Will Climate Change, Genetic and Demographic Variation or Rat Predation Pose the Greatest Risk for Persistence of an Altitudinally Distributed Island Endemic?
Abstract
:1. Introduction
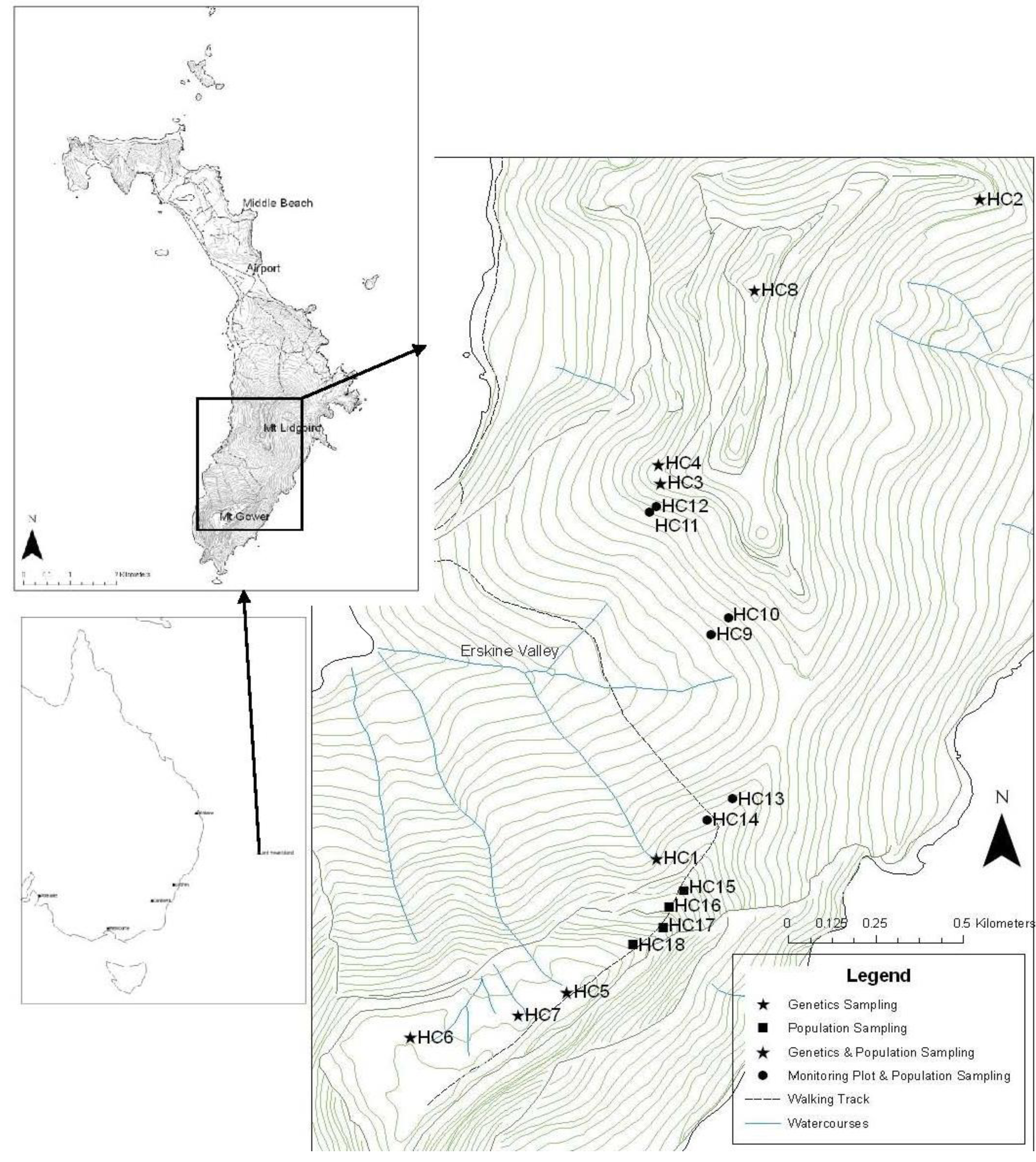
2. Results and Discussion
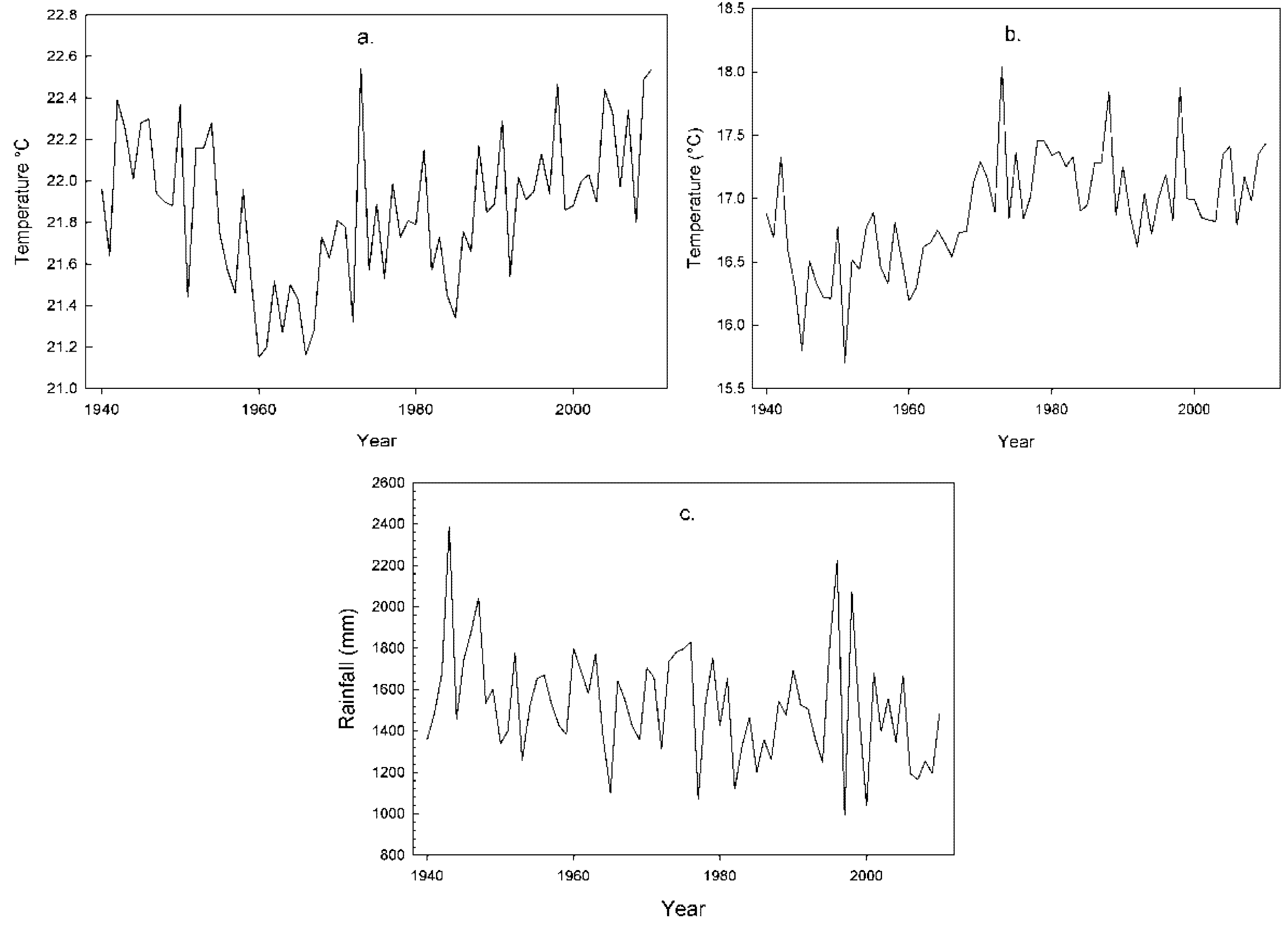
2.1. Climatic Conditions
| Mean Max. Temperature | Mean Min. Temperature | |||
|---|---|---|---|---|
| decade (rs) | decades (f) | decade (rs) | decade (f) | |
| January | −0.050 ** | 16.525 ** | 0.199 ** | 23.580 ** |
| February | −0.005 | 18.309 ** | 0.212 ** | 20.365 ** |
| March | −0.063 ** | 18.37 ** | 0.131 ** | 15.877 ** |
| April | 0.029 | 6.683 ** | 0.106 ** | 14.894 ** |
| May | 0.134 ** | 11.679 ** | 0.151 ** | 21.133 ** |
| June | 0.181 ** | 20.095 ** | 0.103 ** | 4.054 ** |
| July | 0.153 ** | 21.768 ** | 0.119 ** | 8.096 ** |
| August | 0.124 ** | 14.156 ** | 0.113 ** | 8.596 ** |
| September | 0.172 ** | 24.201 ** | 0.168 ** | 13.155 ** |
| October | 0.052 ** | 6.541 ** | 0.122 ** | 8.385 ** |
| November | −0.074 ** | 17.621 ** | 0.111 ** | 7.679 ** |
| December | −0.041 | 29.192 ** | 0.158 ** | 16.540 ** |
| Annual | 0.026 ** | 30.058 ** | 0.082 ** | 41.473 ** |
2.2. Population Demography
| Total/mean | Mt Gower | Mt Lidgbird | Altitude (rs) | Aspect (rs) | Mountain (t) | Exposure (t) | Aspect (f) | Altitude (f) | |
|---|---|---|---|---|---|---|---|---|---|
| Non trunked & trunked | |||||||||
| D (ha) | 1,643.6
(0.07) | 2,003
(203) | 1,182 (173) | 0.418 | −0.282 | 2.966 ** | 1.721 | 2.233 | −2.214 * |
| NND1 (m) | 1.2
(0.07) | 1.1
(0.07) | 1.3
(0.107) | −0.338 | 0.338 | −2.055 | −1.532 | 1.626 | 1.343 |
| NND3 (m) | 2.5
(0.15) | 2.1
(0.10) | 2.9
(0.20) | −0.588 * | 0.373 | −4.428 *** | −2.767 * | 1.927 | 2.834 * |
| Trunked | |||||||||
| D (ha) | 1,102.3
(129.59) | 1,423.3
(138.9) | 689.6
(111.5) | 0.655 ** | −0.417 | 3.940 ** | 2.410 * | 4.749 * | −4.212 ** |
| NND1 (m) | 1.7
(0.132) | 2.7
(0.17) | 4.5
(0.52) | −0.674 ** | 0.513 * | −2.683 ** | −1.111 | 3.086 | 2.357 * |
| NND3 (m) | 3.5
(0.33) | 1.4
(0.08) | 1.9
(0.23) | 0.511 * | −0.818 ** | −3.750 ** | −1.968 | 2.018 | 3.013 ** |
| H1 | 2.9
(0.08) | 2.4
(0.094) | 3.5
(0.149) | −0.220 ** | 0.239 ** | −6.466 *** | −8.868 *** | 4.944 *** | 4.055 *** |
| R/m | 43.0
(1.06) | 43.5
(1.52) | 42.4
(1.43) | −0.078 | −0.046 | 0.539 | 0.415 | 5.495 *** | −2.454 * |
| DBH | 9.2
