Polar Microalgae: New Approaches towards Understanding Adaptations to an Extreme and Changing Environment
Abstract
:1. Introduction
2. Polar Significance
3. Microalgal Mechanisms to Thrive
3.1. Membrane Fluidity
3.2. Enzyme Kinetics
3.3. Compatible Solutes and Cryoprotectants
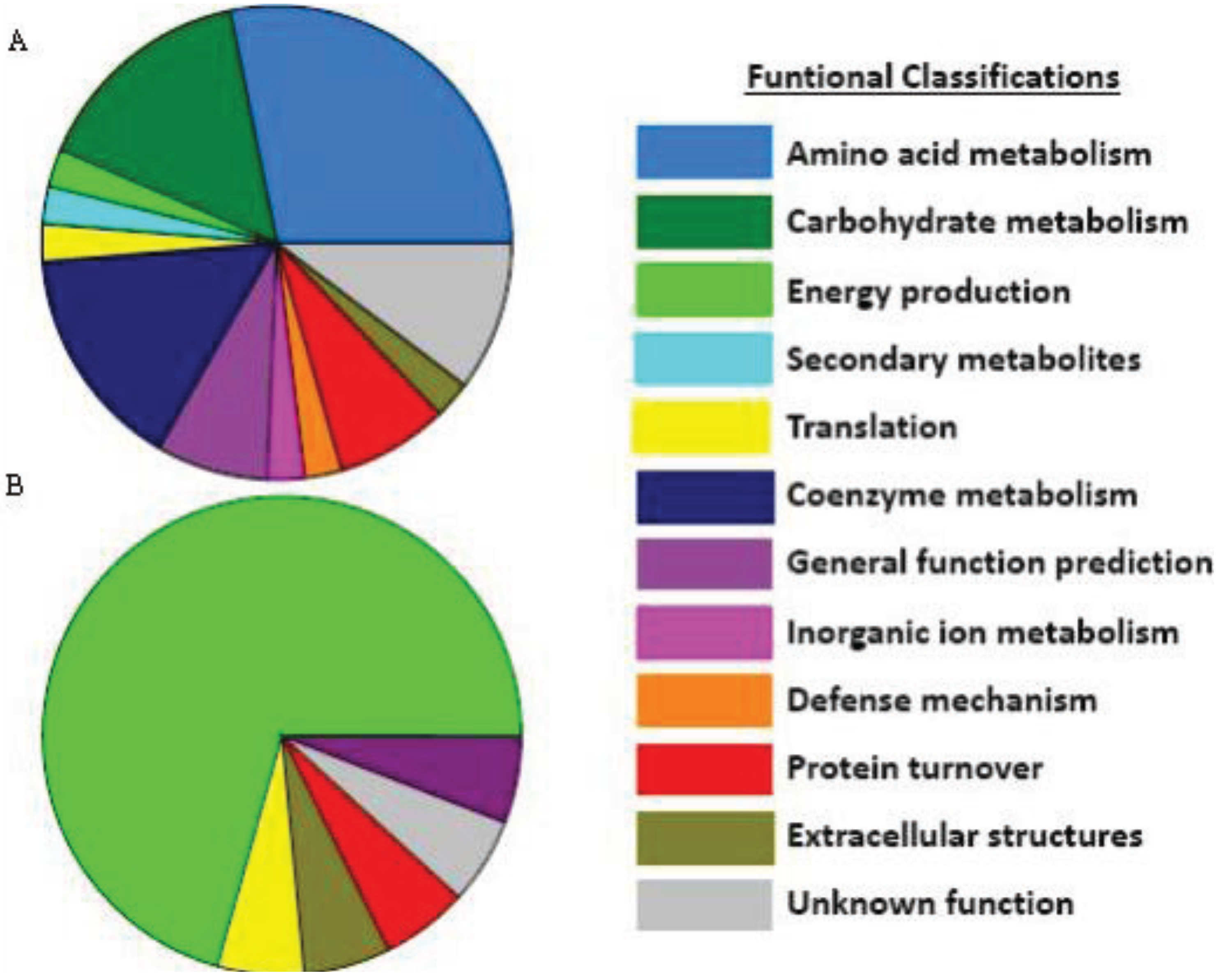
3.4. Extracellular Compounds
3.5. Light Acclimation
3.6. Antioxidants
3.7. Dark Adaptation
4. Using Systems Biology to Understand a Changing World
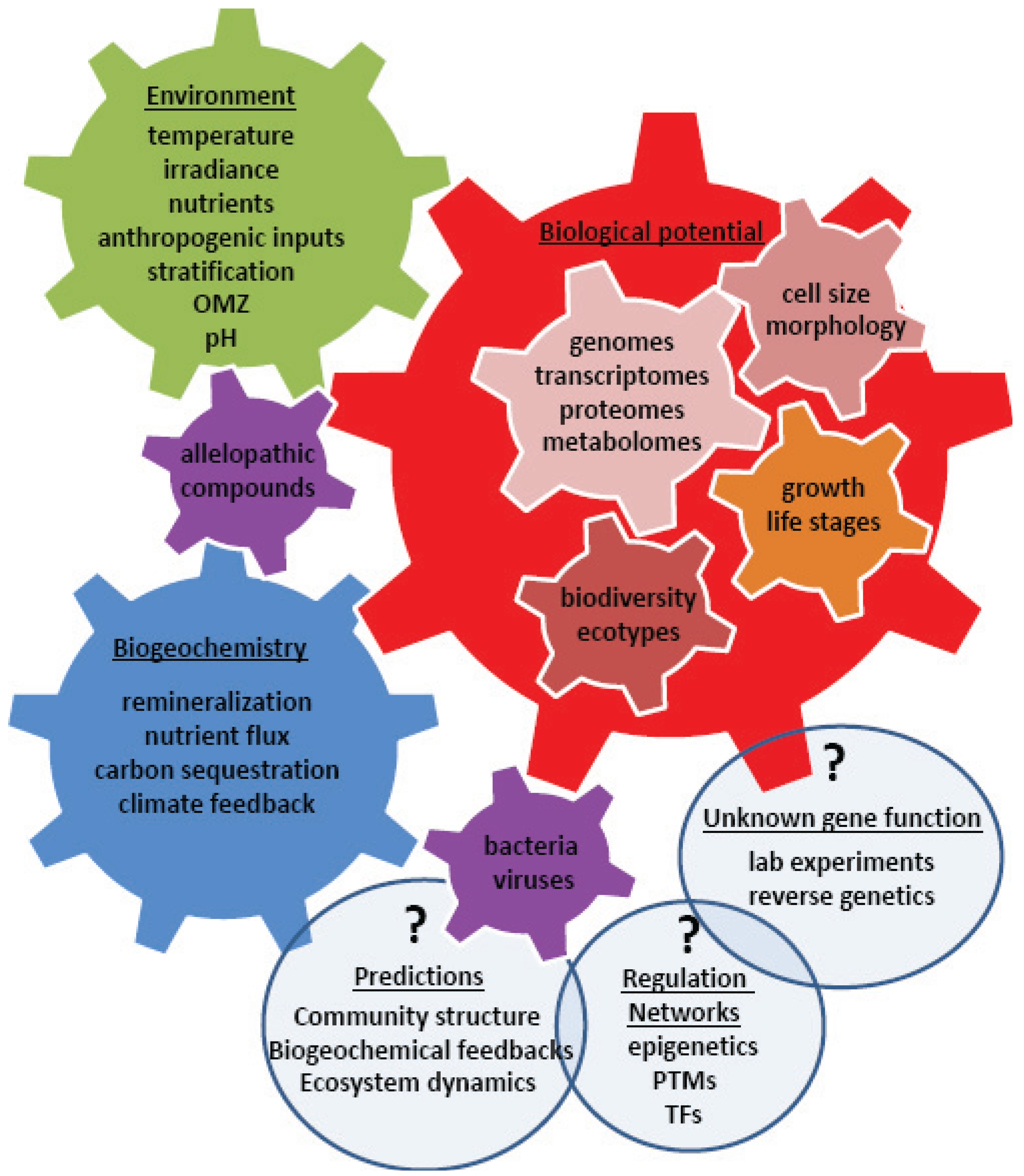
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Smith, W.O.; Lancelot, C. Bottom-up versus top-down control in phytoplankton of the Southern Ocean. Antarct. Sci. 2004, 16, 531–539. [Google Scholar] [CrossRef]
- Kirst, G.O.; Wiencke, C. Ecophysiology of polar algae. J. Phycol. 1995, 31, 181–199. [Google Scholar]
- Thomas, D.N.; Dieckmann, G.S. Antarctic Sea ice—A habitat for extremophiles. Science 2002, 295, 641–644. [Google Scholar] [CrossRef] [Green Version]
- Mock, T.; Thomas, D.N. Microalgae in Polar Regions: Linking Functional Genomics and Physiology with Environmental Conditions. In Psychrophiles: From Biodiversity to Biotechnology; Margesin, R., Schinner, F., Marx, J.-C., Gerday, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 285–312. [Google Scholar]
- Arrigo, K.R.; Mock, T.; Lizotte, M.P. Primary production in sea ice. In Sea Ice: An Introduction to Its Physics, Chemistry, Biology and Geology; Thomas, D., Dieckmann, G., Eds.; Wiley-Blackwell: Malden, MA, USA, 2010; pp. 143–183. [Google Scholar]
- Mock, T.; Thomas, D.N. Recent advances in sea-ice microbiology. Environ. Microbiol. 2005, 7, 605–619. [Google Scholar] [CrossRef]
- Ewert, M.; Deming, J. Sea ice microorganisms: Environmental constraints and extracellular responses. Biology 2013, 2, 603–628. [Google Scholar] [CrossRef]
- Dolhi, J.; Maxwell, D.; Morgan-Kiss, R. Review: The Antarctic Chlamydomonas raudensis: An emerging model for cold adaptation of photosynthesis. Extremophiles 2013, 17, 711–722. [Google Scholar] [CrossRef]
- Morgan-Kiss, R.M.; Priscu, J.C.; Pocock, T.; Gudynaite-Savitch, L.; Huner, N.P.A. Adaptation and acclimation of photosynthetic microorganisms to permanently cold environments. Microbiol. Mol. Biol. Rev. 2006, 70, 222–252. [Google Scholar] [CrossRef]
- Singh, S.M.; Elster, J. Cyanobacteria in Antarctic Lake Environments. In Algae and Cyanobacteria in Extreme Environments; Seckbach, J., Ed.; Springer: Dordrecht, The Netherlands, 2007; Volume 11, pp. 303–320. [Google Scholar]
- Vincent, W. Cold Tolerance in Cyanobacteria and Life in the Cryosphere. In Algae and Cyanobacteria in Extreme Environments; Seckbach, J., Ed.; Springer: Dordrecht, The Netherlands, 2007; Volume 11, pp. 287–301. [Google Scholar]
- Wiencke, C.; Amsler, C.D. Seaweeds and Their Communities in Polar Regions. In Seaweed Biology; Wiencke, C., Bischof, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 219, pp. 265–291. [Google Scholar]
- D’Amico, S. Psychrophilic microorganisms: Challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef]
- Casanueva, A.; Tuffin, M.; Cary, C.; Cowan, D.A. Molecular adaptations to psychrophily: The impact of ‘omic’ technologies. Trends Microbiol. 2010, 18, 374–381. [Google Scholar] [CrossRef]
- Gerday, C. Psychrophily and catalysis. Biology 2013, 2, 719–741. [Google Scholar] [CrossRef]
- Koh, E.Y.; Martin, A.R.; McMinn, A.; Ryan, K.G. Recent Advances and Future Perspectives in Microbial Phototrophy in Antarctic Sea Ice. Biology 2012, 1, 542–556. [Google Scholar] [CrossRef]
- Wilkins, D.; Yau, S.; Williams, T.J.; Allen, M.A.; Brown, M.V.; DeMaere, M.Z.; Lauro, F.M.; Cavicchioli, R. Key microbial drivers in Antarctic aquatic environments. FEMS Microbiol. Rev. 2013, 37, 303–335. [Google Scholar] [CrossRef]
- Parker, M.S.; Mock, T.; Armbrust, E.V. Genomic insights into marine microalgae. Annu. Rev. Genet. 2008, 42, 619–645. [Google Scholar] [CrossRef]
