. Brain specific synaptosomal membrane proteins demonstrated by crossed immunoelectrophoresis. J. Neurochem. 1974, 23, 879–880. [Google Scholar] [CrossRef] [PubMed]
- Rutishauser, U.; Thiery, J.P.; Brackenbury, R.; Sela, B.A.; Edelman, G.M. Mechanisms of adhesion among cells from neural tissues of the chick embryo. Proc. Natl. Acad. Sci. USA 1976, 73, 577–581. [Google Scholar] [CrossRef] [PubMed]
- Doherty, P.; Fruns, M.; Seaton, P.; Dickson, G.; Barton, C.H.; Sears, T.A.; Walsh, F.S. A threshold effect of the major isoforms of NCAM on neurite outgrowth. Nature 1990, 343, 464–466. [Google Scholar] [CrossRef] [PubMed]
- Cremer, H.; Lange, R.; Christoph, A.; Plomann, M.; Vopper, G.; Roes, J.; Brown, R.; Baldwin, S.; Kraemer, P.; Scheff, S.; et al. Inactivation of the N-CAM gene in mice results in size reduction of the olfactory bulb and deficits in spatial learning. Nature 1994, 367, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Cremer, H.; Chazal, G.; Goridis, C.; Represa, A. NCAM is essential for axonal growth and fasciculation in the hippocampus. Mol. Cell. Neurosci. 1997, 8, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Watanabe, M.; Rutishauser, U. Effect of polysialic acid on the behavior of retinal ganglion cell axons during growth into the optic tract and tectum. Development 1995, 121, 3439–3446. [Google Scholar] [PubMed]
- Müller, D.; Wang, C.; Skibo, G.; Toni, N.; Cremer, H.; Calaora, V.; Rougon, G.; Kiss, J.Z. PSA-NCAM is required for activity-induced synaptic plasticity. Neuron 1996, 17, 413–422. [Google Scholar] [CrossRef]
- Hu, H.; Tomasiewicz, H.; Magnuson, T.; Rutishauser, U. The role of polysialic acid in migration of olfactory bulb interneuron precursors in the subventricular zone. Neuron 1996, 16, 735–743. [Google Scholar] [CrossRef]
- Chazal, G.; Durbec, P.; Jankovski, A.; Rougon, G.; Cremer, H. Consequences of neural cell adhesion molecule deficiency on cell migration in the rostral migratory stream of the mouse. J. Neurosci. 2000, 20, 1446–1457. [Google Scholar] [PubMed]
- Diestel, S.; Hinkle, C.L.; Schmitz, B.; Maness, P.F. NCAM140 stimulates integrin-dependent cell migration by ectodomain shedding. J. Neurochem. 2005, 95, 1777–1784. [Google Scholar] [CrossRef] [PubMed]
- Hinkle, C.L.; Diestel, S.; Lieberman, J.; Maness, P.F. Metalloprotease-induced ectodomain shedding of neural cell adhesion molecule (NCAM). J. Neurobiol. 2006, 66, 1378–1395. [Google Scholar] [CrossRef] [PubMed]
- Poltorak, M.; Khoja, I.; Hemperly, J.J.; Williams, J.R.; El-Mallakh, R.; Freed, W.J. Disturbances in cell recognition molecules (N-CAM and L1 antigen) in the CSF of patients with schizophrenia. Exp. Neurol. 1995, 131, 266–272. [Google Scholar] [CrossRef]
- Honer, W.G.; Falkai, P.; Young, C.; Wang, T.; Xie, J.; Bonner, J.; Hu, L.; Boulianne, G.L.; Luo, Z.; Trimble, W.S. Cingulate cortex synaptic terminal proteins and neural cell adhesion molecule in schizophrenia. Neuroscience 1997, 78, 99–110. [Google Scholar] [CrossRef]
- Van Kammen, D.P.; Poltorak, M.; Kelley, M.E.; Yao, J.K.; Gurklis, J.A.; Peters, J.L.; Hemperly, J.J.; Wright, R.D.; Freed, W.J. Further studies of elevated cerebrospinal fluid neuronal cell adhesion molecule in schizophrenia. Biol. Psychiatry 1998, 43, 680–686. [Google Scholar] [CrossRef]
- Vawter, M.P.; Howard, A.L.; Hyde, T.M.; Kleinman, J.E.; Freed, W.J. Alterations of hippocampal secreted N-CAM in bipolar disorder and synaptophysin in schizophrenia. Mol. Psychiatry 1999, 4, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Vawter, M.P.; Frye, M.A.; Hemperly, J.J.; VanderPutten, D.M.; Usen, N.; Doherty, P.; Saffell, J.L.; Issa, F.; Post, R.M.; Wyatt, R.J.; et al. Elevated concentration of N-CAM VASE isoforms in schizophrenia. J. Psychiatr. Res. 2000, 34, 25–34. [Google Scholar] [CrossRef]
- Vawter, M.P.; Freed, W.J.; Kleinman, J.E. Neuropathology of bipolar disorder. Biol. Psychiatry 2000, 48, 486–504. [Google Scholar] [CrossRef]
- Panicker, A.K.; Buhusi, M.; Thelen, K.; Maness, P.F. Cellular signalling mechanisms of neural cell adhesion molecules. Front. Biosci. 2003, 8, d900–d911. [Google Scholar] [CrossRef] [PubMed]
- Pillai-Nair, N.; Panicker, A.K.; Rodriguiz, R.M.; Gilmore, K.L.; Demyanenko, G.P.; Huang, J.Z.; Wetsel, W.C.; Maness, P.F. Neural cell adhesion molecule-secreting transgenic mice display abnormalities in GABAergic interneurons and alterations in behavior. J. Neurosci. 2005, 25, 4659–4671. [Google Scholar] [CrossRef] [PubMed]
- Walmod, P.S.; Kolkova, K.; Berezin, V.; Bock, E. Zippers make signals: NCAM-mediated molecular interactions and signal transduction. Neurochem. Res. 2004, 29, 2015–2035. [Google Scholar] [CrossRef] [PubMed]
- Persohn, E.; Pollerberg, G.E.; Schachner, M. Immunoelectron-microscopic localization of the 180 kD component of the neural cell adhesion molecule N-CAM in postsynaptic membranes. J. Comp. Neurol. 1989, 288, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Dityatev, A.; Dityateva, G.; Schachner, M. Synaptic strength as a function of post- versus presynaptic expression of the neural cell adhesion molecule NCAM. Neuron 2000, 26, 207–217. [Google Scholar] [CrossRef]
- Noble, M.; Albrechtsen, M.; Møller, C.; Lyles, J.; Bock, E.; Goridis, C.; Watanabe, M.; Rutishauser, U. Glial cells express N-CAM/D2-CAM-like polypeptides in vitro. Nature 1985, 316, 725–728. [Google Scholar] [CrossRef] [PubMed]
- Andersson, A.M.; Olsen, M.; Zhernosekov, D.; Gaardsvoll, H.; Krog, L.; Linnemann, D.; Bock, E. Age-related changes in expression of the neural cell adhesion molecule in skeletal muscle: A comparative study of newborn, adult and aged rats. Biochem. J. 1993, 290, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Gaardsvoll, H.; Krog, L.; Zhernosekov, D.; Andersson, A.M.; Edvardsen, K.; Olsen, M.; Bock, E.; Linnemann, D. Age-related changes in expression of neural cell adhesion molecule (NCAM) in heart: A comparative study of newborn, adult and aged rats. Eur. J. Cell Biol. 1993, 61, 100–107. [Google Scholar] [PubMed]
- Langley, O.K.; Aletsee-Ufrecht, M.C.; Grant, N.J.; Gratzl, M. Expression of the neural cell adhesion molecule NCAM in endocrine cells. J. Histochem. Cytochem. 1989, 37, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Moller, C.J.; Christgau, S.; Williamson, M.R.; Madsen, O.D.; Niu, Z.P.; Bock, E.; Baekkeskov, S. Differential expression of neural cell adhesion molecule and cadherins in pancreatic islets, glucagonomas, and insulinomas. Mol. Endocrinol. 1992, 6, 1332–1342. [Google Scholar] [PubMed]
- Thor, G.; Probstmeier, R.; Schachner, M. Characterization of the cell adhesion molecules L1, N-CAM and J1 in the mouse intestine. EMBO J. 1987, 6, 2581–2586. [Google Scholar] [PubMed]
- Lanier, L.L.; Le, A.M.; Civin, C.I.; Loken, M.R.; Phillips, J.H. The relationship of CD16 (Leu-11) and Leu-19 (NKH-1) antigen expression on human peripheral blood NK cells and cytotoxic T lymphocytes. J. Immunol. 1986, 136, 4480–4486. [Google Scholar] [PubMed]
- Schmidt, R.E.; Murray, C.; Daley, J.F.; Schlossman, S.F.; Ritz, J. A subset of natural killer cells in peripheral blood displays a mature T cell phenotype. J. Exp. Med. 1986, 164, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Crossin, K.L.; Chuong, C.M.; Edelman, G.M. Expression sequences of cell adhesion molecules. Proc. Natl. Acad. Sci. USA 1985, 82, 6942–6946. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Yoshida, K.; Ikeda, E.; Asou, H.; Inaba, M.; Otani, M.; Kawase, T. Expression of the neural cell adhesion molecule in astrocytic tumors: An inverse correlation with malignancy. Cancer 1998, 82, 1921–1931. [Google Scholar] [CrossRef]
- Krushel, L.A.; Tai, M.H.; Cunningham, B.A.; Edelman, G.M.; Crossin, K.L. Neural cell adhesion molecule (N-CAM) domains and intracellular signaling pathways involved in the inhibition of astrocyte proliferation. Proc. Natl. Acad. Sci. USA 1998, 95, 2592–2596. [Google Scholar] [CrossRef] [PubMed]
- Glüer, S.; Zense, M.; Radtke, E.; von Schweinitz, D. Polysialylated neural cell adhesion molecule in childhood ganglioneuroma and neuroblastoma of different histological grade and clinical stage. Langenbecks Arch. Surg. 1998, 383, 340–344. [Google Scholar] [PubMed]
- Komminoth, P.; Roth, J.; Saremaslani, P.; Matias-Guiu, X.; Wolfe, H.J.; Heitz, P.U. Polysialic acid of the neural cell adhesion molecule in the human thyroid: A marker for medullary thyroid carcinoma and primary C-cell hyperplasia. An immunohistochemical study on 79 thyroid lesions. Am. J. Surg. Pathol. 1994, 18, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Lantuejoul, S.; Moro, D.; Michalides, R.J.; Brambilla, C.; Brambilla, E. Neural cell adhesion molecules (NCAM) and NCAM-PSA expression in neuroendocrine lung tumors. Am. J. Surg. Pathol. 1998, 22, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Trouillas, J.; Daniel, L.; Guigard, M.P.; Tong, S.; Gouvernet, J.; Jouanneau, E.; Jan, M.; Perrin, G.; Fischer, G.; Tabarin, A.; et al. Polysialylated neural cell adhesion molecules expressed in human pituitary tumors and related to extrasellar invasion. J. Neurosurg. 2003, 98, 1084–1093. [Google Scholar] [CrossRef] [PubMed]
- Zecchini, S.; Cavallaro, U. Neural cell adhesion molecule in cancer: Expression and mechanisms. Adv. Exp. Med. Biol. 2010, 663, 319–333. [Google Scholar] [PubMed]
- Zecchini, S.; Bombardelli, L.; Decio, A.; Bianchi, M.; Mazzarol, G.; Sanguineti, F.; Aletti, G.; Maddaluno, L.; Berezin, V.; Bock, E.; et al. The adhesion molecule NCAM promotes ovarian cancer progression via FGFR signalling. EMBO Mol. Med. 2011, 3, 480–494. [Google Scholar] [CrossRef] [PubMed]
- Ćirović, S.; Vještica, J.; Mueller, C.A.; Tatić, S.; Vasiljević, J.; Milenković, S.; Mueller, G.A.; Marković-Lipkovski, J. NCAM and FGFR1 coexpression and colocalization in renal tumors. Int. J. Clin. Exp. Pathol. 2014, 7, 1402–1414. [Google Scholar] [PubMed]
- Johnson, J.P. Cell adhesion molecules of the immunoglobulin supergene family and their role in malignant transformation and progression to metastatic disease. Cancer Metastasis Rev. 1991, 10, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, U.; Auerbach, B.; Oldenburg, M. The neural cell adhesion molecule NCAM in multiple myeloma. Leuk. Lymphoma 1996, 20, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, M.; Hirsch, M.R.; Deagostini-Bazin, H.; Yamada, O.; Tursz, T.; Goridis, C. Characterization of neural cell adhesion molecules (NCAM) expressed by ewing and neuroblastoma cell lines. Int. J. Cancer 1987, 40, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Moolenaar, C.E.; Pieneman, C.; Walsh, F.S.; Mooi, W.J.; Michalides, R.J. Alternative splicing of neural-cell-adhesion molecule mRNA in human small-cell lung-cancer cell line H69. Int. J. Cancer 1992, 51, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Roth, J.; Zuber, C.; Wagner, P.; Taatjes, D.J.; Weisgerber, C.; Heitz, P.U.; Goridis, C.; Bitter-Suermann, D. Reexpression of poly(sialic acid) units of the neural cell adhesion molecule in Wilms tumor. Proc. Natl. Acad. Sci. USA 1988, 85, 2999–3003. [Google Scholar] [CrossRef] [PubMed]
- Jimbo, T.; Nakayama, J.; Akahane, K.; Fukuda, M. Effect of polysialic acid on the tumor xenografts implanted into nude mice. Int. J. Cancer 2001, 94, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Seidenfaden, R.; Krauter, A.; Schertzinger, F.; Gerardy-Schahn, R.; Hildebrandt, H. Polysialic acid directs tumor cell growth by controlling heterophilic neural cell adhesion molecule interactions. Mol. Cell. Biol. 2003, 23, 5908–5918. [Google Scholar] [CrossRef] [PubMed]
- Kiselyov, V.V.; Soroka, V.; Berezin, V.; Bock, E. Structural biology of NCAM homophilic binding and activation of FGFR. J. Neurochem. 2005, 94, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Soroka, V.; Kiryushko, D.; Novitskaya, V.; Ronn, L.C.; Poulsen, F.M.; Holm, A.; Bock, E.; Berezin, V. Induction of neuronal differentiation by a peptide corresponding to the homophilic binding site of the second Ig module of the neural cell adhesion molecule. J. Biol. Chem. 2002, 277, 24676–24683. [Google Scholar] [CrossRef] [PubMed]
- Horstkorte, R.; Schachner, M.; Magyar, J.P.; Vorherr, T.; Schmitz, B. The fourth immunoglobulin-like domain of NCAM contains a carbohydrate recognition domain for oligomannosidic glycans implicated in association with L1 and neurite outgrowth. J. Cell Biol. 1993, 121, 1409–1421. [Google Scholar] [CrossRef] [PubMed]
- Milev, P.; Maurel, P.; Häring, M.; Margolis, R.K.; Margolis, R.U. TAG-1/axonin-1 is a high-affinity ligand of neurocan, phosphacan/protein-tyrosine phosphatase-zeta/beta, and N-CAM. J. Biol. Chem. 1996, 271, 15716–15723. [Google Scholar] [PubMed]