(0.24) | 8.4
(0.30) | 10.1
(0.36) | −0.252 ** | 0.219 ** | −3.570 *** | −3.935 *** | 14.553 *** | 2.269 * |
| Total | Mt Gower | Mt Lidgbird | Altitude (rs) | Altitude (t) | Aspect (rs) | Mountain (t) | Exposure (t) | Aspect (f) | |
|---|---|---|---|---|---|---|---|---|---|
| RI (%) | 44.3 | 41.52 (7.54) | 47.87 (5.77) | 0.165 | −1.971 | 0.357 | 0.704 | −0.14 | 2.229 |
| nS/RI | 0.15 (0.40) | 0.13 (0.045) | 0.17 (0.074) | −0.349 | 1.655 | −0.574 * | 0.489 | −0.037 | 2.564 |
| Inf/RI | 2.07 (0.08) | 2.05 (0.31) | 1.78 (0.16) | 0.165 | −1.971 | 0.357 | 0.704 | −0.140 | 2.229 |
| MRO | 13.130 | 452.2 (255.61) | 608.6 (150.20) | 0.295 ** | −3.499 ** | −0.221 ** | 2.780 ** | 2.748 ** | 5.342 *** |
| mMRO | 79.3 (6.79) | 96.9 (13.79) | 67.6 (6.29) | 0.187 | −3.255 ** | −0.311 | 2.149 * | 2.184 * | 0.729 |
| Developing Splade | 0.48 | 5.97 (5.52) | 0 | 0.112 | 0.668 | −0.240 | 0.946 | 0.714 | 0.322 |
| Half-Sized Green Fruit | 28.47 | 29.06 (7.62) | 11.40 (4.02) | 0.643 * | −5.637 *** | 0.083 | 1.882 | 0.708 | 4.401 * |
| Full-Sized Green Fruit | 39.23 | 28.97 (7.03) | 48.36 (7.66) | −0.416 | 2.563 * | 0.095 | −1.856 | −2.359 * | 2.243 |
| Full-Sized Mottled Fruit | 4.07 | 1.12 (0.57) | 5.30 (2.28) | −0.165 | 0.854 | 0.435 | −1.995 | −3.512 ** | 2.211 |
| Full-Sized Red Fruit | 0.48 | 0.21 (0.21) | 0.36 (0.36) | 0.141 | −0.159 | 0.094 | −0.375 | −0.794 | 0.971 |
| Old | 27.03 | 34.66 (8.13) | 34.05 (10.29) | 0.088 | 0.12 | −0.024 | 0.047 | 1.546 | 1.845 |
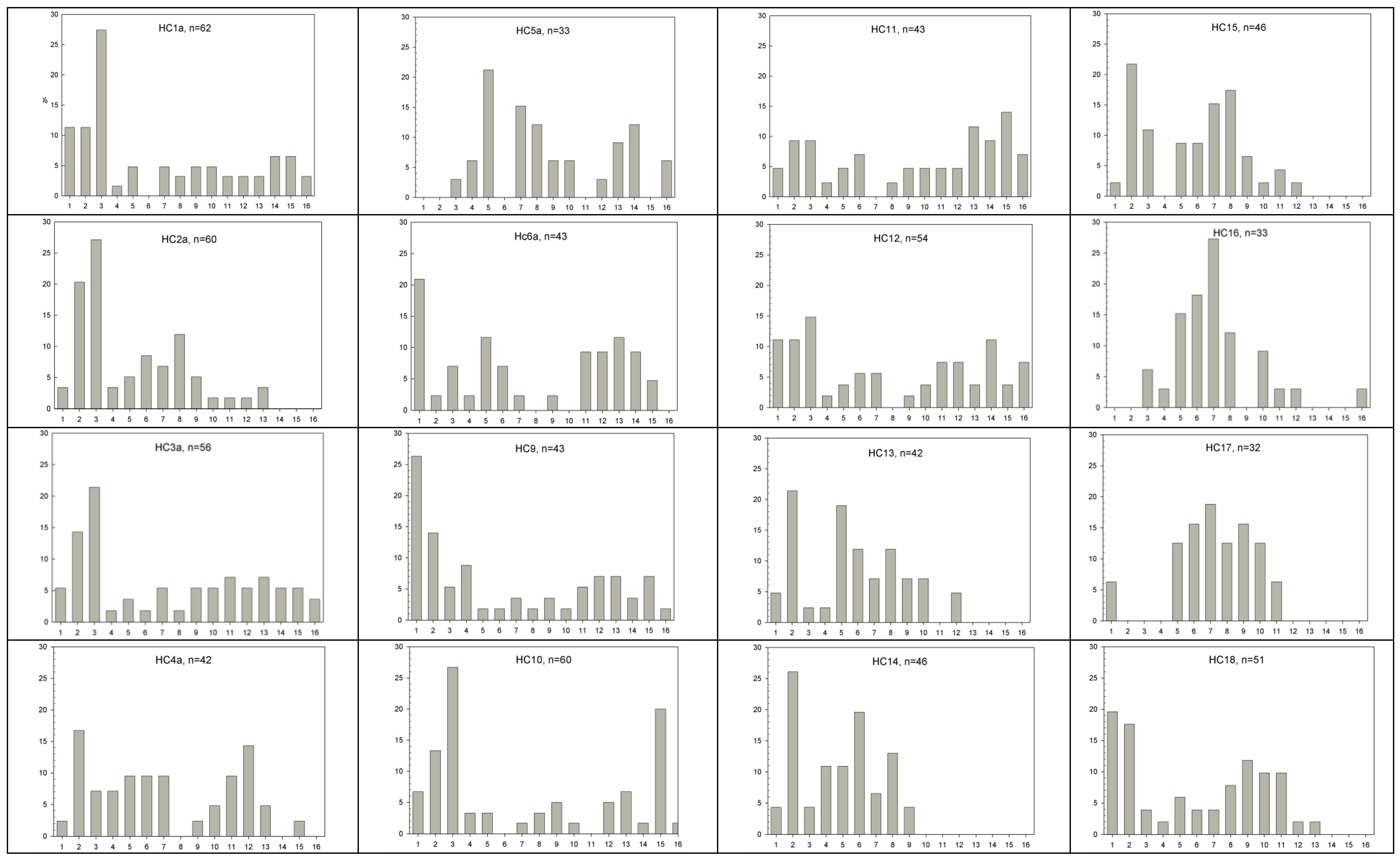
2.3. Fecundity and Phenology

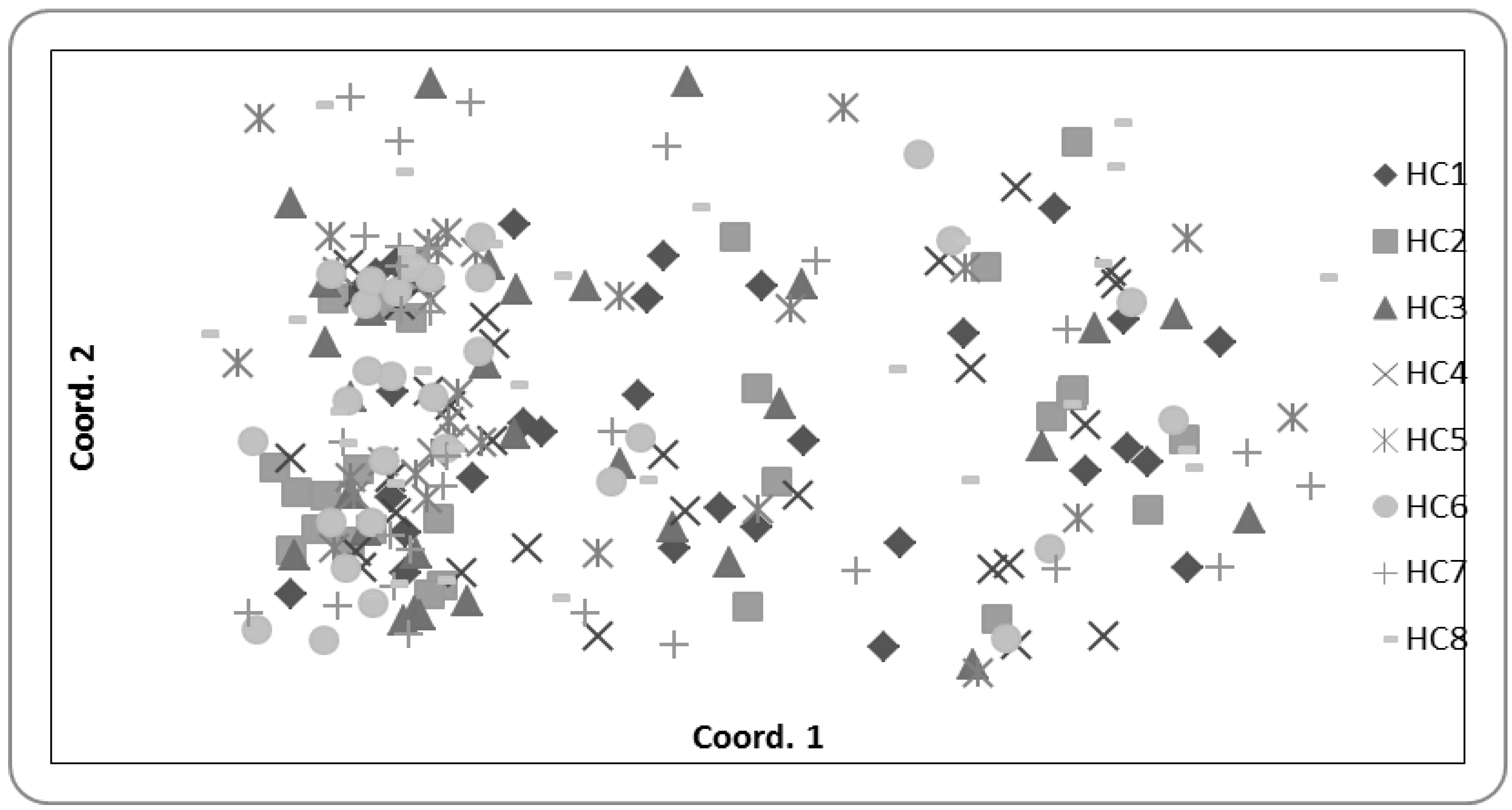
2.4. Population Genetics
| Site | N | A | Ae | P (%) | I | Ho | He | Ap | F |
|---|---|---|---|---|---|---|---|---|---|
| HC1 | 31 | 3.70
(0.616) | 2.22
(0.368) | 100 | 0.814
(0.173) | 0.335
(0.099) | 0.435
(0.083) | 0 | 0.256
(0.174) |
| HC2 | 28 | 4.00
(0.632) | 2.56
(0.541) | 100 | 0.906
(0.179) | 0.310
(0.092) | 0.471
(0.081) | 2 | 0.348
(0.176) |
| HC3 | 30 | 3.80
(0.663) | 2.25
(0.457) | 100 | 0.829
(0.155) | 0.259
(0.067) | 0.443
(0.067) | 2 | 0.389
(0.150) |
| HC4 | 30 | 3.50
(0.671) | 2.30
(0.485) | 90 | 0.789
(0.194) | 0.283
(0.076) | 0.412
(0.091) | 0 | 0.347
(0.160) |
| HC5 | 30 | 3.60
(0.670) | 2.25
(0.410) | 90 | 0.819
(0.178) | 0.269
(0.090) | 0.433
(0.083) | 0 | 0.393
(0.183) |
| HC6 | 29 | 3.70
(0.496) | 2.01
(0.365) | 100 | 0.724
(0.168) | 0.244
(0.070) | 0.371