- Bowler, C.; Vardi, A.; Allen, A.E. Oceanographic and biogeochemical insights from diatom genomes. J. Rev. Mar. Sci. 2010, 2, 333–365. [Google Scholar] [CrossRef]
- Allen, A.E.; LaRoche, J.; Maheswari, U.; Lommer, M.; Schauer, N.; Lopez, P.J.; Finazzi, G.; Fernie, A.R.; Bowler, C. Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. PNAS 2008, 105, 10438–10443. [Google Scholar] [CrossRef]
- Tomanek, L. Environmental proteomics: Changes in the proteome of marine organisms in response to environmental stress, pollutants, infection, symbiosis, and development. J. Rev. Mar. Sci. 2011, 3, 373–399. [Google Scholar] [CrossRef]
- Weber, A.P.M.; Horst, R.J.; Barbier, G.G.; Oesterhelt, C. Metabolism and metabolomics of eukaryotes living under extreme conditions. In International Review of Cytology; Kwang, W.J., Ed.; Academic Press: Salt Lake City, UT, USA, 2007; Volume 256, pp. 1–34. [Google Scholar]
- Bluhm, B.A.; Gebruk, A.V.; Gradinger, R.; Hopcroft, R.R.; Huettmann, F.; Kosobokova, K.N.; Sirenko, B.I.; Weslawski, J.M. Arctic marine biodiversity: An update of species richness and examples of biodiversity change. Oceanography 2011, 24, 232. [Google Scholar] [CrossRef]
- Lizotte, M.P. The contributions of sea ice algae to Antarctic marine primary production. Am. Zool. 2001, 41, 57–73. [Google Scholar] [CrossRef]
- Zachos, J.; Pagani, M.; Sloan, L.; Thomas, E.; Billups, K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 2001, 292, 686–693. [Google Scholar] [CrossRef]
- Comiso, J.C. Large-scale characteristics and variability of the global sea ice cover. In Sea Ice: An Introduction to Its Physics, Chemistry, Biology, and Geology; Thomas, D.N., Dieckmann, G.S., Eds.; Blackwell Science Ltd.: Oxford, UK, 2003; pp. 112–142. [Google Scholar]
- Sabine, C.L.; Feely, R.A.; Gruber, N.; Key, R.M.; Lee, K.; Bullister, J.L.; Wanninkhof, R.; Wong, C.S.; Wallace, D.W.R.; Tilbrook, B.; Millero, F.J.; Peng, T.-H.; Kozyr, A.; Ono, T.; Rios, A.F. The Oceanic Sink for Anthropogenic CO2. Science 2004, 305, 367–371. [Google Scholar] [CrossRef] [Green Version]
- Dittmar, T.; Kattner, G. The biogeochemistry of the river and shelf ecosystem of the Arctic Ocean: a review. Mar. Chem. 2003, 83, 103–120. [Google Scholar] [CrossRef]
- Shakhova, N.; Semiletov, I.; Salyuk, A.; Yusupov, V.; Kosmach, D.; Gustafsson, Ö. Extensive methane venting to the atmosphere from sediments of the East Siberian Arctic Shelf. Science 2010, 327, 1246–1250. [Google Scholar] [CrossRef]
- Kiene, R.; Kieber, D.; Slezak, D.; Toole, D.; del Valle, D.; Bisgrove, J.; Brinkley, J.; Rellinger, A. Distribution and cycling of dimethylsulfide, dimethylsulfoniopropionate, and dimethylsulfoxide during spring and early summer in the Southern Ocean south of New Zealand. Aquat. Sci. 2007, 69, 305–319. [Google Scholar] [CrossRef]
- Gunson, J.R.; Spall, S.A.; Anderson, T.R.; Jones, A.; Totterdell, I.J.; Woodage, M.J. Climate sensitivity to ocean dimethylsulphide emissions. Geophys. Res. Lett. 2006, 33. [Google Scholar] [CrossRef]
- Teoh, M.-L.; Phang, S.-M.; Chu, W.-L. Response of Antarctic, temperate, and tropical microalgae to temperature stress. J. Appl. Phycol. 2012, 1, 1–13. [Google Scholar]
- Thomson, P.G.; Wright, S.W.; Bolch, C.J.S.; Nichols, P.D.; Skerratt, J.H.; McMinn, A. Antarctic distribution, pigment and lipid composition, and molecular identification of the brine dinoflagellate Polarella glacialis (Dinophyceae). J. Phycol. 2004, 40, 867–873. [Google Scholar] [CrossRef]
- Osipova, S.; Dudareva, L.; Bondarenko, N.; Nasarova, A.; Sokolova, N.; Obolkina, L.; Glyzina, O.; Timoshkin, O. Temporal variation in fatty acid composition of Ulothrix Zonata (Chlorophyta) from ice and benthic communities of Lake Baikal. Phycologia 2009, 48, 130–135. [Google Scholar] [CrossRef]
