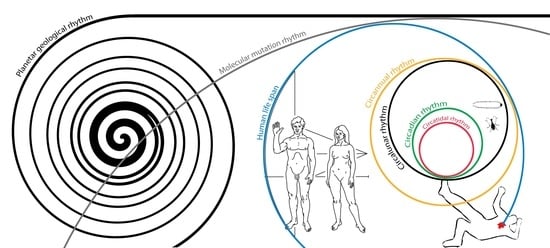
Common Ground between Biological Rhythms and Forensics
Abstract
:Simple Summary
Abstract
1. Introduction
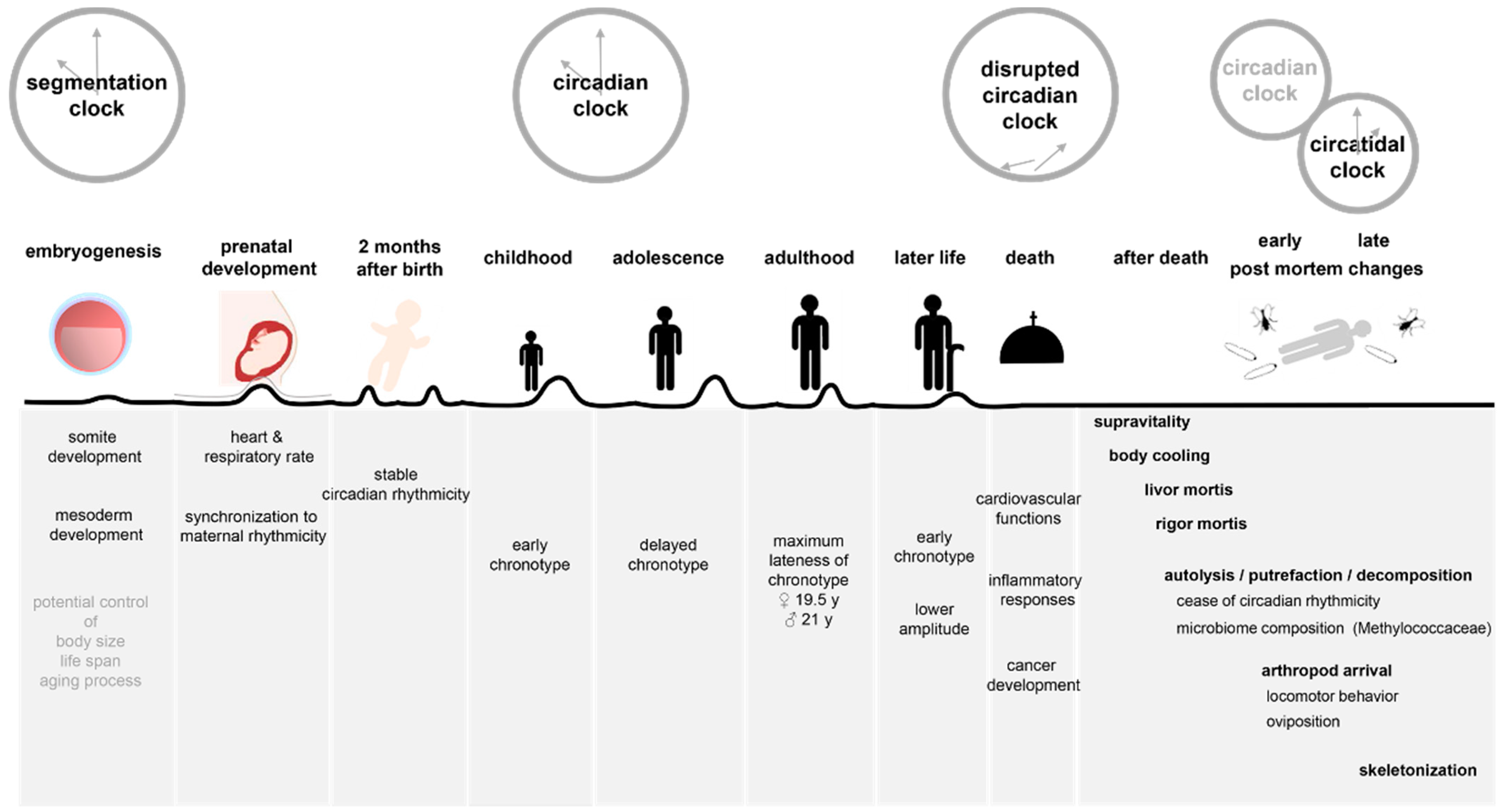
2. Circadian Rhythms after Death
2.1. Assessment of Time of Death and Post-Mortem Interval
2.2. Assessment of Cause of Death
3. Post-Mortem Interval Determination by Biological Rhythms in Forensic Entomology
4. Sexual Dimorphisms of Circadian Rhythms in Human Females and Males
5. Age Determination by Biological Rhythms
5.1. The Epigenetic Clock
5.2. Biological Rhythms in Development
6. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- DeWoskin, D.; Myung, J.; Belle, M.D.C.; Piggins, H.D.; Takumi, T.; Forger, D.B. Distinct roles for GABA across multiple timescales in mammalian circadian timekeeping. Proc. Natl. Acad. Sci. USA 2015, 112, E3911–E3919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- dos Reis, M.; Donoghue, P.C.J.; Yang, Z. Bayesian molecular clock dating of species divergences in the genomics era. Nat. Rev. Genet. 2016, 17, 71–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satoh, A.; Takasu, M.; Yano, K.; Terai, Y. De novo assembly and annotation of the mangrove cricket genome. BMC Res. Notes 2021, 14, 387. [Google Scholar] [CrossRef] [PubMed]
- Roenneberg, T.; Merrow, M. The circadian clock and human health. Curr. Biol. 2016, 26, R432–R443. [Google Scholar] [CrossRef]
- Della Monica, C.; Atzori, G.; Dijk, D.-J. Effects of lunar phase on sleep in men and women in Surrey. J. Sleep Res. 2015, 24, 687–694. [Google Scholar] [CrossRef] [Green Version]
- Hodoglugil, U.; Gunaydin, B.; Yardim, S.; Zengil, H.; Smolensky, M.H. Seasonal variation in the effect of a fixed dose of heparin on activated clotting time in patients prepared for open-heart surgery. Chronobiol. Int. 2001, 18, 865–873. [Google Scholar] [CrossRef]
- Andreatta, G.; Tessmar-Raible, K. The Still Dark Side of the Moon: Molecular Mechanisms of Lunar-Controlled Rhythms and Clocks. J. Mol. Biol. 2020, 432, 3525–3546. [Google Scholar] [CrossRef]
- Rosenwasser, A.M.; Turek, F.W. Neurobiology of circadian rhythm regulation. Sleep Med. Clin. 2015, 10, 403–412. [Google Scholar] [CrossRef]
- Firsov, D.; Bonny, O. Circadian rhythms and the kidney. Nat. Rev. Nephrol. 2018, 14, 626–635. [Google Scholar] [CrossRef]
- Mukherji, A.; Bailey, S.M.; Staels, B.; Baumert, T.F. The circadian clock and liver function in health and disease. J. Hepatol. 2019, 71, 200–211. [Google Scholar] [CrossRef] [Green Version]
- Plikus, M.V.; Van Spyk, E.N.; Pham, K.; Geyfman, M.; Kumar, V.; Takahashi, J.S.; Andersen, B. The circadian clock in skin: Implications for adult stem cells, tissue regeneration, cancer, aging, and immunity. J. Biol. Rhythm. 2015, 30, 163–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, D.; Yang, D.; Lin, L.; Wang, S.; Wu, B. Circadian rhythm in pharmacokinetics and its relevance to chronotherapy. Biochem. Pharmacol. 2020, 178, 114045. [Google Scholar] [CrossRef] [PubMed]
- Verlande, A.; Masri, S. Circadian clocks and cancer: Timekeeping governs cellular metabolism. Trends Endocrinol. Metab. 2019, 30, 445–458. [Google Scholar] [CrossRef]
- Xie, Y.; Tang, Q.; Chen, G.; Xie, M.; Yu, S.; Zhao, J.; Chen, L. New insights into the circadian rhythm and its related diseases. Front. Physiol. 2019, 10, 682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voigt, R.M.; Forsyth, C.B.; Green, S.J.; Engen, P.A.; Keshavarzian, A. Circadian rhythm and the gut microbiome. Int. Rev. Neurobiol. 2016, 131, 193–205. [Google Scholar] [CrossRef]
- Serin, Y.; Acar Tek, N. Effect of circadian rhythm on metabolic processes and the regulation of energy balance. Ann. Nutr. Metab. 2019, 74, 322–330. [Google Scholar] [CrossRef]
- Hergenhan, S.; Holtkamp, S.; Scheiermann, C. Molecular interactions between components of the circadian clock and the immune system. J. Mol. Biol. 2020, 432, 3700–3713. [Google Scholar] [CrossRef]
- Welz, P.-S.; Benitah, S.A. Molecular connections between circadian clocks and aging. J. Mol. Biol. 2020, 432, 3661–3679. [Google Scholar] [CrossRef]