(0.084) | 2 | 0.327
(0.127) |
| HC7 | 29 | 3.60
(0.379) | 2.27
(0.434) | 100 | 0.809
(0.173) | 0.280
(0.092) | 0.435
(0.081) | 1 | 0.318
(0.165) |
| HC8 | 30 | 4.00
(0.650) | 2.18
(0.307) | 100 | 0.874
(0.158) | 0.274
(0.089) | 0.461
(0.068) | 1 | 0.462
(0.185) |
| Total/Mean | 29.52 | 3.74 | 2.26 | 97.5 | 0.820 | 0.282 | 0.433 | 8 | 0.355 |
| Statistic | Value | S.E | |
|---|---|---|---|
| FIS | 0.348 | 0.159 | |
| FIT | 0.366 | 0.155 | |
| FST | 0.03 | 0.002 | |
| Nm | 8.446 | 0.64 | |
| As | 3.738 | ||
| Variation among populations | 2% | ||
| Variation within populations | 98% | ||
| PhiPT | 0.015 *** | ||
2.5. Population Growth Models
| Model | Low altitude | High altitude | Low altitude rat control | High altitude rat control |
|---|---|---|---|---|
| Starting population (/Ha) | 1,886 | 1,109 | 1,886 | 1,109 |
| Final population (n + t) (/Ha) | 652 (195–1,109) | 85 (0–171) | 1,246 (234–2,258) | 560 (78–1,042) |
| % remaining @200 years | 55% | 8% | 66% | 50% |
| K (Ha) | 2,220 | 3,770 | 2,220 | 3,770 |
| λ | 0.9968 | 0.9721 | 1.0108 | 0.9835 |
| Quasi extinction probability | 0.030 (0.000–0.119) | 0.690 (0.601–0.779) | 0.020 (0.000–0.109) | 0.080 (0.000–0.168) |
| Time to Quasi extinction | >200 years (p = 0.01) | 181.1 years (p = 0.02) | >200 years (p = 0.01) | >200 years (p = 0.01) |
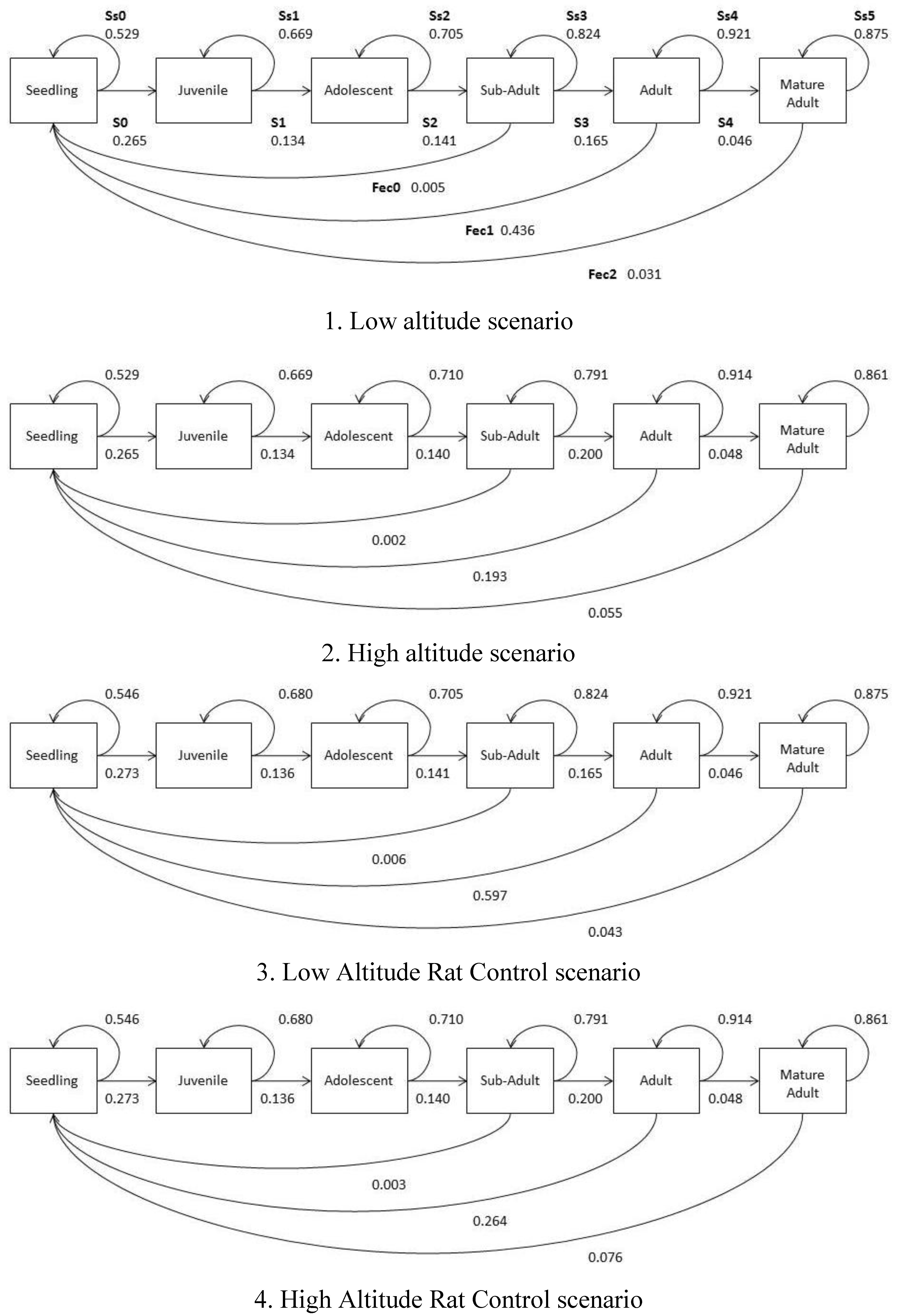
2.6. Climate Change and In Situ Persistence

3. Experimental Section
3.1. Field Methods
3.2. Genetic Analysis
3.3. Statistical Analysis
3.4. Population Growth Models
4. Conclusions
Acknowledgements
References
- MacArthur, R.H.; Wilson, E.O. The Theory of Island Biogeography; Princeton University Press: Princeton, NJ, USA, 1967. [Google Scholar]
- Simberloff, D. Extinction-proneness of island species-Causes and management implications. Raffles Bull. Zool. 2000, 48, 1–9. [Google Scholar]
- Dransfield, J.; Uhl, N.W.; Asmussen, C.B.; Baker, W.J.; Harley, M.M.; Lewis, C.E. Genera Palmarum: The Evolution and Classification of Palms; Royal Botanic Gardens, Kew: Richmond, Surry, UK, 2008. [Google Scholar]
- Kissling, W.D.; Baker, W.J.; Balslev, H.; Barfod, A.S.; Borchsenius, F.; Dransfield, J.; Govaerts, R.; Svenning, J.-C. Quaternary and pre-Quaternary historical legacies in the global distribution of a major tropical plant lineage. Glob. Ecol. Biogeogr. 2012, 21, 909–921. [Google Scholar] [CrossRef]