- Heiland, P.C.; Griffith, L.S.; Lange, R.; Schachner, M.; Hertlein, B.; Traub, O.; Schmitz, B. Tyrosine and serine phosphorylation of the neural cell adhesion molecule L1 is implicated in its oligomannosidic glycan dependent association with NCAM and neurite outgrowth. Eur. J. Cell Biol. 1998, 75, 97–106. [Google Scholar] [CrossRef]
- Cavallaro, U.; Niedermeyer, J.; Fuxa, M.; Christofori, G. N-CAM modulates tumour-cell adhesion to matrix by inducing FGF-receptor signalling. Nat. Cell Biol. 2001, 3, 650–657. [Google Scholar] [CrossRef] [PubMed]
- Francavilla, C.; Loeffler, S.; Piccini, D.; Kren, A.; Christofori, G.; Cavallaro, U. Neural cell adhesion molecule regulates the cellular response to fibroblast growth factor. J. Cell Sci. 2007, 120, 4388–4394. [Google Scholar] [CrossRef] [PubMed]
- Kiselyov, V.V.; Skladchikova, G.; Hinsby, A.M.; Jensen, P.H.; Kulahin, N.; Soroka, V.; Pedersen, N.; Tsetlin, V.; Poulsen, F.M.; Berezin, V.; et al. Structural basis for a direct interaction between FGFR1 and NCAM and evidence for a regulatory role of ATP. Structure 2003, 11, 691–701. [Google Scholar] [CrossRef]
- Francavilla, C.; Cattaneo, P.; Berezin, V.; Bock, E.; Ami, D.; de Marco, A.; Christofori, G.; Cavallaro, U. The binding of NCAM to FGFR1 induces a specific cellular response mediated by receptor trafficking. J. Cell Biol. 2009, 187, 1101–1116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brennaman, L.H.; Zhang, X.; Guan, H.; Triplett, J.W.; Brown, A.; Demyanenko, G.P.; Manis, P.B.; Landmesser, L.; Maness, P.F. Polysialylated NCAM and EphrinA/EphA regulate synaptic development of GABAergic interneurons in prefrontal cortex. Cereb. Cortex 2013, 23, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Brennaman, L.H.; Moss, M.L.; Maness, P.F. EphrinA/EphA-induced ectodomain shedding of neural cell adhesion molecule regulates growth cone repulsion through ADAM10 metalloprotease. J. Neurochem. 2014, 128, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Paratcha, G.; Ledda, F.; Ibáñez, C.F. The neural cell adhesion molecule NCAM is an alternative signaling receptor for GDNF family ligands. Cell 2003, 113, 867–879. [Google Scholar] [CrossRef]
- Cole, G.J.; Schubert, D.; Glaser, L. Cell-substratum adhesion in chick neural retina depends upon protein-heparan sulfate interactions. J. Cell Biol. 1985, 100, 1192–1199. [Google Scholar] [CrossRef] [PubMed]
- Burg, M.A.; Halfter, W.; Cole, G.J. Analysis of proteoglycan expression in developing chicken brain: Characterization of a heparan sulfate proteoglycan that interacts with the neural cell adhesion molecule. J. Neurosci. Res. 1995, 41, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Herndon, M.E.; Stipp, C.S.; Lander, A.D. Interactions of neural glycosaminoglycans and proteoglycans with protein ligands: Assessment of selectivity, heterogeneity and the participation of core proteins in binding. Glycobiology 1999, 9, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Santuccione, A.; Sytnyk, V.; Leshchyns’ka, I.; Schachner, M. Prion protein recruits its neuronal receptor NCAM to lipid rafts to activate p59fyn and to enhance neurite outgrowth. J. Cell Biol. 2005, 169, 341–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prodromidou, K.; Papastefanaki, F.; Sklaviadis, T.; Matsas, R. Functional cross-talk between the cellular prion protein and the neural cell adhesion molecule NCAM is critical for neuronal differentiation of neural stem/precursor cells. Stem Cells 2014, 32, 1674–1687. [Google Scholar] [CrossRef] [PubMed]
- Dzhandzhugazyan, K.; Bock, E. Demonstration of an extracellular ATP-binding site in NCAM: Functional implications of nucleotide binding. Biochemistry 1997, 36, 15381–15395. [Google Scholar] [CrossRef] [PubMed]
- Pollerberg, G.E.; Schachner, M.; Davoust, J. Differentiation state-dependent surface mobilities of two forms of the neural cell adhesion molecule. Nature 1986, 324, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Leshchyns’ka, I.; Sytnyk, V.; Morrow, J.S.; Schachner, M. Neural cell adhesion molecule (NCAM) association with PKCbeta2 via betaI spectrin is implicated in NCAM-mediated neurite outgrowth. J. Cell Biol. 2003, 161, 625–639. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.M.; Ron, D.; Touhara, K.; Chen, C.H.; Mochly-Rosen, D. RACK1, a protein kinase C anchoring protein, coordinates the binding of activated protein kinase C and select pleckstrin homology domains in vitro. Biochemistry 1999, 38, 13787–13794. [Google Scholar] [CrossRef] [PubMed]
- Kolkova, K.; Novitskaya, V.; Pedersen, N.; Berezin, V.; Bock, E. Neural cell adhesion molecule-stimulated neurite outgrowth depends on activation of protein kinase C and the Ras-mitogen-activated protein kinase pathway. J. Neurosci. 2000, 20, 2238–2246. [Google Scholar] [PubMed]
- He, Q.; Meiri, K.F. Isolation and characterization of detergent-resistant microdomains responsive to NCAM-mediated signaling from growth cones. Mol. Cell. Neurosci. 2002, 19, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Büttner, B.; Kannicht, C.; Reutter, W.; Horstkorte, R. Novel cytosolic binding partners of the neural cell adhesion molecule: Mapping the binding domains of PLCγ, LANP, TOAD-64, Syndapin, PP1, and PP2A. Biochemistry 2005, 44, 6938–6947. [Google Scholar] [CrossRef] [PubMed]
- Büttner, B.; Kannicht, C.; Reutter, W.; Horstkorte, R. The neural cell adhesion molecule is associated with major components of the cytoskeleton. Biochem. Biophys. Res. Commun. 2003, 310, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Wobst, H.; Schmitz, B.; Schachner, M.; Diestel, S.; Leshchyns’ka, I.; Sytnyk, V. Kinesin-1 promotes post-Golgi trafficking of NCAM140 and NCAM180 to the cell surface. J. Cell Sci. 2015, 128, 2816–2829. [Google Scholar] [CrossRef] [PubMed]
- Schuch, U.; Lohse, M.J.; Schachner, M. Neural cell adhesion molecules influence second messenger systems. Neuron 1989, 3, 13–20. [Google Scholar] [CrossRef]
- Doherty, P.; Ashton, S.V.; Moore, S.E.; Walsh, F.S. Morphoregulatory activities of NCAM and N-cadherin can be accounted for by G protein-dependent activation of L- and N-type neuronal Ca2+ channels. Cell 1991, 67, 21–33. [Google Scholar] [CrossRef]
- Frei, T.; von Bohlen und Halbach, F.; Wille, W.; Schachner, M. Different extracellular domains of the neural cell adhesion molecule (N-CAM) are involved in different functions. J. Cell Biol. 1992, 118, 177–194. [Google Scholar] [CrossRef] [PubMed]
- Jessen, U.; Novitskaya, V.; Pedersen, N.; Serup, P.; Berezin, V.; Bock, E. The transcription factors CREB and c-Fos play key roles in NCAM-mediated neuritogenesis in PC12-E2 cells. J. Neurochem. 2001, 79, 1149–1160. [Google Scholar] [CrossRef] [PubMed]
- Beggs, H.E.; Baragona, S.C.; Hemperly, J.J.; Maness, P.F. NCAM140 interacts with the focal adhesion kinase p125(fak) and the SRC-related tyrosine kinase p59(fyn). J. Biol. Chem. 1997, 272, 8310–8319. [Google Scholar] [CrossRef] [PubMed]
- Bodrikov, V.; Leshchyns’ka, I.; Sytnyk, V.; Overvoorde, J.; den Hertog, J.; Schachner, M. RPTPalpha is essential for NCAM-mediated p59fyn activation and neurite elongation. J. Cell Biol. 2005, 168, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Schmid, R.S.; Graff, R.D.; Schaller, M.D.; Chen, S.; Schachner, M.; Hemperly, J.J.; Maness, P.F. NCAM stimulates the Ras-MAPK pathway and CREB phosphorylation in neuronal cells. J. Neurobiol. 1999, 38, 542–558. [Google Scholar] [CrossRef]
- Niethammer, P.; Delling, M.; Sytnyk, V.; Dityatev, A.; Fukami, K.; Schachner, M. Cosignaling of NCAM via lipid rafts and the FGF receptor is required for neuritogenesis. J. Cell Biol. 2002, 157, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Little, E.B.; Edelman, G.M.; Cunningham, B.A. Palmitoylation of the cytoplasmic domain of the neural cell adhesion molecule N-CAM serves as an anchor to cellular membranes. Cell Adhes. Commun. 1998, 6, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Sorkin, B.C.; Hoffman, S.; Edelman, G.M.; Cunningham, B.A. Sulfation and phosphorylation of the neural cell adhesion molecule, N-CAM. Science 1984, 225, 1476–1478. [Google Scholar] [CrossRef] [PubMed]
- Pollscheit, J.; Glaubitz, N.; Haller, H.; Horstkorte, R.; Bork, K. Phosphorylation of serine 774 of the neural cell adhesion molecule is necessary for cyclic adenosine monophosphate response element binding protein activation and neurite outgrowth. J. Neurosci. Res. 2012, 90, 1577–1582. [Google Scholar] [CrossRef] [PubMed]
- Mackie, K.; Sorkin, B.C.; Nairn, A.C.; Greengard, P.; Edelman, G.M.; Cunningham, B.A. Identification of two protein kinases that phosphorylate the neural cell-adhesion molecule, N-CAM. J. Neurosci. 1989, 9, 1883–1896. [Google Scholar] [PubMed]
- Diestel, S.; Laurini, C.; Traub, O.; Schmitz, B. Tyrosine 734 of NCAM180 interferes with FGF receptor-dependent signaling implicated in neurite growth. Biochem. Biophys. Res. Commun. 2004, 322, 186–196. [Google Scholar]
- Cassens, C.; Kleene, R.; Xiao, M.F.; Friedrich, C.; Dityateva, G.; Schafer-Nielsen, C.; Schachner, M. Binding of the receptor tyrosine kinase TrkB to the neural cell adhesion molecule (NCAM) regulates phosphorylation of NCAM and NCAM-dependent neurite outgrowth. J. Biol. Chem. 2010, 285, 28959–28967. [Google Scholar] [CrossRef] [PubMed]
- Homrich, M.; Wobst, H.; Laurini, C.; Sabrowski, J.; Schmitz, B.; Diestel, S. Cytoplasmic domain of NCAM140 interacts with ubiquitin-fold modifier-conjugating enzyme-1 (Ufc1). Exp. Cell Res. 2014, 324, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Albach, C.; Damoc, E.; Denzinger, T.; Schachner, M.; Przybylski, M.; Schmitz, B. Identification of N-glycosylation sites of the murine neural cell adhesion molecule NCAM by MALDI-TOF and MALDI-FTICR mass spectrometry. Anal. Bioanal. Chem. 2004, 378, 1129–1135. [Google Scholar] [CrossRef] [PubMed]
- Rutishauser, U. Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat. Rev. Neurosci. 2008, 9, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Bonfanti, L.; Theodosis, D.T. Polysialic acid and activity-dependent synapse remodeling. Cell Adhes. Migr. 2009, 3, 43–50. [Google Scholar] [CrossRef]
- Rutishauser, U.; Landmesser, L. Polysialic acid in the vertebrate nervous system: A promoter of plasticity in cell-cell interactions. Trends Neurosci. 1996, 19, 422–427. [Google Scholar] [CrossRef]
- Storms, S.D.; Rutishauser, U. A role for polysialic acid in neural cell adhesion molecule heterophilic binding to proteoglycans. J. Biol. Chem. 1998, 273, 27124–27129. [Google Scholar] [CrossRef] [PubMed]
- Hübschmann, M.V.; Skladchikova, G.; Bock, E.; Berezin, V. Neural cell adhesion molecule function is regulated by metalloproteinase-mediated ectodomain release. J. Neurosci. Res. 2005, 80, 826–837. [Google Scholar] [CrossRef] [PubMed]
- Kalus, I.; Bormann, U.; Mzoughi, M.; Schachner, M.; Kleene, R. Proteolytic cleavage of the neural cell adhesion molecule by ADAM17/TACE is involved in neurite outgrowth. J. Neurochem. 2006, 98, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Kleene, R.; Mzoughi, M.; Joshi, G.; Kalus, I.; Bormann, U.; Schulze, C.; Xiao, M.F.; Dityatev, A.; Schachner, M. NCAM-induced neurite outgrowth depends on binding of calmodulin to NCAM and on nuclear import of NCAM and fak fragments. J. Neurosci. 2010, 30, 10784–10798. [Google Scholar] [CrossRef] [PubMed]
- Rathjen, F.G.; Schachner, M. Immunocytological and biochemical characterization of a new neuronal cell surface component (L1 antigen) which is involved in cell adhesion. EMBO J. 1984, 3, 1–10. [Google Scholar] [PubMed]
- Kamiguchi, H.; Long, K.E.; Pendergast, M.; Schaefer, A.W.; Rapoport, I.; Kirchhausen, T.; Lemmon, V. The neural cell adhesion molecule L1 interacts with the AP-2 adaptor and is endocytosed via the clathrin-mediated pathway. J. Neurosci. 1998, 18, 5311–5321. [Google Scholar] [PubMed]
- Jacob, J.; Haspel, J.; Kane-Goldsmith, N.; Grumet, M. L1 mediated homophilic binding and neurite outgrowth are modulated by alternative splicing of exon 2. J. Neurobiol. 2002, 51, 177–189. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, E.; Watkins, A.; Schäfer, M.; Brümmendorf, T.; Kenwrick, S. Disease-associated mutations in L1 CAM interfere with ligand interactions and cell-surface expression. Hum. Mol. Genet. 2002, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nolte, C.; Moos, M.; Schachner, M. Immunolocalization of the neural cell adhesion molecule L1 in epithelia of rodents. Cell Tissue Res. 1999, 298, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Balaian, L.B.; Moehler, T.; Montgomery, A.M. The human neural cell adhesion molecule L1 functions as a costimulatory molecule in T cell activation. Eur. J. Immunol. 2000, 30, 938–943. [Google Scholar] [CrossRef]
- Fields, R.D.; Itoh, K. Neural cell adhesion molecules in activity-dependent development and synaptic plasticity. Trends Neurosci. 1996, 19, 473–480. [Google Scholar] [CrossRef]