- Rijo-Ferreira, F.; Takahashi, J.S. Genomics of circadian rhythms in health and disease. Genome Med. 2019, 11, 82. [Google Scholar] [CrossRef] [Green Version]
- Mauvoisin, D.; Gachon, F. Proteomics in circadian biology. J. Mol. Biol. 2020, 432, 3565–3577. [Google Scholar] [CrossRef]
- Hernández-Rosas, F.; López-Rosas, C.A.; Saavedra-Vélez, M.V. Disruption of the molecular circadian clock and cancer: An epigenetic link. Biochem. Genet. 2020, 58, 189–209. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, U.; Thakkar, N.; Das, P.; Pal Bhadra, M. Evolution of circadian rhythms: From bacteria to human. Sleep Med. 2017, 35, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Martin, P.; Albarrán, C.; Garcia, P.; Fernandez de Simon, L.; Jesús Iturralde, M.; Fernández-Rodriguez, A.; Atienza, I.; Capilla, J.; García-Hirschfeld, J.; et al. Challenges of DNA profiling in mass disaster investigations. Croat. Med. J. 2005, 46, 540–548. [Google Scholar]
- Turingan, R.S.; Brown, J.; Kaplun, L.; Smith, J.; Watson, J.; Boyd, D.A.; Steadman, D.W.; Selden, R.F. Identification of human remains using Rapid DNA analysis. Int. J. Leg. Med. 2020, 134, 863–872. [Google Scholar] [CrossRef] [Green Version]
- Sangwan, P.; Nain, T.; Singal, K.; Hooda, N.; Sharma, N. Soil as a tool of revelation in forensic science: A review. Anal. Methods 2020, 12, 5150–5159. [Google Scholar] [CrossRef] [PubMed]
- Amendt, J.; Richards, C.S.; Campobasso, C.P.; Zehner, R.; Hall, M.J.R. Forensic entomology: Applications and limitations. Forensic Sci. Med. Pathol. 2011, 7, 379–392. [Google Scholar] [CrossRef]
- Barrea, L.; Vetrani, C.; Altieri, B.; Verde, L.; Savastano, S.; Colao, A.; Muscogiuri, G. The Importance of Being a “Lark” in Post-Menopausal Women with Obesity: A Ploy to Prevent Type 2 Diabetes Mellitus? Nutrients 2021, 13, 3762. [Google Scholar] [CrossRef]
- Wu, H.-S.; Gao, F.; Yan, L.; Given, C. Evaluating chronotypically tailored light therapy for breast cancer survivors: Preliminary findings on fatigue and disrupted sleep. Chronobiol. Int. 2022, 39, 221–232. [Google Scholar] [CrossRef]
- Yang, C.-L.; Tucker, R.M. Snacking behavior differs between evening and morning chronotype individuals but no differences are observed in overall energy intake, diet quality, or food cravings. Chronobiol. Int. 2022, 39, 616–625. [Google Scholar] [CrossRef]
- Gohar, A.A.; Knauert, M.; Kalot, M.A.; Khan, A.; Sider, D.; Javed, M.A.; Wooldridge, D.; Eck, L.; Buckhold, F.; Colaco, B.; et al. Influence of medical trainee sleep pattern (chronotype) on burn-out and satisfaction with work schedules: A multicentre observational study. Postgrad. Med. J. 2021. [Google Scholar] [CrossRef]
- Potvin, J.; Ramos Socarras, L.; Forest, G. Sleeping through a Lockdown: How Adolescents and Young Adults Struggle with Lifestyle and Sleep Habits Upheaval during a Pandemic. Behav. Sleep Med. 2022, 20, 304–320. [Google Scholar] [CrossRef] [PubMed]
- Belfry, K.D.; Deibel, S.H.; Kolla, N.J. Time of day matters: An exploratory assessment of chronotype in a forensic psychiatric hospital. Front. Psychiatry 2020, 11, 550597. [Google Scholar] [CrossRef] [PubMed]
- Marchuk, L.; Sciore, P.; Reno, C.; Frank, C.B.; Hart, D.A. Postmortem stability of total RNA isolated from rabbit ligament, tendon and cartilage. Biochim. Biophys. Acta 1998, 1379, 171–177. [Google Scholar] [CrossRef]
- Johnson, S.A.; Morgan, D.G.; Finch, C.E. Extensive postmortem stability of RNA from rat and human brain. J. Neurosci. Res. 1986, 16, 267–280. [Google Scholar] [CrossRef]
- Heinrich, M.; Matt, K.; Lutz-Bonengel, S.; Schmidt, U. Successful RNA extraction from various human postmortem tissues. Int. J. Leg. Med. 2007, 121, 136–142. [Google Scholar] [CrossRef]
- Albrecht, U.; Eichele, G. The mammalian circadian clock. Curr. Opin. Genet. Dev. 2003, 13, 271–277. [Google Scholar] [CrossRef]
- Preitner, N.; Damiola, F.; Lopez-Molina, L.; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef]
- Kimura, A.; Ishida, Y.; Hayashi, T.; Nosaka, M.; Kondo, T. Estimating time of death based on the biological clock. Int. J. Leg. Med. 2011, 125, 385–391. [Google Scholar] [CrossRef]