- Fogliano, V.; Andreoli, C.; Martello, A.; Caiazzo, M.; Lobosco, O.; Formisano, F.; Carlino, P.A.; Meca, G.; Graziani, G.; Rigano, V.D.M.; Vona, V.; Carfagna, S.; Rigano, C. Functional ingredients produced by culture of Koliella antarctica. Aquaculture 2010, 299, 115–120. [Google Scholar] [CrossRef]
- Chen, Z.; He, C.; Hu, H. Temperature responses of growth, photosynthesis, fatty acid and nitrate reductase in Antarctic and temperate Stichococcus. Extremophiles 2012, 16, 127–133. [Google Scholar] [CrossRef]
- Mock, T.; Kroon, B.M.A. Photosynthetic energy conversion under extreme conditions—I: Important role of lipids as structural modulators and energy sink under N-limited growth in Antarctic sea ice diatoms. Phytochemistry 2002, 61, 41–51. [Google Scholar] [CrossRef]
- Mock, T.; Kroon, B.M.A. Photosynthetic energy conversion under extreme conditions—II: The significance of lipids under light limited growth in Antarctic sea ice diatoms. Phytochemistry 2002, 61, 53–60. [Google Scholar] [CrossRef]
- Gray, C.G.; Lasiter, A.D.; Leblond, J.D. Mono- and digalactosyldiacylglycerol composition of dinoflagellates. III. Four cold-adapted, peridinin-containing taxa and the presence of trigalactosyldiacylglycerol as an additional glycolipid. Eur. J. Phycol. 2009, 44, 439–445. [Google Scholar] [CrossRef]
- Morgan-Kiss, R.; Ivanov, A.G.; Williams, J.; Mobashsher, K.; Huner, N.P.A. Differential thermal effects on the energy distribution between photosystem II and photosystem I in thylakoid membranes of a psychrophilic and a mesophilic alga. Biochim. Biophys. Acta 2002, 1561, 251–265. [Google Scholar] [CrossRef]
- Blanc, G.; Agarkova, I.; Grimwood, J.; Kuo, A.; Brueggeman, A.; Dunigan, D.D.; Gurnon, J.; Ladunga, I.; Lindquist, E.; Lucas, S.; Pangilinan, J.; Proschold, T.; Salamov, A.; Schmutz, J.; Weeks, D.; Yamada, T.; Lomsadze, A.; Borodovsky, M.; Claverie, J.M.; Grigoriev, I.V.; Van Etten, J.L. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 2012, 13. [Google Scholar] [CrossRef]
- Suga, K.; Honjoh, K.-I.; Furuya, N.; Shimizu, H.; Nishi, K.; Shinohara, F.; Hirabaru, Y.; Maruyama, I.; Miyamoto, T.; Hatano, S.; Iio, M. Two low-temperature-inducible Chlorella genes for Δ-12 and omega-3 fatty acid desaturase (FAD): Isolation of Δ-12 and omega-3 fad cDNA clones. Biosci. Biotechnol. Biochem. 2002, 66, 1314–1327. [Google Scholar] [CrossRef]
- An, M.; Mou, S.; Zhang, X.; Ye, N.; Zheng, Z.; Cao, S.; Xu, D.; Fan, X.; Wang, Y.; Miao, J. Temperature regulates fatty acid desaturases at a transcriptional level and modulates the fatty acid profile in the Antarctic microalga Chlamydomonas sp. ICE-L. Bioresour. Technol. 2013, 134, 151–157. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, S.; Cong, B.; Wu, G.; Liu, C.; Lin, X.; Shen, J.; Huang, X. A novel omega-3 fatty acid desaturase involved in acclimation processes of polar condition from Antarctic ice algae Chlamydomonas sp. ICE-L. Mar. Biotechnol. 2011, 13, 393–401. [Google Scholar] [CrossRef]
- An, M.; Mou, S.; Zhang, X.; Zheng, Z.; Ye, N.; Wang, D.; Zhang, W.; Miao, J. Expression of fatty acid desaturase genes and fatty acid accumulation in Chlamydomonas sp. ICE-L under salt stress. Bioresour. Technol. 2013, 149, 77–83. [Google Scholar] [CrossRef]
- Priscu, J.; Palmisano, A.; Priscu, L.; Sullivan, C. Temperature dependence of inorganic nitrogen uptake and assimilation in Antarctic sea-ice microalgae. Polar Biol. 1989, 9, 443–446. [Google Scholar] [CrossRef]
- Di Martino Rigano, V.; Vona, V.; Lobosco, O.; Carillo, P.; Lunn, J.E.; Carfagna, S.; Esposito, S.; Caiazzo, M.; Rigano, C. Temperature dependence of nitrate reductase in the psychrophilic unicellular alga Koliella antarctica and the mesophilic alga Chlorella sorokiniana. Plant Cell Environ. 2006, 29, 1400–1409. [Google Scholar] [CrossRef]