- Dihné, M.; Bernreuther, C.; Sibbe, M.; Paulus, W.; Schachner, M. A new role for the cell adhesion molecule L1 in neural precursor cell proliferation, differentiation, and transmitter-specific subtype generation. J. Neurosci. 2003, 23, 6638–6650. [Google Scholar] [PubMed]
- Roonprapunt, C.; Huang, W.; Grill, R.; Friedlander, D.; Grumet, M.; Chen, S.; Schachner, M.; Young, W. Soluble cell adhesion molecule L1-Fc promotes locomotor recovery in rats after spinal cord injury. J. Neurotrauma 2003, 20, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Lutz, D.; Kataria, H.; Kleene, R.; Loers, G.; Chaudhary, H.; Guseva, D.; Wu, B.; Jakovcevski, I.; Schachner, M. Myelin basic protein cleaves cell adhesion molecule L1 and improves regeneration after injury. Mol. Neurobiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Jouet, M.; Moncla, A.; Paterson, J.; McKeown, C.; Fryer, A.; Carpenter, N.; Holmberg, E.; Wadelius, C.; Kenwrick, S. New domains of neural cell-adhesion molecule L1 implicated in X-linked hydrocephalus and MASA syndrome. Am. J. Hum. Genet. 1995, 56, 1304–1314. [Google Scholar] [PubMed]
- Fransen, E.; van Camp, G.; Vits, L.; Willems, P.J. L1-associated diseases: Clinical geneticists divide, molecular geneticists unite. Hum. Mol. Genet. 1997, 6, 1625–1632. [Google Scholar] [CrossRef] [PubMed]
- Weller, S.; Gärtner, J. Genetic and clinical aspects of X-linked hydrocephalus (L1 disease): Mutations in theL1CAM gene. Hum. Mutat. 2001, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kanemura, Y.; Okamoto, N.; Sakamoto, H.; Shofuda, T.; Kamiguchi, H.; Yamasaki, M. Molecular mechanisms and neuroimaging criteria for severe L1 syndrome with X-linked hydrocephalus. J. Neurosurg. 2006, 105, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, M.K.; Altevogt, P. L1CAM malfunction in the nervous system and human carcinomas. Cell. Mol. Life Sci. 2010, 67, 2425–2437. [Google Scholar] [CrossRef] [PubMed]
- Fogel, M.; Gutwein, P.; Mechtersheimer, S.; Riedle, S.; Stoeck, A.; Smirnov, A.; Edler, L.; Ben-Arie, A.; Huszar, M.; Altevogt, P. L1 expression as a predictor of progression and survival in patients with uterine and ovarian carcinomas. Lancet 2003, 362, 869–875. [Google Scholar] [CrossRef]
- Altevogt, P.; Doberstein, K.; Fogel, M. L1CAM in human cancer. Int. J. Cancer 2015. [Google Scholar] [CrossRef] [PubMed]
- Kenwrick, S.; Watkins, A.; de Angelis, E. Neural cell recognition molecule L1: Relating biological complexity to human disease mutations. Hum. Mol. Genet. 2000, 9, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, V.; Farr, K.L.; Lagenaur, C. L1-mediated axon outgrowth occurs via a homophilic binding mechanism. Neuron 1989, 2, 1597–1603. [Google Scholar] [CrossRef]
- Brümmendorf, T.; Hubert, M.; Treubert, U.; Leuschner, R.; Tárnok, A.; Rathjen, F.G. The axonal recognition molecule F11 is a multifunctional protein: Specific domains mediate interactions with Ng-CAM and restrictin. Neuron 1993, 10, 711–727. [Google Scholar] [CrossRef]
- Grumet, M.; Flaccus, A.; Margolis, R.U. Functional characterization of chondroitin sulfate proteoglycans of brain: Interactions with neurons and neural cell adhesion molecules. J. Cell Biol. 1993, 120, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, D.R.; Milev, P.; Karthikeyan, L.; Margolis, R.K.; Margolis, R.U.; Grumet, M. The neuronal chondroitin sulfate proteoglycan neurocan binds to the neural cell adhesion molecules Ng-CAM/L1/NILE and N-CAM, and inhibits neuronal adhesion and neurite outgrowth. J. Cell Biol. 1994, 125, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Milev, P.; Friedlander, D.R.; Sakurai, T.; Karthikeyan, L.; Flad, M.; Margolis, R.K.; Grumet, M.; Margolis, R.U. Interactions of the chondroitin sulfate proteoglycan phosphacan, the extracellular domain of a receptor-type protein tyrosine phosphatase, with neurons, glia, and neural cell adhesion molecules. J. Cell Biol. 1994, 127, 1703–1715. [Google Scholar] [CrossRef] [PubMed]
- Kadmon, G.; Imhof, B.A.; Altevogt, P.; Schachner, M. Adhesive hierarchy involving the cell adhesion molecules L1, CD24, and alpha 6 integrin in murine neuroblastoma N2A cells. Biochem. Biophys. Res. Commun. 1995, 214, 94–101. [Google Scholar] [CrossRef] [PubMed]
- DeBernardo, A.P.; Chang, S. Heterophilic interactions of DM-GRASP: GRASP-NgCAM interactions involved in neurite extension. J. Cell Biol. 1996, 133, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Buchstaller, A.; Kunz, S.; Berger, P.; Kunz, B.; Ziegler, U.; Rader, C.; Sonderegger, P. Cell adhesion molecules NgCAM and axonin-1 form heterodimers in the neuronal membrane and cooperate in neurite outgrowth promotion. J. Cell Biol. 1996, 135, 1593–1607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rader, C.; Kunz, B.; Lierheimer, R.; Giger, R.J.; Berger, P.; Tittmann, P.; Gross, H.; Sonderegger, P. Implications for the domain arrangement of axonin-1 derived from the mapping of its NgCAM binding site. EMBO J. 1996, 15, 2056–2068. [Google Scholar] [PubMed]
- Oleszewski, M.; Beer, S.; Katich, S.; Geiger, C.; Zeller, Y.; Rauch, U.; Altevogt, P. Integrin and neurocan binding to L1 involves distinct Ig domains. J. Biol. Chem. 1999, 274, 24602–24610. [Google Scholar] [CrossRef] [PubMed]
- Oleszewski, M.; Gutwein, P.; von der Lieth, W.; Rauch, U.; Altevogt, P. Characterization of the L1-neurocan-binding site. Implications for L1-L1 homophilic binding. J. Biol. Chem. 2000, 275, 34478–34485. [Google Scholar] [CrossRef] [PubMed]
- Mechtersheimer, S.; Gutwein, P.; Agmon-Levin, N.; Stoeck, A.; Oleszewski, M.; Riedle, S.; Postina, R.; Fahrenholz, F.; Fogel, M.; Lemmon, V.; et al. Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J. Cell Biol. 2001, 155, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Castellani, V.; Chédotal, A.; Schachner, M.; Faivre-Sarrailh, C.; Rougon, G. Analysis of the L1-deficient mouse phenotype reveals cross-talk between Sema3A and L1 signaling pathways in axonal guidance. Neuron 2000, 27, 237–249. [Google Scholar] [CrossRef]
- Castellani, V.; de Angelis, E.; Kenwrick, S.; Rougon, G. Cis and trans interactions of L1 with neuropilin-1 control axonal responses to semaphorin 3A. EMBO J. 2002, 21, 6348–6357. [Google Scholar] [CrossRef] [PubMed]
- Thelen, K.; Kedar, V.; Panicker, A.K.; Schmid, R.S.; Midkiff, B.R.; Maness, P.F. The neural cell adhesion molecule L1 potentiates integrin-dependent cell migration to extracellular matrix proteins. J. Neurosci. 2002, 22, 4918–4931. [Google Scholar] [PubMed]
- Kulahin, N.; Li, S.; Hinsby, A.; Kiselyov, V.; Berezin, V.; Bock, E. Fibronectin type III (FN3) modules of the neuronal cell adhesion molecule L1 interact directly with the fibroblast growth factor (FGF) receptor. Mol. Cell. Neurosci. 2008, 37, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Kadmon, G.; Kowitz, A.; Altevogt, P.; Schachner, M. The neural cell adhesion molecule N-CAM enhances L1-dependent cell-cell interactions. J. Cell Biol. 1990, 110, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Simon, H.; Klinz, S.; Fahrig, T.; Schachner, M. Molecular association of the neural adhesion molecules L1 and N-CAM in the surface membrane of neuroblastoma cells is shown by chemical cross-linking. Eur. J. Neurosci. 1991, 3, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Dickson, T.C.; Mintz, C.D.; Benson, D.L.; Salton, S.R. Functional binding interaction identified between the axonal CAM L1 and members of the ERM family. J. Cell Biol. 2002, 157, 1105–1112. [Google Scholar] [CrossRef] [PubMed]
- Dahlin-Huppe, K.; Berglund, E.O.; Ranscht, B.; Stallcup, W.B. Mutational analysis of the L1 neuronal cell adhesion molecule identifies membrane-proximal amino acids of the cytoplasmic domain that are required for cytoskeletal anchorage. Mol. Cell. Neurosci. 1997, 9, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Itoh, K.; Lemmon, V. L1-mediated branching is regulated by two ezrin-radixin-moesin (ERM)-binding sites, the RSLE region and a novel juxtamembrane ERM-binding region. J. Neurosci. 2005, 25, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, T.; Gil, O.D.; Whittard, J.D.; Gazdoiu, M.; Joseph, T.; Wu, J.; Waksman, A.; Benson, D.L.; Salton, S.R.; Felsenfeld, D.P. Interactions between the L1 cell adhesion molecule and ezrin support traction-force generation and can be regulated by tyrosine phosphorylation. J. Neurosci. Res. 2008, 86, 2602–2614. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.V.; Schaefer, A.W.; Landreth, G.; Lemmon, V. Involvement of p90rsk in neurite outgrowth mediated by the cell adhesion molecule L1. J. Biol. Chem. 1996, 271, 18217–18223. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, A.W.; Kamiguchi, H.; Wong, E.V.; Beach, C.M.; Landreth, G.; Lemmon, V. Activation of the MAPK signal cascade by the neural cell adhesion molecule L1 requires L1 internalization. J. Biol. Chem. 1999, 274, 37965–37973. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, A.W.; Kamei, Y.; Kamiguchi, H.; Wong, E.V.; Rapoport, I.; Kirchhausen, T.; Beach, C.M.; Landreth, G.; Lemmon, S.K.; Lemmon, V. L1 endocytosis is controlled by a phosphorylation-dephosphorylation cycle stimulated by outside-in signaling by L1. J. Cell Biol. 2002, 157, 1223–1232. [Google Scholar] [CrossRef] [PubMed]
- Schultheis, M.; Diestel, S.; Schmitz, B. The role of cytoplasmic serine residues of the cell adhesion molecule L1 in neurite outgrowth, endocytosis, and cell migration. Cell. Mol. Neurobiol. 2007, 27, 11–31. [Google Scholar] [CrossRef] [PubMed]
- Kamiguchi, H.; Lemmon, V. Recycling of the cell adhesion molecule L1 in axonal growth cones. J. Neurosci. 2000, 20, 3676–3686. [Google Scholar] [PubMed]
- Long, K.E.; Asou, H.; Snider, M.D.; Lemmon, V. The role of endocytosis in regulating L1-mediated adhesion. J. Biol. Chem. 2001, 276, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Kamiguchi, H.; Yoshihara, F. The role of endocytic l1 trafficking in polarized adhesion and migration of nerve growth cones. J. Neurosci. 2001, 21, 9194–9203. [Google Scholar] [PubMed]
- Yap, C.C.; Wisco, D.; Kujala, P.; Lasiecka, Z.M.; Cannon, J.T.; Chang, M.C.; Hirling, H.; Klumperman, J.; Winckler, B. The somatodendritic endosomal regulator NEEP21 facilitates axonal targeting of L1/NgCAM. J. Cell Biol. 2008, 180, 827–842. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, H.; Lippé, R.; McBride, H.M.; Rubino, M.; Woodman, P.; Stenmark, H.; Rybin, V.; Wilm, M.; Ashman, K.; Mann, M.; et al. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell 1997, 90, 1149–1159. [Google Scholar] [CrossRef]
- Aikawa, Y. Rabex-5 Protein Regulates the Endocytic Trafficking Pathway of Ubiquitinated Neural Cell Adhesion Molecule L1. J. Biol. Chem. 2012, 287, 32312–32323. [Google Scholar] [CrossRef] [PubMed]
- Bennett, V.; Chen, L. Ankyrins and cellular targeting of diverse membrane proteins to physiological sites. Curr. Opin. Cell Biol. 2001, 13, 61–67. [Google Scholar] [CrossRef]
- Davis, J.Q.; Bennett, V. Ankyrin binding activity shared by the neurofascin/L1/NrCAM family of nervous system cell adhesion molecules. J. Biol. Chem. 1994, 269, 27163–27166. [Google Scholar] [PubMed]
- Bennett, V.; Baines, A.J. Spectrin and ankyrin-based pathways: Metazoan inventions for integrating cells into tissues. Physiol. Rev. 2001, 81, 1353–1392. [Google Scholar] [PubMed]
- Hortsch, M.; Nagaraj, K.; Godenschwege, T.A. The interaction between L1-type proteins and ankyrins—A master switch for L1-type CAM function. Cell. Mol. Biol. Lett. 2009, 14, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Yoshihara, F.; Tojima, T.; Ooashi, N.; Yoon, W.; Mikoshiba, K.; Bennett, V.; Kamiguchi, H. L1-dependent neuritogenesis involves ankyrinB that mediates L1-CAM coupling with retrograde actin flow. J. Cell Biol. 2003, 163, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Gil, O.D.; Sakurai, T.; Bradley, A.E.; Fink, M.Y.; Cassella, M.R.; Kuo, J.A.; Felsenfeld, D.P. Ankyrin binding mediates L1CAM interactions with static components of the cytoskeleton and inhibits retrograde movement of L1CAM on the cell surface. J. Cell Biol. 2003, 162, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Garver, T.D.; Ren, Q.; Tuvia, S.; Bennett, V. Tyrosine phosphorylation at a site highly conserved in the L1 family of cell adhesion molecules abolishes ankyrin binding and increases lateral mobility of neurofascin. J. Cell Biol. 1997, 137, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Tuvia, S.; Garver, T.D.; Bennett, V. The phosphorylation state of the FIGQY tyrosine of neurofascin determines ankyrin-binding activity and patterns of cell segregation. Proc. Natl. Acad. Sci. USA 1997, 94, 12957–12962. [Google Scholar] [CrossRef] [PubMed]