- Savolainen, V.; Anstett, M.C.; Lexer, C.; Hutton, I.; Clarkson, J.J.; Norup, M.V.; Powell, M.P.; Springate, D.; Salamin, N.; Baker, W.J. Sympatric speciation in palms on an oceanic island. Nature 2006, 441, 210–213. [Google Scholar]
- Babik, W.; Butlin, R.K.; Baker, W.J.; Papadopulos, A.S.T.; Boulesteix, M.; Anstett, M.C.; Lexer, C.; Hutton, I.; Savolainen, V. How sympatric is speciation in the Howea palms of Lord Howe Island? Mol. Ecol. 2009, 18, 3629–3638. [Google Scholar] [CrossRef]
- Still, C.J.; Foster, P.N.; Schneider, S.H. Simulating the effects of climate change on tropical montane cloud forests. Nature 1999, 398, 608–610. [Google Scholar]
- Hamilton, L.S. Mountain Cloud Forest Conservation and Research: A Synopsis. Mt. Res. Dev. 1993, 15, 259–266. [Google Scholar] [CrossRef]
- Bradshaw, A.D. Evolutionary Significance of Phenotypic Plasticity in Plants. Adv. Genet. 1965, 13, 115–155. [Google Scholar] [CrossRef]
- Guo, H.; Mazer, S.J.; Du, G.Z. Geographic variation in seed mass within and among nine species of Pedicularis (Orobanchaceae): Effects of elevation, plant size and seed number per fruit. J. Ecol. 2010, 98, 1232–1242. [Google Scholar] [CrossRef]
- Takahashi, K. Effects of altitude and competition on growth and mortality of the conifer Abies sachalinensis. Ecol. Res. 2010, 25, 801–812. [Google Scholar] [CrossRef]
- Green, P.S. Norfolk Island and Lord Howe Island; Australian Government Publishing Service: Canberra, Australia, 1994; Volume 49 Oceanic Islands 1. [Google Scholar]
- LHIB, Draft Review of the Permanent Park Preserve Plan of Management; Report to the Lord Howe Island Board: Lord Howe Island, Australia, 2002.
- Harris, R.; Cassis, G.; Auld, T.; Hutton, I. Floristics and structure of the mossy cloud forest of Mt Gower summit, Lord Howe Island. Pac. Conserv. Biol. 2005, 11, 246–256. [Google Scholar]
- Hedyscepe canterburyana. IUCN Red List of Threatened Species. Version 2010.4. International Union for Conservation of Nature and Natural Resources. Available online: http://www.iucnredlist.org/ (accessed on 19/08/2011).
- Thomas, C.D.; Cameron, A.; Green, R.E.; Bakkenes, M.; Beaumont, L.J.; Collingham, Y.C.; Erasmus, B.F.N.; Ferreira de Siqueira, M.; Grainger, A.; Hannah, L.; et al. Extinction risk from climate change. Nature 2004, 427, 145–148. [Google Scholar] [Green Version]