- Vona, V.; Di Martino Rigano, V.; Lobosco, O.; Carfagna, S.; Esposito, S.; Rigano, C. Temperature responses of growth, photosynthesis, respiration and NADH: Nitrate reductase in cryophilic and mesophilic algae. New Phytol. 2004, 163, 325–331. [Google Scholar] [CrossRef]
- Ferrara, M.; Guerriero, G.; Cardi, M.; Esposito, S. Purification and biochemical characterisation of a glucose-6-phosphate dehydrogenase from the psychrophilic green alga Koliella antarctica. Extremophiles 2012, 17, 53–62. [Google Scholar]
- Lyon, B.R.; Lee, P.A.; Bennett, J.M.; DiTullio, G.R.; Janech, M.G. Proteomic analysis of a sea-ice diatom: Salinity acclimation provides new insight into the dimethylsulfoniopropionate production pathway. Plant Physiol. 2011, 157, 1926–1941. [Google Scholar] [CrossRef]
- Strauss, J.; Gao, S.; Morrissey, J.; Bowler, C.; Nagel, G.; Mock, T. A light-driven rhodopsin proton pump from the psychrophilic diatom Fragilariopsis cylindrus. In Proceeding of EMBO Workshop: The Molecular Life of Diatoms, Paris, France, 25–28 June 2013.
- Marchetti, A.; Schruth, D.M.; Durkin, C.A.; Parker, M.S.; Kodner, R.B.; Berthiaume, C.T.; Morales, R.; Allen, A.E.; Armbrust, E.V. Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. PNAS 2012, 109, E317–E325. [Google Scholar] [CrossRef]
- Devos, N.; Ingouff, M.; Loppes, R.; Matagne, R.F. RUBISCO adaptation to low temperatures: A comparative study in psychrophilic and mesophilic unicellular algae. J. Phycol. 1998, 34, 655–660. [Google Scholar]
- Morgan, R.M.; Ivanov, A.G.; Priscu, J.C.; Maxwell, D.P.; Huner, N.P.A. Structure and composition of the photochemical apparatus of the antarctic green alga, Chlamydomonas subcaudata. Photosynth. Res. 1998, 56, 303–314. [Google Scholar] [CrossRef]
- Napolitano, M.J.; Shain, D.H. Distinctions in adenylate metabolism among organisms inhabiting temperature extremes. Extremophiles 2005, 9, 93–98. [Google Scholar] [CrossRef]
- Toseland, A.D.S.J.; Clark, J.R.; Kirkham, A.; Strauss, J.; Uhlig, C.; Lenton, T.M.; Valentin, K.; Pearson, G.A.; Moulton, V.; Mock, T. The impact of temperature on marine phytoplankton resource allocation and metabolism. Nat. Clim. Change 2013, 3, 979–984. [Google Scholar] [CrossRef]
- Welsh, D.T. Ecological significance of compatible solute accumulation by micro-organisms: From single cells to global climate. FEMS Microbiol. Rev. 2000, 24, 263–290. [Google Scholar] [CrossRef]
- Krell, A. Salt stress tolerance in the psychrophilic diatom Fragilariopsis cylindrus. Ph.D. Thesis, University of Bremen, Bremen, Germany, 2006. [Google Scholar]
- Waditee, R.; Bhuiyan, M.N.H.; Rai, V.; Aoki, K.; Tanaka, Y.; Hibino, T.; Suzuki, S.; Takano, J.; Jagendorf, A.T.; Takabe, T.; Takabe, T. Genes for direct methylation of glycine provide high levels of glycinebetaine and abiotic-stress tolerance in Synechococcus and Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 1318–1323. [Google Scholar] [CrossRef]
- DiTullio, G.R.; Garrison, D.L.; Mathot, S. Dimethylsulfonioproprionate in sea ice algae from the Ross Sea polynya. In Antarctic Sea Ice: Biological Processes, Interactions and Variability; Arrigo, K.R., Lizotte, M.P., Eds.; American Geophysical Union: Washington, DC, USA, 1998; pp. 139–146. [Google Scholar]
- Keller, M.D.; Bellows, W.K.; Guillard, R.R.L. Dimethyl sulfide production in marine phytoplankton. In Biogenic Sulfur in the Environment; Saltzman, E.S., Cooper, W.J., Eds.; American Chemical Society: Washington, DC, USA, 1989; pp. 131–142. [Google Scholar]
- Nishiguchi, M.K.; Somero, G.N. Temperature- and concentration-dependence of compatibility of the organic osmolyte [beta]-dimethylsulfoniopropionate. Cryobiology 1992, 29, 118–124. [Google Scholar] [CrossRef]
- Gage, D.A.; Rhodes, D.; Nolte, K.D.; Hicks, W.A.; Leustek, T.; Cooper, A.J.L.; Hanson, A.D. A new route for synthesis of dimethylsulphoniopropionate in marine algae. Nature 1997, 387, 891–894. [Google Scholar] [CrossRef]