- Whittard, J.D.; Sakurai, T.; Cassella, M.R.; Gazdoiu, M.; Felsenfeld, D.P. MAP kinase pathway-dependent phosphorylation of the L1-CAM ankyrin binding site regulates neuronal growth. Mol. Biol. Cell 2006, 17, 2696–2706. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Maness, P.F. Perisomatic GABAergic Innervation in Prefrontal Cortex Is Regulated by Ankyrin Interaction with the L1 Cell Adhesion Molecule. Cereb. Cortex 2010, 20, 2684–2693. [Google Scholar] [CrossRef] [PubMed]
- Kizhatil, K.; Wu, Y.X.; Sen, A.; Bennett, V. A new activity of doublecortin in recognition of the phospho-FIGQY tyrosine in the cytoplasmic domain of neurofascin. J. Neurosci. 2002, 22, 7948–7958. [Google Scholar] [PubMed]
- Silletti, S.; Yebra, M.; Perez, B.; Cirulli, V.; McMahon, M.; Montgomery, A.M. Extracellular signal-regulated kinase (ERK)-dependent gene expression contributes to L1 cell adhesion molecule-dependent motility and invasion. J. Biol. Chem. 2004, 279, 28880–28888. [Google Scholar] [CrossRef] [PubMed]
- Schmid, R.S.; Midkiff, B.R.; Kedar, V.P.; Maness, P.F. Adhesion molecule L1 stimulates neuronal migration through Vav2-Pak1 signaling. Neuroreport 2004, 15, 2791–2794. [Google Scholar] [PubMed]
- Lutz, D.; Wolters-Eisfeld, G.; Joshi, G.; Djogo, N.; Jakovcevski, I.; Schachner, M.; Kleene, R. Generation and nuclear translocation of sumoylated transmembrane fragment of cell adhesion molecule L1. J. Biol. Chem. 2012, 287, 17161–17175. [Google Scholar] [CrossRef] [PubMed]
- Hall, H.; Williams, E.J.; Moore, S.E.; Walsh, F.S.; Prochiantz, A.; Doherty, P. Inhibition of FGF-stimulated phosphatidylinositol hydrolysis and neurite outgrowth by a cell-membrane permeable phosphopeptide. Curr. Biol. 1996, 6, 580–587. [Google Scholar] [CrossRef]
- Doherty, P.; Walsh, F.S. CAM-FGF Receptor Interactions: A Model for Axonal Growth. Mol. Cell. Neurosci. 1996, 8, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Brittis, P.A.; Silver, J.; Walsh, F.S.; Doherty, P. Fibroblast growth factor receptor function is required for the orderly projection of ganglion cell axons in the developing mammalian retina. Mol. Cell. Neurosci. 1996, 8, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Saffell, J.L.; Williams, E.J.; Mason, I.J.; Walsh, F.S.; Doherty, P. Expression of a dominant negative FGF receptor inhibits axonal growth and FGF receptor phosphorylation stimulated by CAMs. Neuron 1997, 18, 231–242. [Google Scholar] [CrossRef]
- Walsh, F.S.; Doherty, P. Neural cell adhesion molecules of the immunoglobulin superfamily: Role in axon growth and guidance. Annu. Rev. Cell Dev. Biol. 1997, 13, 425–456. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Masuda, H.; Horii, J.; Kuma, K.; Yokoyama, N.; Ohba, T.; Nishitani, H.; Miyata, T.; Tanaka, M.; Nishimoto, T. When overexpressed, a novel centrosomal protein, RanBPM, causes ectopic microtubule nucleation similar to gamma-tubulin. J. Cell Biol. 1998, 143, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Lemmon, S.; Lemmon, V. RanBPM is an L1-interacting protein that regulates L1-mediated mitogen-activated protein kinase activation. J. Neurochem. 2005, 94, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
- Bechara, A.; Nawabi, H.; Moret, F.; Yaron, A.; Weaver, E.; Bozon, M.; Abouzid, K.; Guan, J.L.; Tessier-Lavigne, M.; Lemmon, V.; et al. FAK-MAPK-dependent adhesion disassembly downstream of L1 contributes to semaphorin3A-induced collapse. EMBO J. 2008, 27, 1549–1562. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.V.; Schaefer, A.W.; Landreth, G.; Lemmon, V. Casein kinase II phosphorylates the neural cell adhesion molecule L1. J. Neurochem. 1996, 66, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Kamiguchi, H.; Lemmon, V. A neuronal form of the cell adhesion molecule L1 contains a tyrosine-based signal required for sorting to the axonal growth cone. J. Neurosci. 1998, 18, 3749–3756. [Google Scholar] [PubMed]
- Beer, S.; Oleszewski, M.; Gutwein, P.; Geiger, C.; Altevogt, P. Metalloproteinase-mediated release of the ectodomain of L1 adhesion molecule. J. Cell Sci. 1999, 112, 2667–2675. [Google Scholar] [PubMed]
- Gutwein, P.; Oleszewski, M.; Mechtersheimer, S.; Agmon-Levin, N.; Krauss, K.; Altevogt, P. Role of Src kinases in the ADAM-mediated release of L1 adhesion molecule from human tumor cells. J. Biol. Chem. 2000, 275, 15490–15497. [Google Scholar] [CrossRef] [PubMed]
- Gutwein, P.; Mechtersheimer, S.; Riedle, S.; Stoeck, A.; Gast, D.; Joumaa, S.; Zentgraf, H.; Fogel, M.; Altevogt, D.P. ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. FASEB J. 2003, 17, 292–294. [Google Scholar] [CrossRef] [PubMed]
- Kalus, I.; Schnegelsberg, B.; Seidah, N.G.; Kleene, R.; Schachner, M. The proprotein convertase PC5A and a metalloprotease are involved in the proteolytic processing of the neural adhesion molecule L1. J. Biol. Chem. 2003, 278, 10381–10388. [Google Scholar] [CrossRef] [PubMed]
- Maretzky, T.; Schulte, M.; Ludwig, A.; Rose-John, S.; Blobel, C.; Hartmann, D.; Altevogt, P.; Saftig, P.; Reiss, K. L1 Is Sequentially processed by two differently activated metalloproteases and presenilin/-secretase and regulates neural cell adhesion, cell migration, and neurite outgrowth. Mol. Cell. Biol. 2005, 25, 9040–9053. [Google Scholar] [CrossRef] [PubMed]
- Riedle, S.; Kiefel, H.; Gast, D.; Bondong, S.; Wolterink, S.; Gutwein, P.; Altevogt, P. Nuclear translocation and signalling of L1-CAM in human carcinoma cells requires ADAM10 and presenilin/γ-secretase activity. Biochem. J. 2009, 420, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Lutz, D.; Loers, G.; Kleene, R.; Oezen, I.; Kataria, H.; Katagihallimath, N.; Braren, I.; Harauz, G.; Schachner, M. Myelin basic protein cleaves cell adhesion molecule L1 and promotes neuritogenesis and cell survival. J. Biol. Chem. 2014, 289, 13503–13518. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, J.M.; Holzmann, B.; Breitbart, E.W.; Schmiegelow, P.; Riethmüller, G.; Johnson, J.P. Discrimination between benign and malignant cells of melanocytic lineage by two novel antigens, a glycoprotein with a molecular weight of 113,000 and a protein with a molecular weight of 76,000. Cancer Res. 1987, 47, 841–845. [Google Scholar] [PubMed]
- Johnson, J.P.; Rummel, M.M.; Rothbächer, U.; Sers, C. MUC18: A cell adhesion molecule with a potential role in tumor growth and tumor cell dissemination. Curr. Top. Microbiol. Immunol. 1996, 213, 95–105. [Google Scholar] [PubMed]
- Shih, I.M. The role of CD146 (Mel-CAM) in biology and pathology. J. Pathol. 1999, 189, 4–11. [Google Scholar] [CrossRef]
- Shih, I.M.; Speicher, D.; Hsu, M.Y.; Levine, E.; Herlyn, M. Melanoma cell-cell interactions are mediated through heterophilic Mel-CAM/ligand adhesion. Cancer Res. 1997, 57, 3835–3840. [Google Scholar] [PubMed]
- Sorrentino, A.; Ferracin, M.; Castelli, G.; Biffoni, M.; Tomaselli, G.; Baiocchi, M.; Fatica, A.; Negrini, M.; Peschle, C.; Valtieri, M. Isolation and characterization of CD146+ multipotent mesenchymal stromal cells. Exp. Hematol. 2008, 36, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, W.; Kapila, Y.; Lotz, J.; Kapila, S. Multiple differentiation capacity of STRO-1+/CD146+ PDL mesenchymal progenitor cells. Stem Cells Dev. 2009, 18, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Lv, F.J.; Tuan, R.S.; Cheung, K.M.; Leung, V.Y. Concise review: The surface markers and identity of human mesenchymal stem cells. Stem Cells 2014, 32, 1408–1419. [Google Scholar] [CrossRef] [PubMed]
- Gruenloh, W.; Kambal, A.; Sondergaard, C.; McGee, J.; Nacey, C.; Kalomoiris, S.; Pepper, K.; Olson, S.; Fierro, F.; Nolta, J.A. Characterization and in vivo testing of mesenchymal stem cells derived from human embryonic stem cells. Tissue Eng. A 2011, 17, 1517–1525. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yan, X. CD146, a multi-functional molecule beyond adhesion. Cancer Lett. 2013, 330, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.J.; Wu, M.W.; Wang, S.W.; Liu, Z.; Qu, P.; Peng, Q.; Yang, H.; Varma, V.A.; Sun, Q.C.; Petros, J.A.; et al. Isolation and characterization of the major form of human MUC18 cDNA gene and correlation of MUC18 over-expression in prostate cancer cell lines and tissues with malignant progression. Gene 2001, 279, 17–31. [Google Scholar] [CrossRef]
- Shih, I.; Wang, T.; Wu, T.; Kurman, R.J.; Gearhart, J.D. Expression of Mel-CAM in implantation site intermediate trophoblastic cell line, IST-1, limits its migration on uterine smooth muscle cells. J. Cell Sci. 1998, 111, 2655–2664. [Google Scholar] [PubMed]
- Lehmann, J.M.; Riethmüller, G.; Johnson, J.P. MUC18, a marker of tumor progression in human melanoma, shows sequence similarity to the neural cell adhesion molecules of the immunoglobulin superfamily. Proc. Natl. Acad. Sci. USA 1989, 86, 9891–9895. [Google Scholar] [CrossRef] [PubMed]
- Vainio, O.; Dunon, D.; Aïssi, F.; Dangy, J.P.; McNagny, K.M.; Imhof, B.A. HEMCAM, an adhesion molecule expressed by c-kit+ hemopoietic progenitors. J. Cell Biol. 1996, 135, 1655–1668. [Google Scholar] [CrossRef] [PubMed]
- Kohama, K.; Tsukamoto, Y.; Furuya, M.; Okamura, K.; Tanaka, H.; Miki, N.; Taira, E. Molecular cloning and analysis of the mouse gicerin gene. Neurochem. Int. 2005, 46, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Guezguez, B.; Vigneron, P.; Lamerant, N.; Kieda, C.; Jaffredo, T.; Dunon, D. Dual role of melanoma cell adhesion molecule (MCAM)/CD146 in lymphocyte endothelium interaction: MCAM/CD146 promotes rolling via microvilli induction in lymphocyte and is an endothelial adhesion receptor. J. Immunol. 2007, 179, 6673–6685. [Google Scholar] [CrossRef] [PubMed]
- Taira, E.; Nagino, T.; Taniura, H.; Takaha, N.; Kim, C.H.; Kuo, C.H.; Li, B.S.; Higuchi, H.; Miki, N. Expression and functional analysis of a novel isoform of gicerin, an immunoglobulin superfamily cell adhesion molecule. J. Biol. Chem. 1995, 270, 28681–28687. [Google Scholar] [CrossRef] [PubMed]
- Bardin, N.; Reumaux, D.; Geboes, K.; Colombel, J.F.; Blot-Chabaud, M.; Sampol, J.; Duthilleul, P.; Dignat-George, F. Increased expression of CD146, a new marker of the endothelial junction in active inflammatory bowel disease. Inflamm. Bowel Dis. 2006, 12, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Bardin, N.; Blot-Chabaud, M.; Despoix, N.; Kebir, A.; Harhouri, K.; Arsanto, J.P.; Espinosa, L.; Perrin, P.; Robert, S.; Vely, F.; et al. CD146 and its soluble form regulate monocyte transendothelial migration. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 746–753. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, M.J.; Müller, N.; Körschenhausen, D.; Kirsch, K.H.; Penning, R.; Ackenheil, M.; Johnson, J.P.; Hampel, H. Melanoma-associated adhesion molecule MUC18/MCAM (CD146) and transcriptional regulator mader in normal human CNS. Neuroimmunomodulation 1998, 5, 270–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, S.; Taniura, H.; Taira, E.; Miki, N. Involvement of a receptor for neurite outgrowth factor (NOFR) in cerebellar neurogenesis. Neurosci. Lett. 1992, 140, 78–80. [Google Scholar] [CrossRef]
- Hiroi, S.; Taira, E.; Ogawa, K.; Tsukamoto, Y. Neurite extension of DRG neurons by gicerin expression is enhanced by nerve growth factor. Int. J. Mol. Med. 2005, 16, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Hiroi, S.; Tsukamoto, Y.; Sasaki, F.; Miki, N.; Taira, E. Involvement of gicerin, a cell adhesion molecule, in development and regeneration of chick sciatic nerve. FEBS Lett. 2003, 554, 311–314. [Google Scholar] [CrossRef]
- Bardin, N.; Moal, V.; Anfosso, F.; Daniel, L.; Brunet, P.; Sampol, J.; Dignat George, F. Soluble CD146, a novel endothelial marker, is increased in physiopathological settings linked to endothelial junctional alteration. Thromb. Haemost. 2003, 90, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Daniel, L.; Bardin, N.; Moal, V.; Dignat-George, F.; Berland, Y.; Figarella-Branger, D. Tubular CD146 expression in nephropathies is related to chronic renal failure. Nephron Exp. Nephrol. 2005, 99, e105–e111. [Google Scholar] [CrossRef] [PubMed]
- Małyszko, J.; Małyszko, J.S.; Brzosko, S.; Wołczynski, S.; Myśliwiec, M. Markers of endothelial cell activation/injury: CD146 and thrombomodulin are related to adiponectin in kidney allograft recipients. Am. J. Nephrol. 2005, 25, 203–210. [Google Scholar] [PubMed]
- Małyszko, J.; Małyszko, J.S.; Brzosko, S.; Wołczynski, S.; Myśliwiec, M. Adiponectin is related to CD146, a novel marker of endothelial cell activation/injury in chronic renal failure and peritoneally dialyzed patients. J. Clin. Endocrinol. Metab. 2004, 89, 4620–4627. [Google Scholar] [CrossRef] [PubMed]