- Schwartz, M.W.; Iverson, L.R.; Prasad, A.M.; Matthews, S.N.; O’Connor, R.J. Predicting extinctions as a result of climate change. Ecology 2006, 87, 1611–1615. [Google Scholar] [CrossRef]
- Huntley, B. How Plants Respond to Climate Change: Migration Rates, Individualism and the Consequences for Plant Communities. Ann. Bot. (Lond.) 1991, 67, 15–22. [Google Scholar]
- Lenoir, J.; Gégout, J.C.; Pierrat, J.C.; Bontemps, J.D.; Dhôte, J.F. Differences between tree species seedling and adult altitudinal distribution in mountain forests during the recent warm period (1986-2006). Ecography 2009, 32, 765–777. [Google Scholar] [CrossRef]
- Chen, I.C.; Shiu, H.J.; Benedick, S.; Holloway, J.D.; Chey, V.K.; Barlow, H.S.; Hill, J.K.; Thomas, C.D. Elevation increases in moth assemblages over 42 years on a tropical mountain. Proc. Natl. Acad. Sci. USA 2009, 106, 1479–1483. [Google Scholar]
- Walther, G.R.; Gritti, E.S.; Berger, S.; Hickler, T.; Tang, Z.; Sykes, M.T. Palms tracking climate change. Glob. Ecol. Biogeogr. 2007, 16, 801–809. [Google Scholar] [CrossRef]
- Le Roux, P.C.; McGeoch, M.A. Rapid range expansion and community reorganization in response to warming. Glob. Change Biol. 2008, 14, 2950–2962. [Google Scholar] [CrossRef]
- Dang, H.S.; Zhang, Y.J.; Jiang, M.X.; Zhang, Q.F. Growth performance and range shift of the subalpine fir (Abies fargesii) in the Qinling Mountains, China. Int. J. Sustain. Dev. World Ecol. 2010, 17, 162–171. [Google Scholar] [CrossRef]
- Auld, T.D.; Hutton, I.; Ooi, M.K.J.; Denham, A.J. Disruption of recruitment in two endemic palms on Lord Howe Island by invasive rats. Biol. Invasions 2010, 12, 1–11. [Google Scholar] [CrossRef]
- Markham, A. Potential impacts of climate change on tropical forest ecosystems. Clim. Change 1998, 39, 141–143. [Google Scholar] [CrossRef]
- Foster, P. The potential impacts of global climate change on tropical montane cloud forests. Earth Sci. Rev. 2001, 55, 73–106. [Google Scholar] [CrossRef]
- Auld, T.D.; Hutton, I. Conservation Issues for the vascular flora of Lord Howe Island. Cunninghamia 2004, 8, 490–500. [Google Scholar]
- Implications of climate change for Australia’s World Heritage properties: A preliminary assessment. In Technical Report for Department of the Environment, Water, Heritage and the Arts: Fenner School of Environment and Society, the Australian National University, Canberra, Australia, June 2009.