- Lim, A.S.P.; Myers, A.J.; Yu, L.; Buchman, A.S.; Duffy, J.F.; De Jager, P.L.; Bennett, D.A. Sex difference in daily rhythms of clock gene expression in the aged human cerebral cortex. J. Biol. Rhythm. 2013, 28, 117–129. [Google Scholar] [CrossRef]
- Ackermann, K.; Dehghani, F.; Bux, R.; Kauert, G.; Stehle, J.H. Day-night expression patterns of clock genes in the human pineal gland. J. Pineal Res. 2007, 43, 185–194. [Google Scholar] [CrossRef]
- Lech, K.; Liu, F.; Ackermann, K.; Revell, V.L.; Lao, O.; Skene, D.J.; Kayser, M. Evaluation of mRNA markers for estimating blood deposition time: Towards alibi testing from human forensic stains with rhythmic biomarkers. Forensic Sci. Int. Genet. 2016, 21, 119–125. [Google Scholar] [CrossRef] [Green Version]
- Na, Y.J.; Sung, J.H.; Lee, S.C.; Lee, Y.J.; Choi, Y.J.; Park, W.Y.; Shin, H.S.; Kim, J.H. Comprehensive analysis of microRNA-mRNA co-expression in circadian rhythm. Exp. Mol. Med. 2009, 41, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Odriozola, A.; Riancho, J.A.; de la Vega, R.; Agudo, G.; García-Blanco, A.; de Cos, E.; Fernández, F.; Sañudo, C.; Zarrabeitia, M.T. miRNA analysis in vitreous humor to determine the time of death: A proof-of-concept pilot study. Int. J. Leg. Med. 2013, 127, 573–578. [Google Scholar] [CrossRef]
- Maiese, A.; Scatena, A.; Costantino, A.; Di Paolo, M.; La Russa, R.; Turillazzi, E.; Frati, P.; Fineschi, V. Micrornas as useful tools to estimate time since death. A systematic review of current literature. Diagnostics 2021, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Manetti, A.C.; Maiese, A.; Baronti, A.; Mezzetti, E.; Frati, P.; Fineschi, V.; Turillazzi, E. Mirnas as new tools in lesion vitality evaluation: A systematic review and their forensic applications. Biomedicines 2021, 9, 1731. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Z.; Bunney, B.G.; Meng, F.; Hagenauer, M.H.; Walsh, D.M.; Vawter, M.P.; Evans, S.J.; Choudary, P.V.; Cartagena, P.; Barchas, J.D.; et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc. Natl. Acad. Sci. USA 2013, 110, 9950–9955. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-Y.; Logan, R.W.; Ma, T.; Lewis, D.A.; Tseng, G.C.; Sibille, E.; McClung, C.A. Effects of aging on circadian patterns of gene expression in the human prefrontal cortex. Proc. Natl. Acad. Sci. USA 2016, 113, 206–211. [Google Scholar] [CrossRef] [Green Version]
- Angerpointner, T.A.; Fischer, M.; Hecker, W.C.; Wolf, H. Time of death in diseases with lethal outcomes in a pediatric surgery department and a pediatric hospital. Mon. Kinderheilkd 1984, 132, 608–611. [Google Scholar]
- Shan, L.; Hofman, M.A.; van Wamelen, D.J.; Van Someren, E.J.W.; Bao, A.-M.; Swaab Dick, F. Diurnal fluctuation in histidine decarboxylase expression, the rate limiting enzyme for histamine production, and its disorder in neurodegenerative diseases. Sleep 2012, 35, 713–715. [Google Scholar] [CrossRef]
- Horne, J.A.; Ostberg, O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. Int. J. Chronobiol. 1976, 4, 97–110. [Google Scholar]
- Lim, A.S.P.; Chang, A.-M.; Shulman, J.M.; Raj, T.; Chibnik, L.B.; Cain, S.W.; Rothamel, K.; Benoist, C.; Myers, A.J.; Czeisler, C.A.; et al. A common polymorphism near PER1 and the timing of human behavioral rhythms. Ann. Neurol. 2012, 72, 324–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, T.H.; Gomez, A.; Singh, H.; Nelson, K.E.; Brinkac, L.M. Integrating the microbiome as a resource in the forensics toolkit. Forensic Sci. Int. Genet. 2017, 30, 141–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olakanye, A.O.; Nelson, A.; Ralebitso-Senior, T.K. A comparative in situ decomposition study using still born piglets and leaf litter from a deciduous forest. Forensic Sci. Int. 2017, 276, 85–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, D.O.; Metcalf, J.L.; Bibat, A.; Knight, R. Seasonal variation of postmortem microbial communities. Forensic Sci. Med. Pathol. 2015, 11, 202–207. [Google Scholar] [CrossRef]
- Douma, L.G.; Gumz, M.L. Circadian clock-mediated regulation of blood pressure. Free Radic. Biol. Med. 2018, 119, 108–114. [Google Scholar] [CrossRef]