- Şeftalioğlu, A.; Karakoç, L. Expression of CD146 adhesion molecules (MUC18 or MCAM) in the thymic microenvironment. Acta Histochem. 2000, 102, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, Y.; Matsumoto, T.; Kotani, T.; Taira, E.; Takaha, N.; Miki, N.; Yamate, J.; Sakuma, S. The expression of gicerin, a cell adhesion molecule, in regenerating process of collecting ducts and ureters of the chicken kidney following infection with a nephrotropic strain of infectious bronchitis virus. Avian Pathol. 1997, 26, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, Y.; Taira, E.; Nakane, Y.; Tsudzuki, M.; Kohama, K.; Amin, H.; Miki, N.; Sasaki, F. Expression of gicerin, a cell adhesion molecule, in the abnormal retina in silver plumage color mutation of Japanese quail (Coturnix japonica). Neurosci. Lett. 1999, 266, 53–56. [Google Scholar] [CrossRef]
- Holzmann, B.; Bröcker, E.B.; Lehmann, J.M.; Ruiter, D.J.; Sorg, C.; Riethmüller, G.; Johnson, J.P. Tumor progression in human malignant melanoma: Five stages defined by their antigenic phenotypes. Int. J. Cancer 1987, 39, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Sers, C.; Kirsch, K.; Rothbächer, U.; Riethmüller, G.; Johnson, J.P. Genomic organization of the melanoma-associated glycoprotein MUC18: Implications for the evolution of the immunoglobulin domains. Proc. Natl. Acad. Sci. USA 1993, 90, 8514–8518. [Google Scholar] [CrossRef] [PubMed]
- Melnikova, V.O.; Bar-Eli, M. Bioimmunotherapy for melanoma using fully human antibodies targeting MCAM/MUC18 and IL-8. Pigment Cell Res. 2006, 19, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Kraus, A.; Masat, L.; Johnson, J.P. Analysis of the expression of intercellular adhesion molecule-1 and MUC18 on benign and malignant melanocytic lesions using monoclonal antibodies directed against distinct epitopes and recognizing denatured, non-glycosylated antigen. Melanoma Res. 1997, 7, S75–S81. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Chen, X.; Xu, Y.; Qiao, Y.; Zhang, X.; Wang, Y.; Guan, Y.; Sun, F.; Wang, J. CD166 positively regulates MCAM via inhibition to ubiquitin E3 ligases Smurf1 and βTrCP through PI3K/AKT and c-Raf/MEK/ERK signaling in Bel-7402 hepatocellular carcinoma cells. Cell. Signal. 2015, 27, 1694–1702. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.J.; Wu, M.W.; Wang, C.; Liu, Y. Enforced expression of METCAM/MUC18 increases tumorigenesis of human prostate cancer LNCaP cells in nude mice. J. Urol. 2011, 185, 1504–1512. [Google Scholar] [CrossRef] [PubMed]
- Satyamoorthy, K.; Muyrers, J.; Meier, F.; Patel, D.; Herlyn, M. Mel-CAM-specific genetic suppressor elements inhibit melanoma growth and invasion through loss of gap junctional communication. Oncogene 2001, 20, 4676–4684. [Google Scholar] [CrossRef] [PubMed]
- Mills, L.; Tellez, C.; Huang, S.; Baker, C.; McCarty, M.; Green, L.; Gudas, J.M.; Feng, X.; Bar-Eli, M. Fully human antibodies to MCAM/MUC18 inhibit tumor growth and metastasis of human melanoma. Cancer Res. 2002, 62, 5106–5114. [Google Scholar] [PubMed]
- Dagur, P.K.; McCoy, J.P. Endothelial-binding, proinflammatory T cells identified by MCAM (CD146) expression: Characterization and role in human autoimmune diseases. Autoimmun. Rev. 2015, 14, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Taira, E.; Takaha, N.; Taniura, H.; Kim, C.H.; Miki, N. Molecular Cloning and Functional Expression of Gicerin, a Novel Cell Adhesion Molecule That Binds to Neurite Outgrowth Factor. Neuron 1994, 12, 861–872. [Google Scholar] [CrossRef]
- Taira, E.; Nagino, T.; Tsukamoto, Y.; Okumura, S.; Muraoka, O.; Sakuma, F.; Miki, N. Cytoplasmic domain is not essential for the cell adhesion activities of gicerin, an Ig-superfamily molecule. Exp. Cell Res. 1999, 253, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Taira, E.; Kohama, K.; Tsukamoto, Y.; Okumura, S.; Miki, N. Gicerin/CD146 is involved in neurite extension of NGF-treated PC12 cells. J. Cell. Physiol. 2005, 204, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Taira, E.; Kohama, K.; Tsukamoto, Y.; Okumura, S.; Miki, N. Characterization of Gicerin/MUC18/CD146 in the rat nervous system. J. Cell. Physiol. 2004, 198, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Taira, E.; Nagino, T.; Tsukamoto, Y.; Ding, Y.; Sakuma, S.; Miki, N. Neurite promotion from ciliary ganglion neurons by gicerin. Neurochem. Int. 1998, 32, 23–29. [Google Scholar] [CrossRef]
- Bardin, N.; Francès, V.; Lesaule, G.; Horschowski, N.; George, F.; Sampol, J. Identification of the S-Endo 1 endothelial-associated antigen. Biochem. Biophys. Res. Commun. 1996, 218, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.P.; Bar-Eli, M.; Jansen, B.; Markhof, E. Melanoma progression-associated glycoprotein MUC18/MCAM mediates homotypic cell adhesion through interaction with a heterophilic ligand. Int. J. Cancer 1997, 73, 769–774. [Google Scholar] [CrossRef]
- Bardin, N.; Anfosso, F.; Massé, J.M.; Cramer, E.; Sabatier, F.; Le Bivic, A.; Sampol, J.; Dignat-George, F. Identification of CD146 as a component of the endothelial junction involved in the control of cell-cell cohesion. Blood 2001, 98, 3677–3684. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zheng, C.; Zhang, J.; Lu, D.; Zhuang, J.; Xing, S.; Feng, J.; Yang, D.; Yan, X. Recognition of CD146 as an ERM-binding protein offers novel mechanisms for melanoma cell migration. Oncogene 2012, 31, 306–321. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Kalabis, J.; Xu, X.; Meier, F.; Oka, M.; Bogenrieder, T.; Herlyn, M. Reciprocal regulation of MelCAM and AKT in human melanoma. Oncogene 2003, 22, 6891–6899. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Simpson, S.E.; Scialla, T.J.; Bagg, A.; Carroll, M. Survival of acute myeloid leukemia cells requires PI3 kinase activation. Blood 2003, 102, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Anfosso, F.; Bardin, N.; Frances, V.; Vivier, E.; Camoin-Jau, L.; Sampol, J.; Dignat-George, F. Activation of human endothelial cells via S-Endo-1 antigen (CD146) stimulates the tyrosine phosphorylation of focal adhesion kinase p125FAK. J. Biol. Chem. 1998, 273, 26852–26856. [Google Scholar] [CrossRef] [PubMed]
- Anfosso, F.; Bardin, N.; Vivier, E.; Sabatier, F.; Sampol, J.; Dignat-George, F. Outside-in signaling pathway linked to CD146 engagement in human endothelial cells. J. Biol. Chem. 2001, 276, 1564–1569. [Google Scholar] [CrossRef] [PubMed]
- Bu, P.; Zhuang, J.; Feng, J.; Yang, D.; Shen, X.; Yan, X. Visualization of CD146 dimerization and its regulation in living cells. Biochim. Biophys. Acta Mol. Cell Res. 2007, 1773, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Bu, P.; Gao, L.; Zhuang, J.; Feng, J.; Yang, D.; Yan, X. Anti-CD146 monoclonal antibody AA98 inhibits angiogenesis via suppression of nuclear factor-kappaB activation. Mol. Cancer Ther. 2006, 5, 2872–2878. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tang, X.; Weng, W.; Qiao, Y.; Lin, J.; Liu, W.; Liu, R.; Ma, L.; Yu, W.; Yu, Y.; et al. The membrane protein melanoma cell adhesion molecule (MCAM) is a novel tumor marker that stimulates tumorigenesis in hepatocellular carcinoma. Oncogene 2015, 34, 5781–5795. [Google Scholar] [CrossRef] [PubMed]
- Burns, F.R.; von Kannen, S.; Guy, L.; Raper, J.A.; Kamholz, J.; Chang, S. DM-GRASP, a novel immunoglobulin superfamily axonal surface protein that supports neurite extension. Neuron 1991, 7, 209–220. [Google Scholar] [CrossRef]
- Tanaka, H.; Obata, K. Developmental changes in unique cell surface antigens of chick embryo spinal motoneurons and ganglion cells. Dev. Biol. 1984, 106, 26–37. [Google Scholar] [CrossRef]
- Pourquié, O.; Coltey, M.; Thomas, J.L.; le Douarin, N.M. A widely distributed antigen developmentally regulated in the nervous system. Development 1990, 109, 743–752. [Google Scholar] [PubMed]
- Bowen, M.A.; Patel, D.D.; Li, X.; Modrell, B.; Malacko, A.R.; Wang, W.C.; Marquardt, H.; Neubauer, M.; Pesando, J.M.; Francke, U.; et al. Cloning, mapping, and characterization of activated leukocyte-cell adhesion molecule (ALCAM), a CD6 ligand. J. Exp. Med. 1995, 181, 2213–2220. [Google Scholar] [CrossRef] [PubMed]
- Denzinger, T.; Diekmann, H.; Bruns, K.; Laessing, U.; Stuermer, C.A.; Przybylski, M. Isolation, primary structure characterization and identification of the glycosylation pattern of recombinant goldfish neurolin, a neuronal cell adhesion protein. J. Mass Spectrom. 1999, 34, 435–446. [Google Scholar] [CrossRef]
- Swart, G.W. Activated leukocyte cell adhesion molecule (CD166/ALCAM): Developmental and mechanistic aspects of cell clustering and cell migration. Eur. J. Cell Biol. 2002, 81, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Quertermous, T. Molecular isolation and characterization of a soluble isoform of activated leukocyte cell adhesion molecule that modulates endothelial cell function. J. Biol. Chem. 2004, 279, 55315–55323. [Google Scholar] [CrossRef] [PubMed]
- Bech-Serra, J.J.; Santiago-Josefat, B.; Esselens, C.; Saftig, P.; Baselga, J.; Arribas, J.; Canals, F. Proteomic identification of desmoglein 2 and activated leukocyte cell adhesion molecule as substrates of ADAM17 and ADAM10 by difference gel electrophoresis. Mol. Cell. Biol. 2006, 26, 5086–5095. [Google Scholar] [CrossRef] [PubMed]
- Rosso, O.; Piazza, T.; Bongarzone, I.; Rossello, A.; Mezzanzanica, D.; Canevari, S.; Orengo, A.M.; Puppo, A.; Ferrini, S.; Fabbi, M. The ALCAM Shedding by the Metalloprotease ADAM17/TACE Is Involved in Motility of Ovarian Carcinoma Cells. Mol. Cancer Res. 2007, 5, 1246–1253. [Google Scholar] [CrossRef] [PubMed]
- Pourquié, O.; Corbel, C.; le Caer, J.P.; Rossier, J.; le Douarin, N.M. BEN, a surface glycoprotein of the immunoglobulin superfamily, is expressed in a variety of developing systems. Proc. Natl. Acad. Sci. USA 1992, 89, 5261–5265. [Google Scholar] [CrossRef] [PubMed]
- Diekmann, H.; Stuermer, C.A. Zebrafish neurolin-a and -b, orthologs of ALCAM, are involved in retinal ganglion cell differentiation and retinal axon pathfinding. J. Comp. Neurol. 2009, 513, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Fraboulet, S.; Schmidt-Petri, T.; Dhouailly, D.; Pourquié, O. Expression of DM-GRASP/BEN in the developing mouse spinal cord and various epithelia. Mech. Dev. 2000, 95, 221–224. [Google Scholar] [CrossRef]
- Hirata, H.; Murakami, Y.; Miyamoto, Y.; Tosaka, M.; Inoue, K.; Nagahashi, A.; Jakt, L.M.; Asahara, T.; Iwata, H.; Sawa, Y.; et al. ALCAM (CD166) is a surface marker for early murine cardiomyocytes. Cells Tissues Organs 2006, 184, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Swart, G.W.; Lunter, P.C.; Kilsdonk, J.W.; Kempen, L.C. Activated leukocyte cell adhesion molecule (ALCAM/CD166): Signaling at the divide of melanoma cell clustering and cell migration? Cancer Metastasis Rev. 2005, 24, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.G.; Swart, G.W.; Zijlstra, A. ALCAM: Basis Sequence: Mouse. AFCS Nat. Mol. Pages 2011. [Google Scholar] [CrossRef] [PubMed]
- Prat-Vidal, C.; Roura, S.; Farré, J.; Gálvez, C.; Llach, A.; Molina, C.E.; Hove-Madsen, L.; Garcia, J.; Cinca, J.; Bayes-Genis, A. Umbilical cord blood-derived stem cells spontaneously express cardiomyogenic traits. Transplant. Proc. 2007, 39, 2434–2437. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Akiyama, Y.; Tai, S.; Maruyama, K.; Kawaguchi, Y.; Muramatsu, K.; Yamaguchi, K. Changes in the expression of CD106, osteogenic genes, and transcription factors involved in the osteogenic differentiation of human bone marrow mesenchymal stem cells. J. Bone Miner. Metab. 2008, 26, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, R.; Griparic, L.; Vargas, V.; Burgee, K.; Santacruz, P.; Anderson, R.; Schiewe, M.; Silva, F.; Patel, A. A putative mesenchymal stem cells population isolated from adult human testes. Biochem. Biophys. Res. Commun. 2009, 385, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Hua, J.; Yu, H.; Dong, W.; Yang, C.; Gao, Z.; Lei, A.; Sun, Y.; Pan, S.; Wu, Y.; Dou, Z. Characterization of mesenchymal stem cells (MSCs) from human fetal lung: Potential differentiation of germ cells. Tissue Cell 2009, 41, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Risbud, M.V.; Guttapalli, A.; Tsai, T.T.; Lee, J.Y.; Danielson, K.G.; Vaccaro, A.R.; Albert, T.J.; Gazit, Z.; Gazit, D.; Shapiro, I.M. Evidence for skeletal progenitor cells in the degenerate human intervertebral disc. Spine 2007, 32, 2537–2544. [Google Scholar] [CrossRef] [PubMed]