- Enright, N.J.; Watson, A.D. Population dynamics of the nikau palm, Rhopalostylis sapida (Wendl. et Drude), in a temperate forest remnant near Auckalnd, New Zealand. N. Z. J. Bot. 1992, 30, 29–43. [Google Scholar] [CrossRef]
- Rodríguez-Buriticá, S.; Orjuela, M.A.; Galeano, G. Demography and life history of Geonoma orbignyana: An understory palm used as foliage in Colombia. For. Ecol. Manag. 2005, 211, 329–340. [Google Scholar] [CrossRef]
- Widyatmoko, D.; Burgman, M.A.; Guhardja, E.; Mogea, J.P.; Walujo, E.B.; Setiadi, D. Population status, demography and habitat preferences of the threatened lipstick palm Cyrtostachys renda Blume in Kerumutan Reserve, Sumatr. Acta Oecol. 2005, 28, 107–118. [Google Scholar] [CrossRef]
- Gienapp, P.; Teplitsky, C.; Alho, J.S.; Mills, J.A.; Merilä, J. Climate change and evolution: Disentangling environmental and genetic responses. Mol. Ecol. 2008, 17, 167–178. [Google Scholar] [CrossRef]
- Lande, R. Genetics and demography in biological conservation. Science 1988, 241, 1455–1460. [Google Scholar]
- Jump, A.S.; Marchant, R.; Peñuelas, J. Environmental change and the option value of genetic diversity. Trends Plant Sci. 2009, 14, 51–58. [Google Scholar] [CrossRef]
- Bradshaw, A.D.; McNeilly, T. Evolutionary Response to Global Climatic Change. Ann. Bot. (Lond.) 1991, 67, 5–14. [Google Scholar]
- Jump, A.S.; Peñuelas, J. Running to stand still: Adaptation and the response of plants to rapid climate change. Ecol. Lett. 2005, 8, 1010–1020. [Google Scholar] [CrossRef]
- Davis, M.B.; Shaw, R.G. Range shifts and adaptive responses to quaternary climate change. Science 2001, 292, 673–679. [Google Scholar] [CrossRef]
- Pfennig, D.W.; Wund, M.A.; Snell-Rood, E.C.; Cruickshank, T.; Schlichting, C.D.; Moczek, A.P. Phenotypic plasticity’s impacts on diversification and speciation. Trends Ecol. Evol. 2010, 25, 459–467. [Google Scholar] [CrossRef]
- De Jong, G. Evolution of phenotypic plasticity: Patterns of plasticity and the emergence of ecotypes. New Phytol. 2005, 166, 101–118. [Google Scholar] [CrossRef]
- Zizumbo-Villarreal, D.; Fernández-Barrera, M.; Torres-Hernández, N.; Colunga-GarcíaMarín, P. Morphological variation of fruit in Mexican populations of Cocos nucifera L. (Arecaceae) under in situ and ex situ conditions. Genet. Res. Crop Evol. 2005, 52, 421–434. [Google Scholar] [CrossRef]
- Fleischer-Dogley, F.; Kettle, C.J.; Edwards, P.J.; Ghazoul, J.; Määttänen, K.; Kaiser-Bunbury, C.N. Morphological and genetic differentiation in populations of the dispersal-limited coco de mer (Lodoicea maldivica): Implications for management and conservation. Divers. Distrib. 2011, 17, 235–243. [Google Scholar] [CrossRef]
- Crimmins, T.M.; Crimmins, M.A.; Bertelsen, C.D. Flowering range changes across an elevation gradient in response to warming summer temperatures. Glob. Change Biol. 2009, 15, 1141–1152. [Google Scholar] [CrossRef]
- Sander, T.; König, S.; Rothe, G.M.; Janßen, A.; Weisgerber, H. Genetic variation of European beech (Fagus sylvatica L.) along an altitudinal transect at mount Vogelsberg in Hesse, Germany. Mol. Ecol. 2000, 9, 1349–1361. [Google Scholar] [CrossRef]
- Bellusci, F.; Pellegrino, G.; Palermo, A.M.; Gargano, D.; Musacchio, A. Genetic differentiation of the endemic orophyte Campanula pollinensis along an altitudinal gradient. Plant Biosyst. 2005, 139, 349–356. [Google Scholar] [CrossRef]
- Walther, G.R.; Post, E.; Convey, P.; Menzel, A.; Parmesan, C.; Beebee, T.J.C.; Fromentin, J.M.; Hoegh-Guldberg, O.; Bairlein, F. Ecological responses to recent climate change. Nature 2002, 416, 389–395. [Google Scholar]
- Minorsky, P.V. The Hot and the Classic. Plant Physiol. 2002, 129, 1421–1422. [Google Scholar] [CrossRef]
- Frankham, R. Genetics and extinction. Biol. Conserv. 2005, 126, 131–140. [Google Scholar] [CrossRef]
- Pullin, A.S. Conservation Biology; Cambridge University Press: Cambridge, New York ,NY, USA, 2002. [Google Scholar]
- Caujapé-Castells, J.; Tye, A.; Crawford, D.J.; Santos-Guerra, A.; Sakai, A.; Beaver, K.; Lobin, W.; Vincent Florens, F.B.; Moura, M.; Jardim, R.; et al. Conservation of oceanic island floras: Present and future global challenges. Perspect. Plant Ecol. 2010, 12, 107–129. [Google Scholar] [CrossRef]
- Campbell, D.J.; Atkinson, I.A.E. Effects of kiore (Rattus exulans Peale) on recruitment of indigenous coastal trees on northern offshore islands of New Zealand. J. Roy. Soc. N. Z. 1999, 29, 265–290. [Google Scholar] [CrossRef]
- Campbell, D.J.; Atkinson, I.A.E. Depression of tree recruitment by the Pacific rat (Rattus exulans Peale) on New Zealand’s northern offshore islands. Biol. Conserv. 2002, 107, 19–35. [Google Scholar] [CrossRef]
- Chapin, M.H. Rat Damage on Native Hawaiian Palms. Palms 2004, 48, 153–155. [Google Scholar]
- Meyer, J.Y.; Butaud, J.F. The impacts of rats on the endangered native flora of french Polynesia (Pacific Islands): Drivers of plant extinction or coup de grâce species? Biol. Invasions 2009, 11, 1569–1585. [Google Scholar] [CrossRef]
- Billing, J.; Harden, B. Control of introduced Rattus rattus L. on Lord Howe Island. I. The response of mouse populations to warfarin bait used to control rats. Wildl. Res. 2000, 27, 655–658. [Google Scholar] [CrossRef]
- Williams, S.E.; Shoo, L.P.; Isaac, J.L.; Hoffmann, A.A.; Langham, G. Towards an integrated framework for assessing the vulnerability of species to climate change. PLoS Biol. 2008, 6, e325. [Google Scholar] [CrossRef]
- Thomas, C.D. Translocation of species, climate change, and the end of trying to recreate past ecological communities. Trends Ecol. Evol. 2011, 26, 216–221. [Google Scholar] [CrossRef]
- Climate Change 2007: Synthesis Report. Contribution of Working Groups I, II and III to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. In Intergovernmental Panel on Climate Change; Core Writing Team, Pachauri, R.K.; Resinger, A. (Eds.) IPCC: Geneva, Switzerland, 2007.