- Hartikainen, J.; Tarkiainen, I.; Tahvanainen, K.; Mäntysaari, M.; Länsimies, E.; Pyörälä, K. Circadian variation of cardiac autonomic regulation during 24-h bed rest. Clin. Physiol. 1993, 13, 185–196. [Google Scholar] [CrossRef]
- Scheer, F.A.J.L.; Shea, S.A. Human circadian system causes a morning peak in prothrombotic plasminogen activator inhibitor-1 (PAI-1) independent of the sleep/wake cycle. Blood 2014, 123, 590–593. [Google Scholar] [CrossRef] [Green Version]
- Muller, J.E.; Stone, P.H.; Turi, Z.G.; Rutherford, J.D.; Czeisler, C.A.; Parker, C.; Poole, W.K.; Passamani, E.; Roberts, R.; Robertson, T. Circadian variation in the frequency of onset of acute myocardial infarction. N. Engl. J. Med. 1985, 313, 1315–1322. [Google Scholar] [CrossRef]
- Sakelliadis, E.I.; Katsos, K.D.; Zouzia, E.I.; Vlachodimitropoulos, D.G.; Goutas, N.D.; Spiliopoulou, C.A. Biological rhythms of fatal myocardial infarction in Greece: An autopsy study. Acta Cardiol. 2021, 76, 1092–1099. [Google Scholar] [CrossRef]
- Anderson, T.M.; Allen, K.; Ramirez, J.-M.; Mitchell, E.A. Circadian variation in sudden unexpected infant death in the United States. Acta Paediatr. 2021, 110, 1498–1504. [Google Scholar] [CrossRef]
- Sulli, G.; Lam, M.T.Y.; Panda, S. Interplay between Circadian Clock and Cancer: New Frontiers for Cancer Treatment. Trends Cancer 2019, 5, 475–494. [Google Scholar] [CrossRef] [PubMed]
- Nakahata, Y.; Kaluzova, M.; Grimaldi, B.; Sahar, S.; Hirayama, J.; Chen, D.; Guarente, L.P.; Sassone-Corsi, P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 2008, 134, 329–340. [Google Scholar] [CrossRef] [Green Version]
- Yao, H.; Sundar, I.K.; Huang, Y.; Gerloff, J.; Sellix, M.T.; Sime, P.J.; Rahman, I. Disruption of Sirtuin 1-Mediated Control of Circadian Molecular Clock and Inflammation in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Cell Mol. Biol. 2015, 53, 782–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amendt, J.; Campobasso, C.P.; Gaudry, E.; Reiter, C.; LeBlanc, H.N.; Hall, M.J.R.; European Association for Forensic Entomology. Best practice in forensic entomology--standards and guidelines. Int. J. Leg. Med. 2007, 121, 90–104. [Google Scholar] [CrossRef] [PubMed]
- Cascallares, G.; Riva, S.; Franco, D.L.; Risau-Gusman, S.; Gleiser, P.M. Role of the circadian clock in the statistics of locomotor activity in Drosophila. PLoS ONE 2018, 13, e0202505. [Google Scholar] [CrossRef]
- Ulgherait, M.; Midoun, A.M.; Park, S.J.; Gatto, J.A.; Tener, S.J.; Siewert, J.; Klickstein, N.; Canman, J.C.; Ja, W.W.; Shirasu-Hiza, M. Circadian autophagy drives iTRF-mediated longevity. Nature 2021, 598, 353–358. [Google Scholar] [CrossRef]
- Ikeno, T.; Numata, H.; Goto, S.G. Circadian clock genes period and cycle regulate photoperiodic diapause in the bean bug Riptortus pedestris males. J. Insect Physiol. 2011, 57, 935–938. [Google Scholar] [CrossRef]
- Ikeno, T.; Tanaka, S.I.; Numata, H.; Goto, S.G. Photoperiodic diapause under the control of circadian clock genes in an insect. BMC Biol. 2010, 8, 116. [Google Scholar] [CrossRef] [Green Version]
- Hardin, P.E.; Hall, J.C.; Rosbash, M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 1990, 343, 536–540. [Google Scholar] [CrossRef]
- Vosshall, L.B.; Price, J.L.; Sehgal, A.; Saez, L.; Young, M.W. Block in nuclear localization of period protein by a second clock mutation, timeless. Science 1994, 263, 1606–1609. [Google Scholar] [CrossRef] [Green Version]
- Numata, H.; Miyazaki, Y.; Ikeno, T. Common features in diverse insect clocks. Zool. Lett. 2015, 1, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brady, D.; Saviane, A.; Cappellozza, S.; Sandrelli, F. The circadian clock in lepidoptera. Front. Physiol. 2021, 12, 776826. [Google Scholar] [CrossRef]