- Karaöz, E.; Doğan, B.N.; Aksoy, A.; Gacar, G.; Akyüz, S.; Ayhan, S.; Genç, Z.S.; Yürüker, S.; Duruksu, G.; Demircan, P.C.; et al. Isolation and in vitro characterisation of dental pulp stem cells from natal teeth. Histochem. Cell Biol. 2010, 133, 95–112. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Kobayashi, M.; Wang, J.; Habelhah, H.; Okada, F.; Hamada, J.; Moriuchi, T.; Totsuka, Y.; Hosokawa, M. Activated leukocyte cell adhesion molecule (ALCAM) and annexin II are involved in the metastatic progression of tumor cells after chemotherapy with Adriamycin. Clin. Exp. Metastasis 2000, 18, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Lunter, P.C.; van Kilsdonk, J.W.; van Beek, H.; Cornelissen, I.M.; Bergers, M.; Willems, P.H.; van Muijen, G.N.; Swart, G.W. Activated leukocyte cell adhesion molecule (ALCAM/CD166/MEMD), a novel actor in invasive growth, controls matrix metalloproteinase activity. Cancer Res. 2005, 65, 8801–8808. [Google Scholar] [CrossRef] [PubMed]
- Van Kempen, L.C.; Meier, F.; Egeblad, M.; Kersten-Niessen, M.J.; Garbe, C.; Weidle, U.H.; van Muijen, G.N.; Herlyn, M.; Bloemers, H.P.; Swart, G.W. Truncation of activated leukocyte cell adhesion molecule: A gateway to melanoma metastasis. J. Investig. Dermatol. 2004, 122, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Ofori-Acquah, S.F.; King, J.A. Activated leukocyte cell adhesion molecule: A new paradox in cancer. Transl. Res. 2008, 151, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, G.; Pilarsky, C.; Wissmann, C.; Kaiser, S.; Bruemmendorf, T.; Roepcke, S.; Dahl, E.; Hinzmann, B.; Specht, T.; Pervan, J.; et al. Expression profiling of microdissected matched prostate cancer samples reveals CD166/MEMD and CD24 as new prognostic markers for patient survival. J. Pathol. 2005, 205, 359–376. [Google Scholar] [CrossRef] [PubMed]
- Weichert, W.; Knösel, T.; Bellach, J.; Dietel, M.; Kristiansen, G. ALCAM/CD166 is overexpressed in colorectal carcinoma and correlates with shortened patient survival. J. Clin. Pathol. 2004, 57, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Davies, S.; Jiang, W.G. ALCAM, activated leukocyte cell adhesion molecule, influences the aggressive nature of breast cancer cells, a potential connection to bone metastasis. Anticancer Res. 2010, 30, 1163–1168. [Google Scholar] [PubMed]
- Davies, S.R.; Dent, C.; Watkins, G.; King, J.A.; Mokbel, K.; Jiang, W.G. Expression of the cell to cell adhesion molecule, ALCAM, in breast cancer patients and the potential link with skeletal metastasis. Oncol. Rep. 2008, 19, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Ihnen, M.; Köhler, N.; Kersten, J.F.; Milde-Langosch, K.; Beck, K.; Höller, S.; Müller, V.; Witzel, I.; Jänicke, F.; Kilic, E. Expression levels of Activated Leukocyte Cell Adhesion Molecule (ALCAM/CD166) in primary breast carcinoma and distant breast cancer metastases. Dis. Markers 2010, 28, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Sawhney, M.; Matta, A.; Macha, M.A.; Kaur, J.; DattaGupta, S.; Shukla, N.K.; Ralhan, R. Cytoplasmic accumulation of activated leukocyte cell adhesion molecule is a predictor of disease progression and reduced survival in oral cancer patients. Int. J. Cancer 2009, 124, 2098–2105. [Google Scholar] [CrossRef] [PubMed]
- Van den Brand, M.; Takes, R.P.; Blokpoel-deRuyter, M.; Slootweg, P.J.; van Kempen, L.C. Activated leukocyte cell adhesion molecule expression predicts lymph node metastasis in oral squamous cell carcinoma. Oral Oncol. 2010, 46, 393–398. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, C.; Weber, H.; Mogler, C.; Bergmann, F.; Schirmacher, P.; Kenngott, H.G.; Matterne, U.; Mollberg, N.; Rahbari, N.N.; Hinz, U.; et al. Increased expression of ALCAM/CD166 in pancreatic cancer is an independent prognostic marker for poor survival and early tumour relapse. Br. J. Cancer 2009, 101, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Corrias, M.V.; Gambini, C.; Gregorio, A.; Croce, M.; Barisione, G.; Cossu, C.; Rossello, A.; Ferrini, S.; Fabbi, M. Different subcellular localization of ALCAM molecules in neuroblastoma: Association with relapse. Cell. Oncol. 2010, 32, 77–86. [Google Scholar] [PubMed]
- Mezzanzanica, D.; Fabbi, M.; Bagnoli, M.; Staurengo, S.; Losa, M.; Balladore, E.; Alberti, P.; Lusa, L.; Ditto, A.; Ferrini, S.; et al. Subcellular localization of activated leukocyte cell adhesion molecule is a molecular predictor of survival in ovarian carcinoma patients. Clin. Cancer Res. 2008, 14, 1726–1733. [Google Scholar] [CrossRef] [PubMed]
- Van Kempen, L.C.; van den Oord, J.J.; van Muijen, G.N.; Weidle, U.H.; Bloemers, H.P.; Swart, G.W. Activated Leukocyte Cell Adhesion Molecule/CD166, a Marker of Tumor Progression in Primary Malignant Melanoma of the Skin. Am. J. Pathol. 2000, 156, 769–774. [Google Scholar]
- Tanaka, H.; Matsui, T.; Agata, A.; Tomura, M.; Kubota, I.; McFarland, K.C.; Kohr, B.; Lee, A.; Phillips, H.S.; Shelton, D.L. Molecular cloning and expression of a novel adhesion molecule, SC1. Neuron 1991, 7, 535–545. [Google Scholar]
- DeBernardo, A.P.; Chang, S. Native and recombinant DM-GRASP selectively support neurite extension from neurons that express GRASP. Dev. Biol. 1995, 169, 65–75. [Google Scholar] [PubMed]
- Pollerberg, G.E.; Mack, T.G. Cell adhesion molecule SC1/DMGRASP is expressed on growing axons of retina ganglion cells and is involved in mediating their extension on axons. Dev. Biol. 1994, 165, 670–687. [Google Scholar] [CrossRef] [PubMed]
- Avci, H.X.; Zelina, P.; Thelen, K.; Pollerberg, G.E. Role of cell adhesion molecule DM-GRASP in growth and orientation of retinal ganglion cell axons. Dev. Biol. 2004, 271, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Weiner, J.A.; Koo, S.J.; Nicolas, S.; Fraboulet, S.; Pfaff, S.L.; Pourquié, O.; Sanes, J.R. Axon fasciculation defects and retinal dysplasias in mice lacking the immunoglobulin superfamily adhesion molecule BEN/ALCAM/SC1. Mol. Cell. Neurosci. 2004, 27, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Heffron, D.S.; Golden, J.A. DM-GRASP is necessary for nonradial cell migration during chick diencephalic development. J. Neurosci. 2000, 20, 2287–2294. [Google Scholar] [PubMed]
- Stephan, J.P.; Bald, L.; Roberts, P.E.; Lee, J.; Gu, Q.; Mather, J.P. Distribution and function of the adhesion molecule BEN during rat development. Dev. Biol. 1999, 212, 264–277. [Google Scholar] [CrossRef] [PubMed]
- Chédotal, A.; Pourquié, O.; Ezan, F.; San Clemente, H.; Sotelo, C. BEN as a presumptive target recognition molecule during the development of the olivocerebellar system. J. Neurosci. 1996, 16, 3296–3310. [Google Scholar] [PubMed]
- Ott, H.; Diekmann, H.; Stuermer, C.A.; Bastmeyer, M. Function of Neurolin (DM-GRASP/SC-1) in Guidance of Motor Axons during Zebrafish Development. Dev. Biol. 2001, 235, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Buhusi, M.; Demyanenko, G.P.; Jannie, K.M.; Dalal, J.; Darnell, E.P.; Weiner, J.A.; Maness, P.F. ALCAM regulates mediolateral retinotopic mapping in the superior colliculus. J. Neurosci. 2009, 29, 15630–15641. [Google Scholar] [CrossRef] [PubMed]
- Thelen, K.; Maier, B.; Faber, M.; Albrecht, C.; Fischer, P.; Pollerberg, G.E. Translation of the cell adhesion molecule ALCAM in axonal growth cones—Regulation and functional importance. J. Cell Sci. 2012, 125, 1003–1014. [Google Scholar] [CrossRef] [PubMed]
- Thelen, K.; Jaehrling, S.; Spatz, J.P.; Pollerberg, G.E. Depending on Its Nano-Spacing, ALCAM Promotes Cell Attachment and Axon Growth. PLoS ONE 2012, 7, e40493. [Google Scholar] [CrossRef] [PubMed]
- Wade, A.; Thomas, C.; Kalmar, B.; Terenzio, M.; Garin, J.; Greensmith, L.; Schiavo, G. Activated leukocyte cell adhesion molecule modulates neurotrophin signaling. J. Neurochem. 2012, 121, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Bilinska, M.; Pokryszko-Dragan, A.; Sobczynski, M.; Cyrul, M.; Kusnierczyk, P.; Jasek, M. ALCAM and CD6—Multiple sclerosis risk factors. J. Neuroimmunol. 2014, 276, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Du, L.F.; Lv, G.Q.; Yu, X.M.; Gu, Y.L.; Li, J.P.; Zhang, C. Functional polymorphisms in CD166/ALCAM gene associated with increased risk for breast cancer in a Chinese population. Breast Cancer Res. Treat. 2011, 128, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Van Kempen, L.C.; Nelissen, J.M.; Degen, W.G.; Torensma, R.; Weidle, U.H.; Bloemers, H.P.; Figdor, C.G.; Swart, G.W. Molecular Basis for the Homophilic Activated Leukocyte Cell Adhesion Molecule (ALCAM)-ALCAM Interaction. J. Biol. Chem. 2001, 276, 25783–25790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowen, M.A.; Bajorath, J.; Siadak, A.W.; Modrell, B.; Malacko, A.R.; Marquardt, H.; Nadler, S.G.; Aruffo, A. The amino-terminal immunoglobulin-like domain of activated leukocyte cell adhesion molecule binds specifically to the membrane-proximal scavenger receptor cysteine-rich domain of CD6 with a 1:1 stoichiometry. J. Biol. Chem. 1996, 271, 17390–17396. [Google Scholar] [CrossRef] [PubMed]
- Hassan, N.J.; Barclay, A.N.; Brown, M.H. Frontline: Optimal T cell activation requires the engagement of CD6 and CD166. Eur. J. Immunol. 2004, 34, 930–940. [Google Scholar] [CrossRef] [PubMed]
- Te Riet, J.; Zimmerman, A.W.; Cambi, A.; Joosten, B.; Speller, S.; Torensma, R.; van Leeuwen, F.N.; Figdor, C.G.; de Lange, F. Distinct kinetic and mechanical properties govern ALCAM-mediated interactions as shown by single-molecule force spectroscopy. J. Cell Sci. 2007, 120, 3965–3976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimferrer, I.; Calvo, M.; Mittelbrunn, M.; Farnós, M.; Sarrias, M.R.; Enrich, C.; Vives, J.; Sánchez-Madrid, F.; Lozano, F. Relevance of CD6-mediated interactions in T cell activation and proliferation. J. Immunol. 2004, 173, 2262–2270. [Google Scholar] [CrossRef] [PubMed]
- Hassan, N.J.; Simmonds, S.J.; Clarkson, N.G.; Hanrahan, S.; Puklavec, M.J.; Bomb, M.; Barclay, A.N.; Brown, M.H. CD6 regulates T-cell responses through activation-dependent recruitment of the positive regulator SLP-76. Mol. Cell. Biol. 2006, 26, 6727–6738. [Google Scholar] [CrossRef] [PubMed]
- Singer, N.G.; Richardson, B.C.; Powers, D.; Hooper, F.; Lialios, F.; Endres, J.; Bott, C.M.; Fox, D.A. Role of the CD6 glycoprotein in antigen-specific and autoreactive responses of cloned human T lymphocytes. Immunology 1996, 88, 537–543. [Google Scholar] [PubMed]
- Zimmerman, A.W.; Joosten, B.; Torensma, R.; Parnes, J.R.; van Leeuwen, F.N.; Figdor, C.G. Long-term engagement of CD6 and ALCAM is essential for T-cell proliferation induced by dendritic cells. Blood 2006, 107, 3212–3220. [Google Scholar] [CrossRef] [PubMed]
- Cayrol, R.; Wosik, K.; Berard, J.L.; Dodelet-Devillers, A.; Ifergan, I.; Kebir, H.; Haqqani, A.S.; Kreymborg, K.; Krug, S.; Moumdjian, R.; et al. Activated leukocyte cell adhesion molecule promotes leukocyte trafficking into the central nervous system. Nat. Immunol. 2008, 9, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Bowen, M.A.; Aruffo, A.A.; Bajorath, J. Cell surface receptors and their ligands: In vitro analysis of CD6-CD166 interactions. Proteins 2000, 40, 420–428. [Google Scholar] [CrossRef]
- Zimmerman, A.W.; Nelissen, J.M.; van Emst-de Vries, S.E.; Willems, P.H.; de Lange, F.; Collard, J.G.; van Leeuwen, F.N.; Figdor, C.G. Cytoskeletal restraints regulate homotypic ALCAM-mediated adhesion through PKC independently of Rho-like GTPases. J. Cell Sci. 2004, 117, 2841–2852. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; van Bokhoven, A.; Jansen, C.F.; Bussemakers, M.J.; Schalken, J.A. Coordinate recruitment of E-cadherin and ALCAM to cell-cell contacts by alpha-catenin. Biochem. Biophys. Res. Commun. 2000, 267, 870–874. [Google Scholar] [CrossRef] [PubMed]
- Mertens, A.E.; Roovers, R.C.; Collard, J.G. Regulation of Tiam1-Rac signalling. FEBS Lett. 2003, 546, 11–16. [Google Scholar] [CrossRef]
- Uhlenbrock, K.; Eberth, A.; Herbrand, U.; Daryab, N.; Stege, P.; Meier, F.; Friedl, P.; Collard, J.G.; Ahmadian, M.R. The RacGEF Tiam1 inhibits migration and invasion of metastatic melanoma via a novel adhesive mechanism. J. Cell Sci. 2004, 117, 4863–4871. [Google Scholar] [CrossRef] [PubMed]
- Piazza, T.; Cha, E.; Bongarzone, I.; Canevari, S.; Bolognesi, A.; Polito, L.; Bargellesi, A.; Sassi, F.; Ferrini, S.; Fabbi, M. Internalization and recycling of ALCAM/CD166 detected by a fully human single-chain recombinant antibody. J. Cell Sci. 2005, 118, 1515–1525. [Google Scholar] [CrossRef] [PubMed]
- Finne, J.; Finne, U.; Deagostini-Bazin, H.; Goridis, C. Occurrence of alpha 2–8 linked polysialosyl units in a neural cell adhesion molecule. Biochem. Biophys. Res. Commun. 1983, 112, 482–487. [Google Scholar] [CrossRef]
- Hoffman, S.; Sorkin, B.C.; White, P.C.; Brackenbury, R.; Mailhammer, R.; Rutishauser, U.; Cunningham, B.A.; Edelman, G.M. Chemical characterization of a neural cell adhesion molecule purified from embryonic brain membranes. J. Biol. Chem. 1982, 257, 7720–7729. [Google Scholar] [PubMed]
- Faissner, A.; Teplow, D.B.; Kübler, D.; Keilhauer, G.; Kinzel, V.; Schachner, M. Biosynthesis and membrane topography of the neural cell adhesion molecule L1. EMBO J. 1985, 4, 3105–3313. [Google Scholar] [PubMed]