- CSIRO, BOM State of the Climate; CSIRO & Australian Bureau of Meteorology: Canberra, Australia, 2012.
- Rotstayn, L.D.; Collier, M.A.; Dix, M.R.; Feng, Y.; Gordon, H.B.; O’Farrell, S.P.; Smith, I.N.; SSyktus, J. Improved simulation of Australian climate and ENSO-related rainfall variability in a global climate model with an interactive aerosol treatment. Int. J. Climatol. 2010, 30, 1067–1088. [Google Scholar]
- Loope, L.L.; Giambelluca, T.W. Vulnerability of island tropical montane cloud forests to climate change, with special reference to East Maui, Hawaii. Clim. Change 1998, 39, 503–517. [Google Scholar] [CrossRef]
- Raxworthy, C.J.; Pearson, R.G.; Rabibisoa, N.; Rakotondrazafy, A.M.; Ramanamanjato, J.B.; Raselimanana, A.P.; Wu, S.; Nussbaum, R.A.; Stone, D.A. Extinction vulnerability of tropical montane endemism from warming and upslope displacement: A preliminary appraisal for the highest massif in Madagascar. Glob. Change Biol. 2008, 14, 1703–1720. [Google Scholar] [CrossRef]
- Whittaker, R.J.; Bush, M.B.; Richards, K. Plant recolonization and vegetation succession on the Krakatau Islands, Indonesia. Ecol. Monogr. 1989, 59, 59–123. [Google Scholar] [CrossRef]
- Pounds, J.A.; Fogden, M.P.L.; Campbell, J.H. Biological response to climate change on a tropical mountain. Nature 1999, 398, 611–615. [Google Scholar]
- Pettit, N.E.; Dowe, J.L. Distribution and population structure of the vulnerable riparian palm Livistona lanuginosa A. N. Rodd (Arecaceae) in the Burdekin River catchment, north Queensland. Pac. Conserv. Biol. 2003, 9, 207–214. [Google Scholar]
- Wilson, L. Personal communication, Lord Howe Island: NSW, Australia, 2010.
- Svenning, J.C. Environmental heterogeneity, recruitment limitation and the mesoscale distribution of palms in a tropical montane rain forest (Maquipucuna, Ecuador). J. Trop. Ecol. 2001, 17, 97–113. [Google Scholar] [CrossRef]
- Widyatmoko, D.; Burgman, M. Influences of edaphic factors on the distribution and abundances of a rare palm (Cyrtostachys renda) in a peat swamp forest in eastern Sumatra, Indonesia. Aust. Ecol. 2006, 31, 964–974. [Google Scholar] [CrossRef]
- Tomlinson, P.B. The Structural Biology of Palms; Oxford University Press: Oxford, UK, 1990. [Google Scholar]
- Blach-Overgaard, A.; Svenning, J.C.; Dransfield, J.; Greve, M.; Balslev, H. Determinants of palm species distributions across Africa: The relative roles of climate, non-climatic environmental factors, and spatial constraints. Ecography 2010, 33, 380–391. [Google Scholar]
- Otero-Arnaiz, A.; Oyama, K. Reproductive phenology, seed-set and pollination in Chamaedorea alternans, an understorey dioecious palm in a rain forest in Mexico. J. Trop. Ecol. 2001, 17, 745–754. [Google Scholar]
- Adler, G.H.; Lambert, T.D. Spatial and temporal variation in the fruiting phenology of palms in isolated stands. Plant Species Biol. 2008, 23, 9–17. [Google Scholar] [CrossRef]
- Shapcott, A.; Hutton, I.; Baker, W.J.; Auld, T. Conservation genetics and ecology of an endemic montane palm on Lord Howe Islandand potential for resilience. Conserv. Genet. 2012, 13, 257–270. [Google Scholar] [CrossRef]
- Menzel, A.; Sparks, T.H.; Estrella, N.; Roy, D.B. Altered geographic and temporal variability in phenology in response to climate change. Glob. Ecol. Biogeogr. 2006, 15, 498–504. [Google Scholar]
- Bradshaw, W.E.; Holzapfel, C.M. Genetic response to rapid climate change: It’s seasonal timing that matters. Mol. Ecol. 2008, 17, 157–166. [Google Scholar] [CrossRef]
- Dowe, J.L.; Benzie, J.; Ballment, E. Ecology and genetics of Carpoxylon macrospermum H Wendl & Drude (Arecaceae), an endangered palm from Vanuatu. Biol. Conserv. 1997, 79, 205–216. [Google Scholar] [CrossRef]
- Eguiarté, L.E.; Perez-Nasser, N.; Piñero, D. Genetic structure, outcrossing rate and heterosis in Astrocaryum mexicanum (tropical palm): Implications for evolution and conservation. Heredity 1992, 69, 217–228. [Google Scholar] [CrossRef]
- Luna, R.; Epperson, B.K.; Oyama, K. High levels of genetic variability and inbreeding in two Neotropical dioecious plams with contrasting life histories. Heredity 2007, 99, 466–476. [Google Scholar] [CrossRef]
- Shapcott, A. Comparison of the population genetics and densities of five Pinanga palm species at Kuala Belalong, Brunei. Mol. Ecol. 1999, 8, 1641–1654. [Google Scholar] [CrossRef]
- Navascues, M.; Vendramin, G.G.; Emerson, B.C. The Effect of Altitude on the Pattern of Gene Flow in the Endemic Canary Island Pine, Pinus canariensis. Silvae Genet. 2008, 57, 357–363. [Google Scholar]
- Hartl, D.L. A Primer of Population Genetics, 3rd ed; Sinauer Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- Cibrián-Jaramillo, A.; Bacon, C.D.; Garwood, N.C.; Bateman, R.M.; Thomas, M.M.; Russell, S.; Bailey, C.D.; Hahn, W.J.; Bridgewater, S.G.M.; DeSalle, R. Population genetics of the understory fishtail palm Chamaedorea ernesti-augusti in Belize: High genetic connectivity with local differentiation. BMC Genet. 2009, 10, 65. [Google Scholar]