- Grzywacz, A.; Hall, M.J.R.; Pape, T.; Szpila, K. Muscidae (Diptera) of forensic importance-an identification key to third instar larvae of the western Palaearctic region and a catalogue of the muscid carrion community. Int. J. Leg. Med. 2017, 131, 855–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moffatt, C.; Heaton, V.; De Haan, D. The distribution of blow fly (Diptera: Calliphoridae) larval lengths and its implications for estimating post mortem intervals. Int. J. Leg. Med. 2016, 130, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Saunders, D. A comparative study of circadian rhythmicity and photoperiodism in closely related species of blow flies: External coincidence, maternal induction, and diapause at northern latitudes. J. Biol. Rhythm. 2021, 36, 532–547. [Google Scholar] [CrossRef] [PubMed]
- Kenny, N.A.; Saunders, D.S. Adult locomotor rhythmicity as “hands” of the maternal photoperiodic clock regulating larval diapause in the blowfly, Calliphora vicina. J. Biol. Rhythm. 1991, 6, 217–233. [Google Scholar] [CrossRef]
- Saunders, D.S.; Cymborowski, B. Selection for high diapause incidence in blow flies (Calliphora vicina) maintained under long days increases the maternal critical daylength: Some consequences for the photoperiodic clock. J. Insect Physiol. 2003, 49, 777–784. [Google Scholar] [CrossRef]
- Cymborowski, B. Effects of 5,7-dihydroxytriptamine (5,7-DHT) on circadian locomotor activity of the blow fly, Calliphora vicina. J. Insect Sci. 2003, 3, 14. [Google Scholar] [CrossRef]
- Bostock, E.; Green, E.W.; Kyriacou, C.P.; Vanin, S. Chronobiological studies on body search, oviposition and emergence of Megaselia scalaris (Diptera, Phoridae) in controlled conditions. Forensic Sci. Int. 2017, 275, 155–159. [Google Scholar] [CrossRef]
- Short, C.A.; Meuti, M.E.; Zhang, Q.; Denlinger, D.L. Entrainment of eclosion and preliminary ontogeny of circadian clock gene expression in the flesh fly, Sarcophaga crassipalpis. J. Insect Physiol. 2016, 93–94, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Miyazaki, Y.; Goto, S.G.; Tanaka, K.; Saito, O.; Watari, Y. Thermoperiodic regulation of the circadian eclosion rhythm in the flesh fly, Sarcophaga crassipalpis. J. Insect Physiol. 2011, 57, 1249–1258. [Google Scholar] [CrossRef]
- Yamamoto, M.; Nishimura, K.; Shiga, S. Clock and Hormonal Controls of an Eclosion Gate in the Flesh Fly Sarcophaga crassipalpis. Zool. Sci. 2017, 34, 151–160. [Google Scholar] [CrossRef]
- Prohaska, F.; Joplin, K.H.; Moore, D. Effects of gender, age, and nutrition on circadian locomotor activity rhythms in the flesh fly Sarcophaga crassipalpis. J. Insect Physiol. 2018, 107, 265–275. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Goto, S.G. Distinct physiological mechanisms induce latitudinal and sexual differences in the photoperiodic induction of diapause in a fly. J. Biol. Rhythm. 2019, 34, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Chaika, V.; Pikula, K.; Vshivkova, T.; Zakharenko, A.; Reva, G.; Drozdov, K.; Vardavas, A.I.; Stivaktakis, P.D.; Nikolouzakis, T.K.; Stratidakis, A.K.; et al. The toxic influence and biodegradation of carbon nanofibers in freshwater invertebrates of the families Gammaridae, Ephemerellidae, and Chironomidae. Toxicol. Rep. 2020, 7, 947–954. [Google Scholar] [CrossRef] [PubMed]
- González Medina, A.; Soriano Hernando, Ó.; Jiménez Ríos, G. The Use of the Developmental Rate of the Aquatic Midge Chironomus riparius (Diptera, Chironomidae) in the Assessment of the Postsubmersion Interval. J. Forensic Sci. 2015, 60, 822–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalal, J.; Sharma, S.; Bhardwaj, T.; Dhattarwal, S.K.; Verma, K. Seasonal study of the decomposition pattern and insects on a submerged pig cadaver. J. Forensic Leg. Med. 2020, 74, 102023. [Google Scholar] [CrossRef]
- Kobelkova, A.; Goto, S.G.; Peyton, J.T.; Ikeno, T.; Lee, R.E.; Denlinger, D.L. Continuous activity and no cycling of clock genes in the Antarctic midge during the polar summer. J. Insect Physiol. 2015, 81, 90–96. [Google Scholar] [CrossRef] [Green Version]
- Kaiser, T.S.; Poehn, B.; Szkiba, D.; Preussner, M.; Sedlazeck, F.J.; Zrim, A.; Neumann, T.; Nguyen, L.-T.; Betancourt, A.J.; Hummel, T.; et al. The genomic basis of circadian and circalunar timing adaptations in a midge. Nature 2016, 540, 69–73. [Google Scholar] [CrossRef] [Green Version]