- Wolff, J.M.; Rathjen, F.G.; Frank, R.; Roth, S. Biochemical characterization of polypeptide components involved in neurite fasciculation and elongation. Eur. J. Biochem. 1987, 168, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, B.; Peter-Katalinic, J.; Egge, H.; Schachner, M. Monoclonal antibodies raised against membrane glycoproteins from mouse brain recognize N-linked oligomannosidic glycans. Glycobiology 1993, 3, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Fahrig, T.; Schmitz, B.; Weber, D.; Kücherer-Ehret, A.; Faissner, A.; Schachner, M. Two monoclonal antibodies recognizing carbohydrate epitopes on neural adhesion molecules interfere with cell interactions. Eur. J. Neurosci. 1990, 2, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Linnemann, D.; Edvardsen, K.; Bock, E. Developmental study of the cell adhesion molecule L1. Dev. Neurosci. 1988, 10, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Rutishauser, U.; Hoffman, S.; Edelman, G.M. Binding properties of a cell adhesion molecule from neuraltissue. Proc. Natl. Acad. Sci. USA 1982, 9, 685–689. [Google Scholar] [CrossRef]
- Angata, K.; Suzuki, M.; Fukuda, M. Differential and cooperative polysialylation of the neural cell adhesion molecule by two polysialyltransferases, PST and STX. J. Biol. Chem. 1998, 273, 28524–28532. [Google Scholar] [CrossRef] [PubMed]
- Ebeling, O.; Duczmal, A.; Aigner, S.; Geiger, C.; Schollhammer, S.; Kemshead, J.T.; Moller, P.; Schwartz-Albiez, R.; Altevogt, P. L1 adhesion molecule on human lymphocytes and monocytes: Expression and involvement in binding to alpha v beta 3 integrin. Eur. J. Immunol. 1996, 26, 2508–2516. [Google Scholar] [CrossRef] [PubMed]
- Jouve, N.; Despoix, N.; Espeli, M.; Gauthier, L.; Cypowyj, S.; Fallague, K.; Schiff, C.; Dignat-George, F.; Vély, F.; Leroyer, A.S. The involvement of CD146 and its novel ligand galectin-1 in apoptotic regulation of endothelial cells. J. Biol. Chem. 2013, 288, 2571–2579. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, K.; Fitzgerald, K.; Baker, J.; Regnstrom, K.; Gardai, S.; Bard, F.; Mocci, S.; Seto, P.; You, M.; Larochelle, C.; et al. Laminin-411 Is a vascular ligand for MCAM and facilitates TH17 cell entry into the CNS. PLoS ONE 2012, 7, e4044. [Google Scholar] [CrossRef] [PubMed]
- Taniura, H.; Kuo, C.H.; Hayashi, Y.; Miki, N. Purification and Characterization of an 82-kD Membrane Protein as a neurite outgrowth factor binding protein: Possible involvement of NOF binding protein in axonal outgrowth in developing retina. J. Cell Biol. 1991, 112, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Zhuang, J.; Duan, H.; Luo, Y.; Zeng, Q.; Fan, K.; Yan, H.; Lu, D.; Ye, Z.; Hao, J.; et al. CD146 is a coreceptor for VEGFR-2 in tumor angiogenesis. Blood 2012, 120, 2330–2339. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Zhang, C.; Tu, T.; Sun, M.; Liu, D.; Lu, D.; Feng, J.; Yang, D.; Liu, F.; Yan, X. Wnt5a uses CD146 as a receptor to regulate cell motility and convergent extension. Nat. Commun. 2013. [Google Scholar] [CrossRef] [PubMed]
- El-Deeb, S.; Thompson, S.C.; Covault, J. Characterization of a cell surface adhesion molecule expressed by a subset of developing chick neurons. Dev. Biol. 1992, 149, 213–227. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M.; Eddins, M.J. Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta 2004, 1695, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Varshavsky, A. The ubiquitin system. Trends Biochem. Sci. 1997, 22, 383–387. [Google Scholar] [CrossRef]
- Ciechanover, A.; Heller, H.; Elias, S.; Haas, A.L.; Hershko, A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc. Natl. Acad. Sci. USA 1980, 77, 1365–1368. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef] [PubMed]
- Hoeller, D.; Dikic, I. Targeting the ubiquitin system in cancer therapy. Nature 2009, 458, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A. Ubiquitin: Roles in protein modification and breakdown. Cell 1983, 34, 11–12. [Google Scholar] [CrossRef]
- Nakayama, K.I.; Nakayama, K. Ubiquitin ligases: Cell-cycle control and cancer. Nat. Rev. Cancer 2006, 6, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Levkowitz, G.; Waterman, H.; Ettenberg, S.A.; Katz, M.; Tsygankov, A.Y.; Alroy, I.; Lavi, S.; Iwai, K.; Reiss, Y.; Ciechanover, A.; et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell 1999, 4, 1029–1040. [Google Scholar] [CrossRef]
- Marchese, A.; Raiborg, C.; Santini, F.; Keen, J.H.; Stenmark, H.; Benovic, J.L. The E3 ubiquitin ligase AIP4 mediates ubiquitination and sorting of the G protein-coupled receptor CXCR4. Dev. Cell 2003, 5, 709–722. [Google Scholar] [CrossRef]
- Piper, R.C.; Dikic, I.; Lukacs, G.L. Ubiquitin-dependent sorting in endocytosis. Cold Spring Harb. Perspect. Biol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Ryu, K.S.; Choi, Y.S.; Ko, J.; Kim, S.O.; Kim, H.J.; Cheong, H.K.; Jeon, Y.H.; Choi, B.S.; Cheong, C. Direct characterization of E2-dependent target specificity and processivity using an artificial p27-linker-E2 ubiquitination system. BMB Rep. 2008, 41, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, F.; Dikic, I. Atypical ubiquitin chains: New molecular signals. “Protein Modifications: Beyond the Usual Suspects” review series. EMBO Rep. 2008, 9, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Haglund, K.; Dikic, I. The role of ubiquitylation in receptor endocytosis and endosomal sorting. J. Cell Sci. 2012, 125, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Clague, M.J.; Urbé, S. Ubiquitin: Same molecule, different degradation pathways. Cell 2010, 143, 682–685. [Google Scholar] [CrossRef] [PubMed]
- Paul, S. Dysfunction of the ubiquitin-proteasome system in multiple disease conditions: Therapeutic approaches. Bioessays 2008, 30, 1172–1184. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Schwartz, D.; Elias, J.E.; Thoreen, C.C.; Cheng, D.; Marsischky, G.; Roelofs, J.; Finley, D.; Gygi, S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003, 21, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M.; Fushman, D. Polyubiquitin chains: Polymeric protein signals. Curr. Opin. Chem. Biol. 2004, 8, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Clague, M.J.; Urbé, S. Endocytosis: The DUB version. Trends Cell Biol. 2006, 16, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Huen, M.S.; Sy, S.M.; Chen, J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat. Rev. Mol. Cell Biol. 2010, 11, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; de Lange, T. 53BP1: Pro choice in DNA repair. Trends Cell Biol. 2014, 24, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Chen, Z.J. Regulation of NFκB by ubiquitination. Curr. Opin. Immunol. 2013, 25, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Spence, J.; Gali, R.R.; Dittmar, G.; Sherman, F.; Karin, M.; Finley, D. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 2000, 102, 67–76. [Google Scholar] [CrossRef]
- Xu, P.; Duong, D.M.; Seyfried, N.T.; Cheng, D.; Xie, Y.; Robert, J.; Rush, J.; Hochstrasser, M.; Finley, D.; Peng, J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 2009, 137, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Dynek, J.N.; Goncharov, T.; Dueber, E.C.; Fedorova, A.V.; Izrael-Tomasevic, A.; Phu, L.; Helgason, E.; Fairbrother, W.J.; Deshayes, K.; Kirkpatrick, D.S.; et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010, 29, 4198–4209. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Williamson, A.; Banerjee, S.; Philipp, I.; Rape, M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 2008, 133, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, S.; Yadam, M.; Matsumoto, M.; Ishida, N.; Nakayama, K.I. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 2001, 276, 33111–33120. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.S.; Ma, P.C.; Ota, I.M.; Varshavsky, A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 1995, 270, 17442–17456. [Google Scholar] [CrossRef] [PubMed]
- Chastagner, P.; Israël, A.; Brou, C. Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep. 2006, 7, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Kölling, R.; Hollenberg, C.P. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 1994, 13, 3261–3271. [Google Scholar] [PubMed]
- Hicke, L. A new ticket for entry into budding vesicles-ubiquitin. Cell 2001, 106, 527–530. [Google Scholar] [CrossRef]
- Geoffroy, M.C.; Hay, R.T. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 2009, 10, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Rahighi, S.; Ikeda, F.; Kawasaki, M.; Akutsu, M.; Suzuki, N.; Kato, R.; Kensche, T.; Uejima, T.; Bloor, S.; Komander, D.; et al. Specific Recognition of Linear Ubiquitin Chains by NEMO Is Important for NF-κB Activation. Cell 2009, 136, 1098–1109. [Google Scholar] [CrossRef] [PubMed]
- Husnjak, K.; Dikic, I. Ubiquitin-binding proteins: Decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 2012, 81, 291–322. [Google Scholar] [CrossRef] [PubMed]
- Sen, A.; Madhivanan, K.; Mukherjee, D.; Aguilar, R.C. The epsin protein family: Coordinators of endocytosis and signaling. Biomol. Concepts 2012, 3, 117–126. [Google Scholar] [CrossRef] [PubMed]
- De Melker, A.A.; van der Horst, G.; Borst, J. Ubiquitin ligase activity of c-Cbl guides the epidermal growth factor receptor into clathrin-coated pits by two distinct modes of Eps15 recruitment. J. Biol. Chem. 2004, 279, 55465–55473. [Google Scholar] [CrossRef] [PubMed]
- Stang, E. Cbl-dependent ubiquitination is required for progression of EGF receptors into clathrin-coated pits. Mol. Biol. Cell 2004, 15, 3591–3604. [Google Scholar] [CrossRef] [PubMed]
- Sigismund, S.; Woelk, T.; Puri, C.; Maspero, E.; Tacchetti, C.; Transidico, P.; di Fiore, P.P.; Polo, S. Clathrin-independent endocytosis of ubiquitinated cargos. Proc. Natl. Acad. Sci. USA 2005, 102, 2760–2765. [Google Scholar] [CrossRef] [PubMed]
- Gucwa, A.L.; Brown, D.A. UIM domain-dependent recruitment of the endocytic adaptor protein Eps15 to ubiquitin-enriched endosomes. BMC Cell Biol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Clague, M.J.; Liu, H.; Urbé, S. Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev. Cell 2012, 23, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Lobert, V.H.; Brech, A.; Pedersen, N.M.; Wesche, J.; Oppelt, A.; Malerød, L.; Stenmark, H. Ubiquitination of α5β1 Integrin Controls Fibroblast Migration through Lysosomal Degradation of Fibronectin-Integrin Complexes. Dev. Cell 2010, 19, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Umebayashi, K. The roles of ubiquitin and lipids in protein sorting along the endocytic pathway. Cell Struct. Funct. 2003, 28, 443–453. [Google Scholar] [PubMed]
- Reyes-Turcu, F.E.; Ventii, K.H.; Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [PubMed]
- Schwarz, L.A.; Patrick, G.N. Ubiquitin-dependent endocytosis, trafficking and turnover of neuronal membrane proteins. Mol. Cell. Neurosci. 2012, 49, 387–393. [Google Scholar] [PubMed]
- Bomberger, J.M.; Barnaby, R.L.; Stanton, B.A. The deubiquitinating enzyme USP10 regulates the post-endocytic sorting of cystic fibrosis transmembrane conductance regulator in airway epithelial cells. J. Biol. Chem. 2009, 284, 18778–18789. [Google Scholar] [PubMed]
- Berthouze, M.; Venkataramanan, V.; Li, Y.; Shenoy, S.K. The deubiquitinases USP33 and USP20 coordinate beta2 adrenergic receptor recycling and resensitization. EMBO J. 2009, 28, 1684–1696. [Google Scholar] [PubMed]
- Butterworth, M.B.; Edinger, R.S.; Ovaa, H.; Burg, D.; Johnson, J.P.; Frizzell, R.A. The deubiquitinating enzyme UCH-L3 regulates the apical membrane recycling of the epithelial sodium channel. J. Biol. Chem. 2007, 282, 37885–37893. [Google Scholar] [CrossRef] [PubMed]
- Diestel, S.; Schaefer, D.; Cremer, H.; Schmitz, B. NCAM is ubiquitylated, endocytosed and recycled in neurons. J. Cell Sci. 2007, 120, 4035–4049. [Google Scholar] [CrossRef] [PubMed]
- Bailey, C.H.; Chen, M.; Keller, F.; Kandel, E.R. Serotonin-mediated endocytosis of apCAM: An early step of learning-related synaptic growth in Aplysia. Science 1992, 256, 645–649. [Google Scholar] [CrossRef] [PubMed]
- Montarolo, P.G.; Goelet, P.; Castellucci, V.F.; Morgan, J.; Kandel, E.R.; Schacher, S. A critical period for macromolecular synthesis in long-term heterosynaptic facilitation in Aplysia. Science 1986, 234, 1249–1254. [Google Scholar] [CrossRef] [PubMed]
- Glanzman, D.L.; Mackey, S.L.; Hawkins, R.D.; Dyke, A.M.; Lloyd, P.E.; Kandel, E.R. Depletion of serotonin in the nervous system of Aplysia reduces the behavioral enhancement of gill withdrawal as well as the heterosynaptic facilitation produced by tail shock. J. Neurosci. 1989, 9, 4200–4213. [Google Scholar] [PubMed]