- Tunnacliffe, A.; Wise, M.J. The continuing conundrum of the LEA proteins. Naturwissenschaften 2007, 94, 791–812. [Google Scholar] [CrossRef]
- Honjoh, K.-I.; Yoshimoto, M.; Joh, T.; Kajiwara, T.; Miyamoto, T.; Hatano, S. Isolation and characterization of hardening-induced proteins in Chlorella vulgaris C-27: Identification of late embryogenesis abundant proteins. Plant Cell Physiol. 1995, 36, 1421–1430. [Google Scholar]
- Liu, X.; Wang, Y.; Gao, H.; Xu, X. Identification and characterization of genes encoding two novel LEA proteins in Antarctic and temperate strains of Chlorella vulgaris. Gene 2011, 482, 51–58. [Google Scholar] [CrossRef]
- Lyon, B.R.; Medical University of South Carolina-Hollings Marine Lab, Charleston, SC, USA. Unpublished work. 2011.
- Mock, T.; Krell, A.; Glockner, G.; Kolukisaoglu, U.; Valentin, K. Analysis of expressed sequence tags (ESTs) from the polar diatom Fragilariopsis cylindrus. J. Phycol. 2005, 42, 78–85. [Google Scholar]
- Raymond, J.A. Algal ice-binding proteins change the structure of sea ice. PNAS 2011, 108, E198. [Google Scholar] [CrossRef]
- Janech, M.G.; Krell, A.; Mock, T.; Kang, J.S.; Raymond, J.A. Ice-binding proteins from sea ice diatoms (Bacillariophyceae). J. Phycol. 2006, 42, 410–416. [Google Scholar] [CrossRef] [Green Version]
- Raymond, J.A.; Kim, H.J. Possible role of horizontal gene transfer in the colonization of sea ice by algae. PLoS ONE 2012, 7, e35968. [Google Scholar] [CrossRef]
- Raymond, J.A.; Morgan-Kiss, R. Separate origins of ice-binding proteins in Antarctic Chlamydomonas species. PLoS ONE 2013, 8, e59186. [Google Scholar] [CrossRef]
- Krembs, C.; Eicken, H.; Junge, K.; Deming, J.W. High concentrations of exopolymeric substances in Arctic winter sea ice: Implications for the polar ocean carbon cycle and cryoprotection of diatoms. Deep Sea Res. Part I 2002, 49, 2163–2181. [Google Scholar]
- Krembs, C.; Eicken, H.; Deming, J.W. Exopolymer alteration of physical properties of sea ice and implications for ice habitability and biogeochemistry in a warmer Arctic. PNAS 2011, 108, 3653–3658. [Google Scholar] [CrossRef]
- Mock, T.; Gradinger, R. Determination of Arctic ice algal production with a new in situ incubation technique. Mar. Ecol. Prog. Ser. 1999, 177, 15–26. [Google Scholar] [CrossRef]
- Cota, G.F. Photoadaptation of high Arctic ice algae. Nature 1985, 315, 219–222. [Google Scholar] [CrossRef]
- Jung, G.; Lee, C.G.; Kang, S.H.; Jin, E. Annotation and expression profile analysis of cDNAs from the Antarctic diatom Chaetoceros neogracile. J. Microbiol. Biotechnol. 2007, 17, 1330–1337. [Google Scholar]
- Morgan-Kiss, R.; Ivanov, A.; Modla, S.; Czymmek, K.; Hüner, N.; Priscu, J.; Lisle, J.; Hanson, T. Identity and physiology of a new psychrophilic eukaryotic green alga, Chlorella sp., strain BI, isolated from a transitory pond near Bratina Island, Antarctica. Extremophiles 2008, 12, 701–711. [Google Scholar] [CrossRef]
- Ralph, P.J.; McMinn, A.; Ryan, K.G.; Ashworth, C. Short-term effect of temperature on the photokinetics of microalgae from the surface layers of Antarctic pack ice. J. Phycol. 2005, 41, 763–769. [Google Scholar] [CrossRef]
- Robinson, D.; Kolber, Z.; Sullivan, C. Photophysiology and photoacclimation in surface sea ice algae from McMurdo Sound, Antarctica. Mar. Ecol. Prog. Ser. 1997, 147, 243–256. [Google Scholar] [CrossRef]
- Lepetit, B.; Sturm, S.; Rogato, A.; Gruber, A.; Sachse, M.; Falciatore, A.; Kroth, P.G.; Lavaud, J. High light acclimation in the secondary plastids containing diatom Phaeodactylum tricornutum is triggered by the redox state of the plastoquinone pool. Plant. Physiol. 2013, 161, 853–865. [Google Scholar] [CrossRef]
- Green, B.; Alami, M.; Zhu, S.; Guo, J.; Maldonado, M. The LHC superfamily and the complex roles of its members in photoacclimation. In Proceedings of EMBO Workshop: The Molecular Life of Diatoms, Paris, France, 25–28 June 2013.