- Kaiser, T.S.; Neumann, D.; Heckel, D.G.; Berendonk, T.U. Strong genetic differentiation and postglacial origin of populations in the marine midge Clunio marinus (Chironomidae, Diptera). Mol. Ecol. 2010, 19, 2845–2857. [Google Scholar] [CrossRef] [Green Version]
- Beer, K.; Helfrich-Förster, C. Model and Non-model Insects in Chronobiology. Front. Behav. Neurosci. 2020, 14, 601676. [Google Scholar] [CrossRef]
- Frederickx, C.; Dekeirsschieter, J.; Verheggen, F.J.; Haubruge, E. The community of Hymenoptera parasitizing necrophagous Diptera in an urban biotope. J. Insect Sci. 2013, 13, 32. [Google Scholar] [CrossRef] [Green Version]
- Rahimi-Kaldeh, S.; Ashouri, A.; Bandani, A.; Tomioka, K. The effect of Wolbachia on diapause, fecundity, and clock gene expression in Trichogramma brassicae (Hymenoptera: Trichogrammatidae). Dev. Genes Evol. 2017, 227, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Ingram, K.K.; Kutowoi, A.; Wurm, Y.; Shoemaker, D.; Meier, R.; Bloch, G. The molecular clockwork of the fire ant Solenopsis invicta. PLoS ONE 2012, 7, e45715. [Google Scholar] [CrossRef] [Green Version]
- Roenneberg, T.; Kuehnle, T.; Juda, M.; Kantermann, T.; Allebrandt, K.; Gordijn, M.; Merrow, M. Epidemiology of the human circadian clock. Sleep Med. Rev. 2007, 11, 429–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roenneberg, T.; Kuehnle, T.; Pramstaller, P.P.; Ricken, J.; Havel, M.; Guth, A.; Merrow, M. A marker for the end of adolescence. Curr. Biol. 2004, 14, R1038–R1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adan, A.; Natale, V. Gender differences in morningness-eveningness preference. Chronobiol. Int. 2002, 19, 709–720. [Google Scholar] [CrossRef]
- Chellappa, S.L.; Steiner, R.; Oelhafen, P.; Cajochen, C. Sex differences in light sensitivity impact on brightness perception, vigilant attention and sleep in humans. Sci. Rep. 2017, 7, 14215. [Google Scholar] [CrossRef]
- Piccinni, A.; Marazziti, D.; Del Debbio, A.; Bianchi, C.; Roncaglia, I.; Mannari, C.; Origlia, N.; Catena Dell’Osso, M.; Massimetti, G.; Domenici, L.; et al. Diurnal variation of plasma brain-derived neurotrophic factor (BDNF) in humans: An analysis of sex differences. Chronobiol. Int. 2008, 25, 819–826. [Google Scholar] [CrossRef]
- Cain, S.W.; Chang, A.-M.; Vlasac, I.; Tare, A.; Anderson, C.; Czeisler, C.A.; Saxena, R. Circadian Rhythms in Plasma Brain-derived Neurotrophic Factor Differ in Men and Women. J. Biol. Rhythm. 2017, 32, 75–82. [Google Scholar] [CrossRef]
- Cain, S.W.; Dennison, C.F.; Zeitzer, J.M.; Guzik, A.M.; Khalsa, S.B.S.; Santhi, N.; Schoen, M.W.; Czeisler, C.A.; Duffy, J.F. Sex differences in phase angle of entrainment and melatonin amplitude in humans. J. Biol. Rhythm. 2010, 25, 288–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Škrlec, I.; Talapko, J.; Juzbašić, M.; Steiner, R. Sex differences in circadian clock genes and myocardial infarction susceptibility. J. Cardiovasc. Dev. Dis. 2021, 8, 53. [Google Scholar] [CrossRef]
- Yang, B.-Z.; Han, S.; Kranzler, H.R.; Palmer, A.A.; Gelernter, J. Sex-specific linkage scans in opioid dependence. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2017, 174, 261–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivkees, S.A. Developing circadian rhythmicity. Basic and clinical aspects. Pediatr. Clin. N. Am. 1997, 44, 467–487. [Google Scholar] [CrossRef]
- Serón-Ferré, M.; Ducsay, C.A.; Valenzuela, G.J. Circadian rhythms during pregnancy. Endocr. Rev. 1993, 14, 594–609. [Google Scholar] [CrossRef]
- Nehme, P.A.; Amaral, F.; Lowden, A.; Skene, D.J.; Cipolla-Neto, J.; Moreno, C.R.C. Reduced melatonin synthesis in pregnant night workers: Metabolic implications for offspring. Med. Hypotheses 2019, 132, 109353. [Google Scholar] [CrossRef]
- Acosta-Rodríguez, V.A.; Rijo-Ferreira, F.; Green, C.B.; Takahashi, J.S. Importance of circadian timing for aging and longevity. Nat. Commun. 2021, 12, 2862. [Google Scholar] [CrossRef]
- Youngstedt, S.D.; Kripke, D.F.; Elliott, J.A.; Klauber, M.R. Circadian abnormalities in older adults. J. Pineal Res. 2001, 31, 264–272. [Google Scholar] [CrossRef]