- Hu, Y.; Barzilai, A.; Chen, M.; Bailey, C.H.; Kandel, E.R. 5-HT and cAMP induce the formation of coated pits and vesicles and increase the expression of clathrin light chain in sensory neurons of aplysia. Neuron 1993, 10, 921–929. [Google Scholar] [CrossRef]
- Miñana, R.; Duran, J.M.; Tomas, M.; Renau-Piqueras, J.; Guerri, C. Neural cell adhesion molecule is endocytosed via a clathrin-dependent pathway. Eur. J. Neurosci. 2001, 13, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Bailey, C.H.; Kaang, B.K.; Chen, M.; Martin, K.C.; Lim, C.S.; Casadio, A.; Kandel, E.R. Mutation in the phosphorylation sites of MAP kinase blocks learning-related internalization of apCAM in Aplysia sensory neurons. Neuron 1997, 18, 913–924. [Google Scholar] [CrossRef]
- Mayford, M.; Barzilai, A.; Keller, F.; Schacher, S.; Kandel, E.R. Modulation of an NCAM-related adhesion molecule with long-term synaptic plasticity in Aplysia. Science 1992, 256, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Bailey, C.H.; Kandel, E.R.; Si, K. The persistence of long-term memory: A molecular approach to self-sustaining changes in learning-induced synaptic growth. Neuron 2004, 44, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Foley, A.G.; Hartz, B.P.; Gallagher, H.C.; Rønn, L.C.; Berezin, V.; Bock, E.; Regan, C.M. A synthetic peptide ligand of neural cell adhesion molecule (NCAM) IgI domain prevents NCAM internalization and disrupts passive avoidance learning. J. Neurochem. 2000, 74, 2607–2613. [Google Scholar] [CrossRef] [PubMed]
- Monzo, H.J.; Park, T.I.; Dieriks, B.V.; Jansson, D.; Faull, R.L.; Dragunow, M.; Curtis, M.A. Insulin and IGF1 modulate turnover of polysialylated neural cell adhesion molecule (PSA-NCAM) in a process involving specific extracellular matrix components. J. Neurochem. 2013, 6, 758–770. [Google Scholar] [CrossRef] [PubMed]
- Baska, K.M.; Manandhar, G.; Feng, D.; Agca, Y.; Tengowski, M.W.; Sutovsky, M.; Yi, Y.J.; Sutovsky, P. Mechanism of extracellular ubiquitination in the mammalian epididymis. J. Cell. Physiol. 2008, 215, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Terrell, J.; Shih, S.; Dunn, R.; Hicke, L. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol. Cell 1998, 1, 193–202. [Google Scholar] [CrossRef]
- Lucero, P.; Peñalver, E.; Vela, L.; Lagunas, R. Monoubiquitination is sufficient to signal internalization of the maltose transporter in Saccharomyces cerevisiae. J. Bacteriol. 2000, 182, 241–243. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, F.; Sakuma, M.; Matsuo, Y.; Arase, H.; Yamasaki, S.; Nakamura, N.; Saito, T.; Ohno, H. A Di-leucine signal in the ubiquitin moiety. Possible involvement in ubiquitination-mediated endocytosis. J. Biol. Chem. 2000, 275, 26213–26219. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.F.; Davis, N.G. Ubiquitination of the PEST-like endocytosis signal of the yeast a-factor receptor. J. Biol. Chem. 2000, 275, 8143–8153. [Google Scholar] [CrossRef] [PubMed]
- Barriere, H.; Nemes, C.; Lechardeur, D.; Khan-Mohammad, M.; Fruh, K.; Lukacs, G.L. Molecular basis of oligoubiquitin-dependent internalization of membrane proteins in Mammalian cells. Traffic 2006, 7, 282–297. [Google Scholar] [CrossRef] [PubMed]
- Hawryluk, M.J.; Keyel, P.A.; Mishra, S.K.; Watkins, S.C.; Heuser, J.E.; Traub, L.M. Epsin 1 is a polyubiquitin-selective clathrin-associated sorting protein. Traffic 2006, 7, 262–281. [Google Scholar] [CrossRef] [PubMed]
- Wobst, H.; Förster, S.; Laurini, C.; Sekulla, A.; Dreiseidler, M.; Höhfeld, J.; Schmitz, B.; Diestel, S. UCHL1 regulates ubiquitination and recycling of the neural cell adhesion molecule NCAM. FEBS J. 2012, 279, 4398–4409. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fallon, L.; Lashuel, H.A.; Liu, Z.; Lansbury, P.T., Jr. The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell 2002, 111, 209–218. [Google Scholar] [CrossRef]
- Larsen, C.N.; Krantz, B.A.; Wilkinson, K.D. Substrate Specificity of Deubiquitinating Enzymes: Ubiquitin C-Terminal Hydrolases. Biochemistry 1998, 37, 3358–3368. [Google Scholar] [CrossRef] [PubMed]
- Osaka, H.; Wang, Y.L.; Takada, K.; Takizawa, S.; Setsuie, R.; Li, H.; Sato, Y.; Nishikawa, K.; Sun, Y.J.; Sakurai, M.; et al. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum. Mol. Genet. 2003, 12, 1945–1958. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, M.K.; Schmitz, B.; Diestel, S. L1CAM ubiquitination facilitates its lysosomal degradation. FEBS Lett. 2010, 584, 4475–4480. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Fujisaki, K.; Watanabe, M. Human L1CAM carrying the missense mutations of the fibronectin-like type III domains is localized in the endoplasmic reticulum and degraded by polyubiquitylation. J. Neurosci. Res. 2011, 89, 1637–1645. [Google Scholar] [CrossRef] [PubMed]
- Castellani, V.; Falk, J.; Rougon, G. Semaphorin3A-induced receptor endocytosis during axon guidance responses is mediated by L1 CAM. Mol. Cell. Neurosci. 2004, 26, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Ewan, L.C.; Jopling, H.M.; Jia, H.; Mittar, S.; Bagherzadeh, A.; Howell, G.J.; Walker, J.H.; Zachary, I.C.; Ponnambalam, S. Intrinsic tyrosine kinase activity is required for vascular endothelial growth factor receptor 2 ubiquitination, sorting and degradation in endothelial Cells. Traffic 2006, 7, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Hicke, L.; Zanolari, B.; Riezman, H. Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J. Cell Biol. 1998, 141, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.G.; Krolewski, J.J.; Fuchs, S.Y. Phosphorylation and specific ubiquitin acceptor sites are required for ubiquitination and degradation of the IFNAR1 subunit of type I interferon receptor. J. Biol. Chem. 2004, 279, 46614–46620. [Google Scholar] [CrossRef] [PubMed]
- Aikawa, Y. Ubiquitination within the membrane-proximal ezrin-radixin-moesin (ERM)-binding region of the L1 cell adhesion molecule. Commun. Integr. Biol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Thelen, K.; Georg, T.; Bertuch, S.; Zelina, P.; Pollerberg, G.E. Ubiquitination and endocytosis of cell adhesion molecule DM-GRASP regulate its cell surface presence and affect its role for axon navigation. J. Biol. Chem. 2008, 283, 32792–32801. [Google Scholar] [CrossRef] [PubMed]
- Dalerba, P.; Dylla, S.J.; Park, I.K.; Liu, R.; Wang, X.; Cho, R.W.; Hoey, T.; Gurney, A.; Huang, E.H.; Simeone, D.M.; et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 10158–10163. [Google Scholar] [CrossRef] [PubMed]
- Levin, T.G.; Powell, A.E.; Davies, P.S.; Silk, A.D.; Dismuke, A.D.; Anderson, E.C.; Swain, J.R.; Wong, M.H. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology 2010, 139, 2072–2082.e5. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Hindoyan, A.; Wang, S.; Tran, L.M.; Goldstein, A.S.; Lawson, D.; Chen, D.; Li, Y.; Guo, C.; Zhang, B.; et al. Identification of CD166 as a surface marker for enriching prostate stem/progenitor and cancer initiating cells. PLoS ONE 2012, 7, e42564. [Google Scholar] [CrossRef] [PubMed]
- Jezierska, A.; Matysiak, W.; Motyl, T. ALCAM/CD166 protects breast cancer cells against apoptosis and autophagy. Med. Sci. Monit. 2006, 12, BR263–BR273. [Google Scholar] [PubMed]
- Ma, L.; Wang, J.; Lin, J.; Pan, Q.; Yu, Y.; Sun, F. Cluster of differentiation 166 CD166) regulated by phosphatidylinositide 3-Kinase (PI3K)/AKT signaling to exert its anti-apoptotic role via yes-associated protein (YAP) in liver cancer. J. Biol. Chem. 2014, 289, 6921–6933. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Pan, Q.; Sun, F.; Yu, Y.; Wang, J. Cluster of differentiation 166 (CD166) regulates cluster of differentiation (CD44) via NF-κB in liver cancer cell line Bel-7402. Biochem. Biophys. Res. Commun. 2014, 451, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Witze, E.S.; Litman, E.S.; Argast, G.M.; Moon, R.T.; Ahn, N.G. Wnt5a control of cell polarity and directional movement by polarized redistribution of adhesion receptors. Science 2008, 320, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Nichols, B.J. GM1-containing lipid rafts are depleted within clathrin-coated pits. Curr. Biol. 2003, 13, 686–690. [Google Scholar] [CrossRef]
- Subtil, A.; Gaidarov, I.; Kobylarz, K.; Lampson, M.A.; Keen, J.H.; McGraw, T.E. Acute cholesterol depletion inhibits clathrin-coated pit budding. Proc. Natl. Acad. Sci. USA 1999, 96, 6775–6780. [Google Scholar] [CrossRef] [PubMed]
- Zuhorn, I.S.; Kalicharan, R.; Hoekstra, D. Lipoplex-mediated transfection of mammalian cells occurs through the cholesterol-dependent clathrin-mediated pathway of endocytosis. J. Biol. Chem. 2002, 277, 18021–18028. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Gruenberg, J. Biogenesis of transport intermediates in the endocytic pathway. FEBS Lett. 1999, 452, 61–66. [Google Scholar] [CrossRef]
- Chen, H.; de Camilli, P. The association of epsin with ubiquitinated cargo along the endocytic pathway is negatively regulated by its interaction with clathrin. Proc. Natl. Acad. Sci. USA 2005, 102, 2766–2771. [Google Scholar] [CrossRef] [PubMed]
- Belouzard, S.; Rouillé, Y. Ubiquitylation of leptin receptor OB-Ra regulates its clathrin-mediated endocytosis. EMBO J. 2006, 25, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Raiborg, C.; Bache, K.G.; Gillooly, D.J.; Madshus, I.H.; Stang, E.; Stenmark, H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 2002, 4, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Piper, R.C.; Luzio, J.P. Ubiquitin-dependent sorting of integral membrane proteins for degradation in lysosomes. Curr. Opin. Cell Biol. 2007, 19, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Biederer, T.; Sara, Y.; Mozhayeva, M.; Atasoy, D.; Liu, X.; Kavalali, E.T.; Südhof, T.C. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science 2002, 297, 1525–1531. [Google Scholar] [CrossRef] [PubMed]
- Stagi, M.; Fogel, A.I.; Biederer, T. SynCAM 1 participates in axo-dendritic contact assembly and shapes neuronal growth cones. Proc. Natl. Acad. Sci. USA 2010, 107, 7568–7573. [Google Scholar] [CrossRef] [PubMed]
- Fogel, A.I.; Stagi, M.; Perez de Arce, K.; Biederer, T. Lateral assembly of the immunoglobulin protein SynCAM 1 controls its adhesive function and instructs synapse formation. EMBO J. 2011, 30, 4728–4738. [Google Scholar] [CrossRef] [PubMed]
- Kidd, T.; Brose, K.; Mitchell, K.J.; Fetter, R.D.; Tessier-Lavigne, M.; Goodman, C.S.; Tear, G. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell 1998, 92, 205–215. [Google Scholar] [CrossRef]
- Seeger, M.; Tear, G.; Ferres-Marco, D.; Goodman, C.S. Mutations affecting growth cone guidance in Drosophila: Genes necessary for guidance toward or away from the midline. Neuron 1993, 10, 409–426. [Google Scholar] [CrossRef]
- Ypsilanti, A.R.; Zagar, Y.; Chédotal, A. Moving away from the midline: New developments for Slit and Robo. Development 2010, 137, 1939–1952. [Google Scholar] [CrossRef] [PubMed]
- Chance, R.K.; Bashaw, G.J. Slit-dependent endocytic trafficking of the robo receptor is required for son of sevenless recruitment and midline axon repulsion. PLoS Genet. 2015, 1, e1005402. [Google Scholar] [CrossRef] [PubMed]
- Yuasa-Kawada, J.; Kinoshita-Kawada, M.; Rao, Y.; Wu, J.Y. Deubiquitinating enzyme USP33/VDU1 is required for Slit signaling in inhibiting breast cancer cell migration. Proc. Natl. Acad. Sci. USA 2009, 106, 14530–14535. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Lee, H.K.; Seo, I.A.; Bae, H.R.; Suh, D.J.; Wu, J.; Rao, Y.; Hwang, K.G.; Park, H.T. Netrin induces down-regulation of its receptor, deleted in colorectal cancer, through the ubiquitin-proteasome pathway in the embryonic cortical neuron. J. Neurochem. 2005, 95, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tian, N.; Leshchyns’ka, I.; Welch, J.H.; Diakowski, W.; Yang, H.; Schachner, M.; Sytnyk, V. Lipid raft-dependent endocytosis of close homolog of adhesion molecule L1 (CHL1) promotes neuritogenesis. J. Biol. Chem. 2012, 287, 44447–44463. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Homrich, M.; Gotthard, I.; Wobst, H.; Diestel, S. Cell Adhesion Molecules and Ubiquitination—Functions and Significance. Biology 2016, 5, 1. https://doi.org/10.3390/biology5010001
Homrich M, Gotthard I, Wobst H, Diestel S. Cell Adhesion Molecules and Ubiquitination—Functions and Significance. Biology. 2016; 5(1):1. https://doi.org/10.3390/biology5010001
Chicago/Turabian StyleHomrich, Mirka, Ingo Gotthard, Hilke Wobst, and Simone Diestel. 2016. "Cell Adhesion Molecules and Ubiquitination—Functions and Significance" Biology 5, no. 1: 1. https://doi.org/10.3390/biology5010001
APA StyleHomrich, M., Gotthard, I., Wobst, H., & Diestel, S. (2016). Cell Adhesion Molecules and Ubiquitination—Functions and Significance. Biology, 5(1), 1. https://doi.org/10.3390/biology5010001