- Scariot, A.; Lleras, E.; Hay, J.D. Reproductive biology of the palm Acrocinua aculeata in Central Brazil. Biotropica 1991, 23, 12–22. [Google Scholar] [CrossRef]
- Ghazoul, J. Pollen and seed dispersal among dispersed plants. Biol. Rev. Camb. Philos. 2005, 80, 413–443. [Google Scholar] [CrossRef]
- Gimenez-Benavides, L.; Albert, M.J.; Iriondo, J.M.; Escudero, A. Demographic processes of upward range contraction in a long-lived Mediterranean high mountain plant. Ecography 2011, 34, 85–93. [Google Scholar]
- Vandermeer, J.H. 377: On the Construction of the Population Projection Matrix for a Population Grouped in Unequal Stages. Biometrics 1975, 31, 239–242. [Google Scholar] [CrossRef]
- Silva Matos, D.M.; Freckleton, R.P.; Watkinson, A.R. The role of density dependence in the population dynamics of a tropical palm. Ecology 1999, 80, 2635–2650. [Google Scholar]
- Sampaio, M.B.; Scariot, A. Effects of stochastic herbivory events on population maintenance of an understorey palm species (Geonoma schottiana) in riparian tropical forest. J. Trop. Ecol. 2010, 26, 151–161. [Google Scholar] [CrossRef]
- Towns, D.R. Eradications as reverse invasions: Lessons from Pacific rat (Rattus exulans) removals on New Zealand islands. Biol. Invasions 2009, 11, 1719–1733. [Google Scholar] [CrossRef]
- Smith, D.G.; Shiinoki, E.K.; VanderWerf, E.A. Recovery of native species following rat eradication on Mokoli'i Island, O'ahu, Hawai'i. Pac. Sci. 2006, 60, 299–303. [Google Scholar] [CrossRef]
- Ratsirarson, J.; Silander, J.A., Jr.; Richard, A.F. Conservation and management of a threatened Madagascar palm species, Neodypsis decaryi, Jumele. Conserv. Biol. 1996, 10, 40–52. [Google Scholar]
- Whittaker, R.J.; Fernández-Palacios, J.M. Island Biogeography: Ecology, Evolution, and Conservation, 2nd ed; Oxford University Press: New York, NY, USA, 2007. [Google Scholar]
- Irvine, I. Late-Pliocene-Quaternary Biostratigraphy and Climatic Changes in DSDP 208, Lord Howe island Rise. Bull. Aust. Soc. Explor. Geophys. 1978, 9, 146–148. [Google Scholar] [CrossRef]
- Bijlsma, R.; Loeschcke, V. Environmental stress, adaptation and evolution: An overview. J. Evol. Biol. 2005, 18, 744–749. [Google Scholar]
- Billotte, N.; Marseillac, N.; Brottier, P.; Noyer, J.L.; Jacquemoud-Collet, J.P.; Moreau, C.; Couvreur, T.; Chevallier, M.H.; Pintaud, J.C.; Risterucci, A.M. Nuclear microsatellite markers for the date palm (Phoenix dactylifera L.): Characterization and utility across the genus Phoenix and in other palm genera. Mol. Ecol. Notes 2004, 4, 256–258. [Google Scholar] [CrossRef]
- Billotte, N.; Risterucci, A.M.; Barcelos, E.; Noyer, J.L.; Amblard, P.; Baurens, F.C. Development, characterisation, and across-taxa utility of oil palm (Elaeis guineensis Jacq.) microsatellite markers. Genome 2001, 44, 413–425. [Google Scholar]
- Gaiotto, F.A.; Brondani, R.P.V.; Grattapaglia, D. Microsatellite markers for heart of palm-Euterpe edulis and E. oleracea Mart. (Arecaceae). Mol. Ecol. Notes 2001, 1, 86–88. [Google Scholar] [CrossRef]
- Montufar, R.; Mariac, C.; Pham, J.L.; Pintaud, J.C. Isolation of 23 polymorphic microsatellite loci in the Neotropical palm Oenocarpus bataua Martius (Arecaceae). Mol. Ecol. Notes 2007, 7, 75–78. [Google Scholar]
- Shimizu, M.; Kosaka, N.; Shimada, T.; Nagahata, T.; Iwasaki, H.; Nagai, H.; Shiba, T.; Emi, M. Universal fluorescent labeling (UFL) method for automated microsatellite analysis. DNA Res. 2002, 9, 173–178. [Google Scholar]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar]
- Frankham, R. Conservation genetics. Annu. Rev. Genet. 1995, 29, 305–327. [Google Scholar] [CrossRef]
- Wright, S. The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Peakall, R.; Ruibal, M.; Lindenmayer, D.B. Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evolution 2003, 57, 1182–1195. [Google Scholar]
- Akçakaya, H. Population viability analyses with demographically and spatially structured models. Ecol. Bull. 2000, 48, 23–38. [Google Scholar]
- Akçakaya, H.; Atwood, J.L. A habitat-based metapopulation model of the California Gnatcatcher. Conserv. Biol. 1997, 11, 422–434. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Simmons, C.L.; Auld, T.D.; Hutton, I.; Baker, W.J.; Shapcott, A. Will Climate Change, Genetic and Demographic Variation or Rat Predation Pose the Greatest Risk for Persistence of an Altitudinally Distributed Island Endemic? Biology 2012, 1, 736-765. https://doi.org/10.3390/biology1030736
Simmons CL, Auld TD, Hutton I, Baker WJ, Shapcott A. Will Climate Change, Genetic and Demographic Variation or Rat Predation Pose the Greatest Risk for Persistence of an Altitudinally Distributed Island Endemic? Biology. 2012; 1(3):736-765. https://doi.org/10.3390/biology1030736
Chicago/Turabian StyleSimmons, Catherine Laura, Tony D. Auld, Ian Hutton, William J. Baker, and Alison Shapcott. 2012. "Will Climate Change, Genetic and Demographic Variation or Rat Predation Pose the Greatest Risk for Persistence of an Altitudinally Distributed Island Endemic?" Biology 1, no. 3: 736-765. https://doi.org/10.3390/biology1030736