- Park, S.; Jung, G.; Hwang, Y.-s.; Jin, E. Dynamic response of the transcriptome of a psychrophilic diatom, Chaetoceros neogracile, to high irradiance. Planta 2010, 231, 349–360. [Google Scholar] [CrossRef]
- Mock, T.; Hoch, N. Long-term temperature acclimation of photosynthesis in steady-state cultures of the polar diatom Fragilariopsis cylindrus. Photosynth. Res. 2005, 85, 307–317. [Google Scholar] [CrossRef]
- Szyszka, B.; Ivanov, A.G.; Huner, N.P. Psychrophily is associated with differential energy partitioning, photosystem stoichiometry and polypeptide phosphorylation in Chlamydomonas raudensis. Biochimica et Biophysica Acta 2007, 1767, 789–800. [Google Scholar] [CrossRef]
- Takizawa, K.; Takahashi, S.; Huner, N.P.; Minagawa, J. Salinity affects the photoacclimation of Chlamydomonas raudensis Ettl UWO241. Photosynth. Res. 2009, 99, 195–203. [Google Scholar] [CrossRef]
- Ryan, K.G.; McMinn, A.; Hegseth, E.N.; Davy, S.K. The effects of ultraviolet-b radiation on Antarctic sea-ice algae. J. Phycol. 2012, 48, 74–84. [Google Scholar] [CrossRef]
- Miao, J.; Li, G.; Hou, X.; Zhang, Y.; Jiang, Y.; Wang, B.; Zhang, B. Study on induced synthesis of anti-UV substances in the Antarctic algae. High. Tech. Lett. 2002, 6, 179–183. [Google Scholar]
- Obertegger, U.; Camin, F.; Guella, G.; Flaim, G. Adaptation of a psychrophilic freshwater dinoflagellate to ultraviolet radiation. J. Phycol. 2011, 47, 811–820. [Google Scholar] [CrossRef]
- Schriek, R. Effects of light and temperature on the enzymatic antioxidative defense systems in the Antarctic ice diatom Entomoneis kufferathii Manguin. Rep. Polar Res. 2000, 349, 1–130. [Google Scholar]
- Janknegt, P.J.; Van De Poll, W.H.; Visser, R.J.W.; Rijstenbil, J.W.; Buma, A.G.J. Oxidative stress responses in the marine antarctic diatom Chaetoceros brevis (bacillariophyceae) during photoacclimation. J. Phycol. 2008, 44, 957–966. [Google Scholar] [CrossRef]
- Janknegt, P.J.; De Graaff, C.M.; Van De Poll, W.H.; Visser, R.J.W.; Rijstenbil, J.W.; Buma, A.G.J. Short-term antioxidative responses of 15 microalgae exposed to excessive irradiance including ultraviolet radiation. Eur. J. Phycol. 2009, 44, 525–539. [Google Scholar] [CrossRef]
- Hwang, Y.-S.; Jung, G.; Jin, E. Transcriptome analysis of acclimatory responses to thermal stress in Antarctic algae. Biochem. Biophys. Res. Commun. 2008, 367, 635–641. [Google Scholar] [CrossRef]
- Sunda, W.; Kieber, D.J.; Kiene, R.P.; Huntsman, S. An antioxidant function for DMSP and DMS in marine algae. Nature 2002, 418, 317–320. [Google Scholar] [CrossRef]
- Chen, C.; Dickman, M.B. Proline suppresses apoptosis in the fungal pathogen Colletotrichum trifolii. Proc. Natl. Acad. Sci. USA 2005, 102, 3459–3464. [Google Scholar] [CrossRef]
- Hünken, M.; Harder, J.; Kirst, G.O. Epiphytic bacteria on the Antarctic ice diatom Amphiprora kufferathii Manguin cleave hydrogen peroxide produced during algal photosynthesis. Plant. Biol. 2008, 10, 519–526. [Google Scholar] [CrossRef]
- Kan, G.-F.; Miao, J.-L.; Shi, C.-J.; Li, G.-Y. Proteomic alterations of Antarctic ice microalga Chlamydomonas sp. under low-temperature stress. J. Integr. Plant Biol. 2006, 48, 965–970. [Google Scholar] [CrossRef]
- Kwon, S.J.; Kwon, S.I.; Bae, M.S.; Cho, E.J.; Park, O.K. Role of the methionine sulfoxide reductase MsrB3 in cold acclimation in Arabidopsis. Plant Cell. Physiol. 2007, 48, 1713–1723. [Google Scholar] [CrossRef]
- Kirch, H.-H.; Bartels, D.; Wei, Y.; Schnable, P.S.; Wood, A.J. The ALDH gene superfamily of Arabidopsis. Trends Plant Sci. 2004, 9, 371–377. [Google Scholar] [CrossRef]
- Peters, E. Prolonged darkness and diatom mortality: II. Marine temperate species. J. Exp. Mar. Biol. Ecol. 1996, 207, 43–58. [Google Scholar] [CrossRef]
- Peters, E.; Thomas, D.N. Prolonged darkness and diatom mortality I: Marine Antarctic species. J. Exp. Mar. Biol. Ecol. 1996, 207, 25–41. [Google Scholar] [CrossRef]
- van Oijen, T.; Leeuwe, M.; Gieskes, W.C. Variation of particulate carbohydrate pools over time and depth in a diatom-dominated plankton community at the Antarctic Polar Front. Polar Biol. 2003, 26, 195–201. [Google Scholar]
- Palmisano, A.; Garrison, D. Microorganisms in Antarctic sea ice. In Antarctic Microbiology; Friedmann, E., Ed.; Wiley-Liss: New York, NY, USA, 1993; pp. 167–218. [Google Scholar]
- Neven, I.A.; Stefels, J.; van Heuven, S.M.A.C.; de Baar, H.J.W.; Elzenga, J.T.M. High plasticity in inorganic carbon uptake by Southern Ocean phytoplankton in response to ambient CO2. Deep Sea Res. Part II 2011, 58, 2636–2646. [Google Scholar] [CrossRef]
- Armbrust, E.V.; Berges, J.A.; Bowler, C.; Green, B.R.; Martinez, D.; Putnam, N.H.; Zhou, S.; Allen, A.E.; Apt, K.E.; Bechner, M.; Brzezinski, M.A.; Chaal, B.K.; Chiovitti, A.; Davis, A.K.; Demarest, M.S.; Detter, J.C.; Glavina, T.; Goodstein, D.; Hadi, M.Z.; Hellsten, U.; Armbrust, E.V. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolis. Science 2004, 306, 79–86. [Google Scholar] [CrossRef]
- Doucette, G.J.; Fryxell, G.A. Thalassiosira antarctica: vegetative and resting stage chemical composition of an ice-related marine diatom. Mar. Biol. 1983, 78, 1–6. [Google Scholar] [CrossRef]
- Mock, T.; Valentin, K. Photosynthesis and cold acclimation: Molecular evidence from a polar diatom. J. Phycol. 2004, 40, 732–741. [Google Scholar] [CrossRef]
- Baldisserotto, C.; Ferroni, L.; Moro, I.; Fasulo, M.P.; Pancaldi, S. Modulations of the thylakoid system in snow xanthophycean alga cultured in the dark for two months: comparison between microspectrofluorimetric responses and morphological aspects. Protoplasma 2005, 226, 125–135. [Google Scholar] [CrossRef]
- Ferroni, L.; Baldisserotto, C.; Zennaro, V.; Soldani, C.; Fasulo, M.P.; Pancaldi, S. Acclimation to darkness in the marine chlorophyte Koliella antarctica cultured under low salinity: hypotheses on its origin in the polar environment. Eur. J. Phycol. 2007, 42, 91–104. [Google Scholar] [CrossRef]
- Zaslavskaia, L.A.; Lippmeier, J.C.; Kroth, P.G.; Grossman, A.R.; Apt, K.E. Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. J. Phycol. 2000, 36, 379–386. [Google Scholar]
- Bachy, C.; Lopez-Garcia, P.; Vereshchaka, A.; Moreira, D. Diversity and vertical distribution of microbial eukaryotes in the snow, sea ice and seawater near the North Pole at the end of the polar night. Front. Microbiol. 2011, 2. [Google Scholar] [CrossRef]
- Duarte, R.T.D.; Nóbrega, F.; Nakayama, C.R.; Pellizari, V.H. Brazilian research on extremophiles in the context of astrobiology. Int. J. Astrobiol. 2012, 11, 325–333. [Google Scholar] [CrossRef]
- Homepage of Fragilariopsis cylindrus Genome. Available online: http://genome.jgi-psf.org/Fracy1/Fracy1.home.html (accessed on 7 September 2013).