- Kripke, D.F.; Youngstedt, S.D.; Elliott, J.A.; Tuunainen, A.; Rex, K.M.; Hauger, R.L.; Marler, M.R. Circadian phase in adults of contrasting ages. Chronobiol. Int. 2005, 22, 695–709. [Google Scholar] [CrossRef]
- Ritz-Timme, S.; Cattaneo, C.; Collins, M.J.; Waite, E.R.; Schütz, H.W.; Kaatsch, H.J.; Borrman, H.I. Age estimation: The state of the art in relation to the specific demands of forensic practise. Int. J. Leg. Med. 2000, 113, 129–136. [Google Scholar] [CrossRef]
- Correia Dias, H.; Cunha, E.; Corte Real, F.; Manco, L. Age prediction in living: Forensic epigenetic age estimation based on blood samples. Leg. Med. 2020, 47, 101763. [Google Scholar] [CrossRef] [PubMed]
- Woźniak, A.; Heidegger, A.; Piniewska-Róg, D.; Pośpiech, E.; Xavier, C.; Pisarek, A.; Kartasińska, E.; Boroń, M.; Freire-Aradas, A.; Wojtas, M.; et al. Development of the VISAGE enhanced tool and statistical models for epigenetic age estimation in blood, buccal cells and bones. Aging 2021, 13, 6459–6484. [Google Scholar] [CrossRef] [PubMed]
- Montesanto, A.; D’Aquila, P.; Lagani, V.; Paparazzo, E.; Geracitano, S.; Formentini, L.; Giacconi, R.; Cardelli, M.; Provinciali, M.; Bellizzi, D.; et al. A new robust epigenetic model for forensic age prediction. J. Forensic Sci. 2020, 65, 1424–1431. [Google Scholar] [CrossRef] [PubMed]
- Orozco-Solis, R.; Sassone-Corsi, P. Circadian clock: Linking epigenetics to aging. Curr. Opin. Genet. Dev. 2014, 26, 66–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.Y.; Lee, S.D.; Shin, K.-J. Forensic DNA methylation profiling from evidence material for investigative leads. BMB Rep. 2016, 49, 359–369. [Google Scholar] [CrossRef] [Green Version]
- Pacheco-Bernal, I.; Becerril-Pérez, F.; Aguilar-Arnal, L. Circadian rhythms in the three-dimensional genome: Implications of chromatin interactions for cyclic transcription. Clin. Epigenetics 2019, 11, 79. [Google Scholar] [CrossRef]
- Crosio, C.; Cermakian, N.; Allis, C.D.; Sassone-Corsi, P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat. Neurosci. 2000, 3, 1241–1247. [Google Scholar] [CrossRef]
- Oh, G.; Ebrahimi, S.; Carlucci, M.; Zhang, A.; Nair, A.; Groot, D.E.; Labrie, V.; Jia, P.; Oh, E.S.; Jeremian, R.H.; et al. Cytosine modifications exhibit circadian oscillations that are involved in epigenetic diversity and aging. Nat. Commun. 2018, 9, 644. [Google Scholar] [CrossRef]
- Nakatome, M.; Orii, M.; Hamajima, M.; Hirata, Y.; Uemura, M.; Hirayama, S.; Isobe, I. Methylation analysis of circadian clock gene promoters in forensic autopsy specimens. Leg. Med. 2011, 13, 205–209. [Google Scholar] [CrossRef]
- Etchegaray, J.-P.; Lee, C.; Wade, P.A.; Reppert, S.M. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 2003, 421, 177–182. [Google Scholar] [CrossRef]
- Koike, N.; Yoo, S.-H.; Huang, H.-C.; Kumar, V.; Lee, C.; Kim, T.-K.; Takahashi, J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpkin, A.J.; Howe, L.D.; Tilling, K.; Gaunt, T.R.; Lyttleton, O.; McArdle, W.L.; Ring, S.M.; Horvath, S.; Smith, G.D.; Relton, C.L. The epigenetic clock and physical development during childhood and adolescence: Longitudinal analysis from a UK birth cohort. Int. J. Epidemiol. 2017, 46, 549–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horvath, S.; Gurven, M.; Levine, M.E.; Trumble, B.C.; Kaplan, H.; Allayee, H.; Ritz, B.R.; Chen, B.; Lu, A.T.; Rickabaugh, T.M.; et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016, 17, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sládek, M.; Kudrnáčová Röschová, M.; Adámková, V.; Hamplová, D.; Sumová, A. Chronotype assessment via a large scale socio-demographic survey favours yearlong Standard time over Daylight Saving Time in central Europe. Sci. Rep. 2020, 10, 1419. [Google Scholar] [CrossRef] [PubMed]
- Oates, A.C.; Morelli, L.G.; Ares, S. Patterning embryos with oscillations: Structure, function and dynamics of the vertebrate segmentation clock. Development 2012, 139, 625–639. [Google Scholar] [CrossRef] [Green Version]