- Marine MIcrobial Eukaryote Transcriptome Sequencing Project. Available online: www.marinemicroeukaryotes.org (accessed on 7 September 2013).
- Yoon, H.S.; Price, D.C.; Stepanauskas, R.; Rajah, V.D.; Sieracki, M.E.; Wilson, W.H.; Yang, E.C.; Duffy, S.; Bhattacharya, D. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science 2011, 332, 714–717. [Google Scholar] [CrossRef]
- Lovejoy, C.; Massana, R.; Pedros-Alio, C. Diversity and distribution of marine microbial eukaryotes in the Arctic Ocean and adjacent seas. Appl. Environ. Microbiol. 2006, 72, 3085–3095. [Google Scholar] [CrossRef] [Green Version]
- Amaral-Zettler, L.A.; McCliment, E.A.; Ducklow, H.W.; Huse, S.M. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE 2009, 4, e6372. [Google Scholar] [CrossRef]
- Charvet, S.; Vincent, W.; Lovejoy, C. Chrysophytes and other protists in High Arctic lakes: molecular gene surveys, pigment signatures and microscopy. Polar Biol. 2012, 35, 733–748. [Google Scholar] [CrossRef]
- Piquet, A.M.T.; Bolhuis, H.; Davidson, A.T.; Thomson, P.G.; Buma, A.G.J. Diversity and dynamics of Antarctic marine microbial eukaryotes under manipulated environmental UV radiation. FEMS Microbiol. Ecol. 2008, 66, 352–366. [Google Scholar] [CrossRef]
- Potvin, M.; Lovejoy, C. PCR-based diversity estimates of artificial and environmental 18S rRNA gene libraries. J. Eukaryotic Microbiol. 2009, 56, 174–181. [Google Scholar] [CrossRef]
- Zhu, F.; Massana, R.; Not, F.; Marie, D.; Vaulot, D. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol. Ecol. 2005, 52, 79–92. [Google Scholar] [CrossRef]
- Malviya, S.; Veluchamy, A.; Bittner, L.; Tanaka, A.; Bowler, C. Comprehensive biogeographical insights into the complexity of marine diatom communities. In EMBO Workshop: The Molecular Life of Diatoms, Paris, France, 25–28 June 2013.
- Poulsen, N.; Chesley, P.M.; Kröger, N. Molecular genetic manipulation of the diatom Thalassiosira pseudonana (bacillariophyceae). J. Phycol. 2006, 42, 1059–1065. [Google Scholar] [CrossRef]
- De Riso, V.; Raniello, R.; Maumus, F.; Rogato, A.; Bowler, C.; Falciatore, A. Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 2009, 37, e96. [Google Scholar] [CrossRef]
- Pocock, T.; Vetterli, A.; Falk, S. Evidence for phenotypic plasticity in the Antarctic extremophile Chlamydomonas raudensis Ettl. UWO 241. J. Exp. Bot. 2011, 62, 1169–1177. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Braff, J.; Karl, D.M.; DeLong, E.F. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific Subtropical Gyre. Appl. Environ. Microbiol. 2009, 75, 5345–5355. [Google Scholar] [CrossRef]
- Veluchamy, A.; Lin, X.; Maumus, F.; Rivarola, M.; Bhavsar, J.; Creasy, T.; O’Brien, K.; Sengamalay, N.A.; Tallon, L.J.; Smith, A.D.; Rayko, E.; Ahmed, I.; Crom, S.L.; Farrant, G.K.; Sgro, J.-Y.; Olson, S.A.; Bondurant, S.S.; Allen, A.; Rabinowicz, P.D.; Sussman, M.R.; Bowler, C.; Tirichine, L. Insights into the role of DNA methylation in diatoms by genome-wide profiling in Phaeodactylum tricornutum. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef]
- Maumus, F.; Allen, A.E.; Mhiri, C.; Hu, H.; Jabbari, K.; Vardi, A.; Grandbastien, M.A.; Bowler, C. Potential impact of stress activated retrotransposons on genome evolution in a marine diatom. BMC Genomics 2009, 10, 624. [Google Scholar] [CrossRef]
- Richards, E.J. Inherited epigenetic variation—Revisiting soft inheritance. Nat. Rev. Genet. 2006, 7, 395–401. [Google Scholar] [CrossRef]
- Norden-Krichmar, T.M.; Allen, A.E.; Gaasterland, T.; Hildebrand, M. Characterization of the small RNA transcriptome of the diatom, Thalassiosira pseudonana. PLoS ONE 2011, 6, e22870. [Google Scholar]
- Anesio, A.M.; Bellas, C.M. Are low temperature habitats hot spots of microbial evolution driven by viruses? Trends Microbiol. 2011, 19, 52–57. [Google Scholar] [CrossRef]
- Pearce, I.; Davidson, A.T.; Bell, E.M.; Wright, S. Seasonal changes in the concentration and metabolic activity of bacteria and viruses at an Antarctic coastal site. Aquat. Microb. Ecol. 2007, 47, 11–23. [Google Scholar] [CrossRef]
- Yau, S.; Lauro, F.M.; DeMaere, M.Z.; Brown, M.V.; Thomas, T.; Raftery, M.J.; Andrews-Pfannkoch, C.; Lewis, M.; Hoffman, J.M.; Gibson, J.A.; Cavicchioli, R. Virophage control of antarctic algal host–virus dynamics. PNAS 2011, 108, 6163–6168. [Google Scholar] [CrossRef]
- Vardi, A.; Haramaty, L.; Van Mooy, B.A.; Fredricks, H.F.; Kimmance, S.A.; Larsen, A.; Bidle, K.D. Host-virus dynamics and subcellular controls of cell fate in a natural coccolithophore population. Proc. Natl. Acad. Sci. USA 2012, 109, 19327–19332. [Google Scholar]
- Kroth, P.; Windler, M.; Leinweber, K.; Schulze, B.; Spiteller, D.; Buhmann, M. Interactions of diatoms and bacteria in biofilms. In Proceedings of EMBO Workshop: The Molecular Life of Diatoms, Paris, France, 25–28 June 2013.
- Amin, S.; Hmelo, L.; Parsek, M.; Armbrust, E.V. Multiple complex interactions between a toxigenic diatom and an associated bacterium revealed using whole cell transcriptomics. In Proceedings of EMBO: The Molecular Life of Diatoms, Paris, France, 25–28 June 2013.
- Vardi, A.; Bidle, K.D.; Kwityn, C.; Hirsh, D.J.; Thompson, S.M.; Callow, J.A.; Falkowski, P.; Bowler, C. A diatom gene regulating nitric-oxide signaling and susceptibility to diatom-derived aldehydes. Curr. Biol. 2008, 18, 895–899. [Google Scholar] [CrossRef]
- Caldwell, G.S. The influence of bioactive oxylipins from marine diatoms on invertebrate reproduction and development. Mar. Drugs 2009, 7, 367–400. [Google Scholar] [CrossRef]
- Smetacek, V.; Nicol, S. Polar ocean ecosystems in a changing world. Nature 2005, 437, 362–368. [Google Scholar] [CrossRef]
- Tremblay, J.-É.; Robert, D.; Varela, D.; Lovejoy, C.; Darnis, G.; Nelson, R.; Sastri, A. Current state and trends in Canadian Arctic marine ecosystems: I. Primary production. Clim. Change 2012, 115, 161–178. [Google Scholar] [CrossRef]
- Arrigo, K.R.; Perovich, D.K.; Pickart, R.S.; Brown, Z.W.; van Dijken, G.L.; Lowry, K.E.; Mills, M.M.; Palmer, M.A.; Balch, W.M.; Bahr, F.; Bates, N.R.; Benitez-Nelson, C.; Bowler, B.; Brownlee, E.; Ehn, J.K.; Frey, K.E.; Garley, R.; Laney, S.R.; Lubelczyk, L.; Mathis, J.; Matsuoka, A.; Mitchell, B.G.; Moore, G.W.K.; Ortega-Retuerta, E.; Pal, S.; Polashenski, C.M.; Reynolds, R.A.; Schieber, B.; Sosik, H.M.; Stephens, M.; Swift, J.H. Massive phytoplankton blooms under Arctic sea ice. Science 2012, 336, 1408. [Google Scholar] [CrossRef]
- Marinov, I.; Doney, S.C.; Lima, I.D. Response of ocean phytoplankton community structure to climate change over the 21st century: partitioning the effects of nutrients, temperature and light. Biogeosciences 2010, 7, 3941–3959. [Google Scholar] [CrossRef]
- Montes-Hugo, M.; Doney, S.C.; Ducklow, H.W.; Fraser, W.; Martinson, D.; Stammerjohn, S.E.; Schofield, O. Recent changes in phytoplankton communities associated with rapid regional climate change along the western Antarctic Peninsula. Science 2009, 323, 1470–1473. [Google Scholar] [CrossRef]
- Karsten, U.; Schlie, C.; Woelfel, J.; Becker, B. Benthic diatoms in Arctic waters -ecological functions and adaptations. Polarforschung 2012, 81, 77–84. [Google Scholar]
- Dunbar, R.B.; Arrigo, K.R.; Lutz, M.; DiTullio, G.R.; Leventer, A.R.; Lizotte, M.P.; Van Woert, M.L.; Robinson, D.H. Non-Redfield production and export of marine organic matter: A recurrent part of the annual cycle in the Ross Sea, Antarctica. Antarct. Sci. Ser. 2003, 78, 179–195. [Google Scholar] [CrossRef]
- Lee, S.H.; Whitledge, T.E.; Kang, S.-H. Spring time production of bottom ice algae in the landfast sea ice zone at Barrow, Alaska. J. Exp. Mar. Biol. Ecol. 2008, 367, 204–212. [Google Scholar] [CrossRef]
- Comeau, A.M.; Li, W.K.W.; Tremblay, J.-É.; Carmack, E.C.; Lovejoy, C. Arctic Ocean microbial community structure before and after the 2007 record sea ice minimum. PLoS ONE 2011, 6, e27492. [Google Scholar]
- Gabric, A.J.; Shephard, J.M.; Knight, J.M.; Jones, G.; Trevena, A.J. Correlations between the satellite-derived seasonal cycles of phytoplankton biomass and aerosol optical depth in the Southern Ocean: Evidence for the influence of sea ice. Glob. Biogeochem. Cycle 2005, 19, 1–10. [Google Scholar]
- Boyd, P.W.; Strzepek, R.; Fu, F.; Hutchinsc, D.A. Environmental control of open-ocean phytoplankton groups: Now and in the future. Limnol. Oceanogr. 2010, 55, 1353–1376. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Lyon, B.R.; Mock, T. Polar Microalgae: New Approaches towards Understanding Adaptations to an Extreme and Changing Environment. Biology 2014, 3, 56-80. https://doi.org/10.3390/biology3010056
Lyon BR, Mock T. Polar Microalgae: New Approaches towards Understanding Adaptations to an Extreme and Changing Environment. Biology. 2014; 3(1):56-80. https://doi.org/10.3390/biology3010056
Chicago/Turabian StyleLyon, Barbara R., and Thomas Mock. 2014. "Polar Microalgae: New Approaches towards Understanding Adaptations to an Extreme and Changing Environment" Biology 3, no. 1: 56-80. https://doi.org/10.3390/biology3010056