- Palmeirim, I.; Henrique, D.; Ish-Horowicz, D.; Pourquié, O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 1997, 91, 639–648. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, M.; Hayashi, H.; Garcia-Ojalvo, J.; Yoshioka-Kobayashi, K.; Kageyama, R.; Yamanaka, Y.; Ikeya, M.; Toguchida, J.; Alev, C.; Ebisuya, M. Species-specific segmentation clock periods are due to differential biochemical reaction speeds. Science 2020, 369, 1450–1455. [Google Scholar] [CrossRef]
- Clark, E.; Peel, A.D.; Akam, M. Arthropod segmentation. Development 2019, 146, dev170480. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, M.; Yamanaka, Y.; Uemura, M.; Osawa, M.; Saito, M.K.; Nagahashi, A.; Nishio, M.; Guo, L.; Ikegawa, S.; Sakurai, S.; et al. Recapitulating the human segmentation clock with pluripotent stem cells. Nature 2020, 580, 124–129. [Google Scholar] [CrossRef]
- Diaz-Cuadros, M.; Wagner, D.E.; Budjan, C.; Hubaud, A.; Tarazona, O.A.; Donelly, S.; Michaut, A.; Al Tanoury, Z.; Yoshioka-Kobayashi, K.; Niino, Y.; et al. In vitro characterization of the human segmentation clock. Nature 2020, 580, 113–118. [Google Scholar] [CrossRef]
- Swovick, K.; Firsanov, D.; Welle, K.A.; Hryhorenko, J.; Wise, J.P.; George, C.; Sformo, T.L.; Seluanov, A.; Gorbunova, V.; Ghaemmaghami, S. Interspecies differences in proteome turnover kinetics are correlated with lifespans and energetic demands. Mol. Cell. Proteom. 2020, 20, 100041. [Google Scholar] [CrossRef] [PubMed]
- Marshall, M. These cellular clocks help explain why elephants are bigger than mice. Nature 2021, 592, 682–684. [Google Scholar] [CrossRef] [PubMed]
- Roenneberg, T.; Chua, E.J.; Bernardo, R.; Mendoza, E. Modelling biological rhythms. Curr. Biol. 2008, 18, R826–R835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikita, E.; Nikitas, P. On the use of machine learning algorithms in forensic anthropology. Leg. Med. 2020, 47, 101771. [Google Scholar] [CrossRef]
- Foster, R.G.; Roenneberg, T. Human responses to the geophysical daily, annual and lunar cycles. Curr. Biol. 2008, 18, R784–R794. [Google Scholar] [CrossRef] [Green Version]
- Zurl, M.; Poehn, B.; Rieger, D.; Krishnan, S.; Rokvic, D.; Veedin Rajan, V.B.; Gerrard, E.; Schlichting, M.; Orel, L.; Ćorić, A.; et al. Two light sensors decode moonlight versus sunlight to adjust a plastic circadian/circalunidian clock to moon phase. Proc. Natl. Acad. Sci. USA 2022, 119, e2115725119. [Google Scholar] [CrossRef]
| Forensic Output | Biomarkers | Tissue Source | Reference |
|---|---|---|---|
| time of death | REV-ERBα:BMAL1 | liver, kidney, heart | Kimura A. et al., 2011 [38] |
| PER2, PER3, BMAL1 | dorsolateral prefrontal cortex | Lim A. S. P. et al., 2013 [39] | |
| daytime of death | melatonin, cortisol, HSPA1B, MKNK2, PER3 | blood | Lech K. et al., 2016 [41] |
| mir-34c, mir-541, mir-888, mir-484, mir-142-5p | vitreous humor | Odriozola A. et al., 2013 [43] | |
| PER1 polymorphism rs7221412 | cerebral cortex, peripheral blood mononuclear cells, CD14+CD16− monocytes | Lim A. S. P. et al., 2012 [51] | |
| post-mortem interval | Methylococcaceae | buried piglets and soil | Olakanye A. O. et al., 2017 [53] |
| locomotor behavior, oviposition | Megaselia scalaris | Bostock E. et al., 2017 [79] | |
| sex dimorphism | PER2, PER3, BMAL1 peak time | dorsolateral prefrontal cortex | Lim A. S. P. et al., 2013 [39] |
| CLOCK polymorphism rs11932595 | peripheral blood lymphocytes | Škrlec I. et al., 2021 [102] | |
| characteristic age span | chronotype | NA | Roenneberg T. et al., 2007 [95] |
| cause of death | methylation status of PER1-3, CRY1-2, BMAL1, CLOCK, CK1e, TIM promotors | blood, heart, lung, liver, kidney, brain | Nakatome M. et al., 2011 [119] |
| myocardial infarction daytime | NA | Sakelliadis E. I. et al., 2021 [59] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janjić, K.; Reisinger, C.; Kanz, F. Common Ground between Biological Rhythms and Forensics. Biology 2022, 11, 1071. https://doi.org/10.3390/biology11071071
Janjić K, Reisinger C, Kanz F. Common Ground between Biological Rhythms and Forensics. Biology. 2022; 11(7):1071. https://doi.org/10.3390/biology11071071
Chicago/Turabian StyleJanjić, Klara, Christoph Reisinger, and Fabian Kanz. 2022. "Common Ground between Biological Rhythms and Forensics" Biology 11, no. 7: 1071. https://doi.org/10.3390/biology11071071