Concepts and Methods to Access Novel Antibiotics from Actinomycetes
Abstract
:1. Introduction
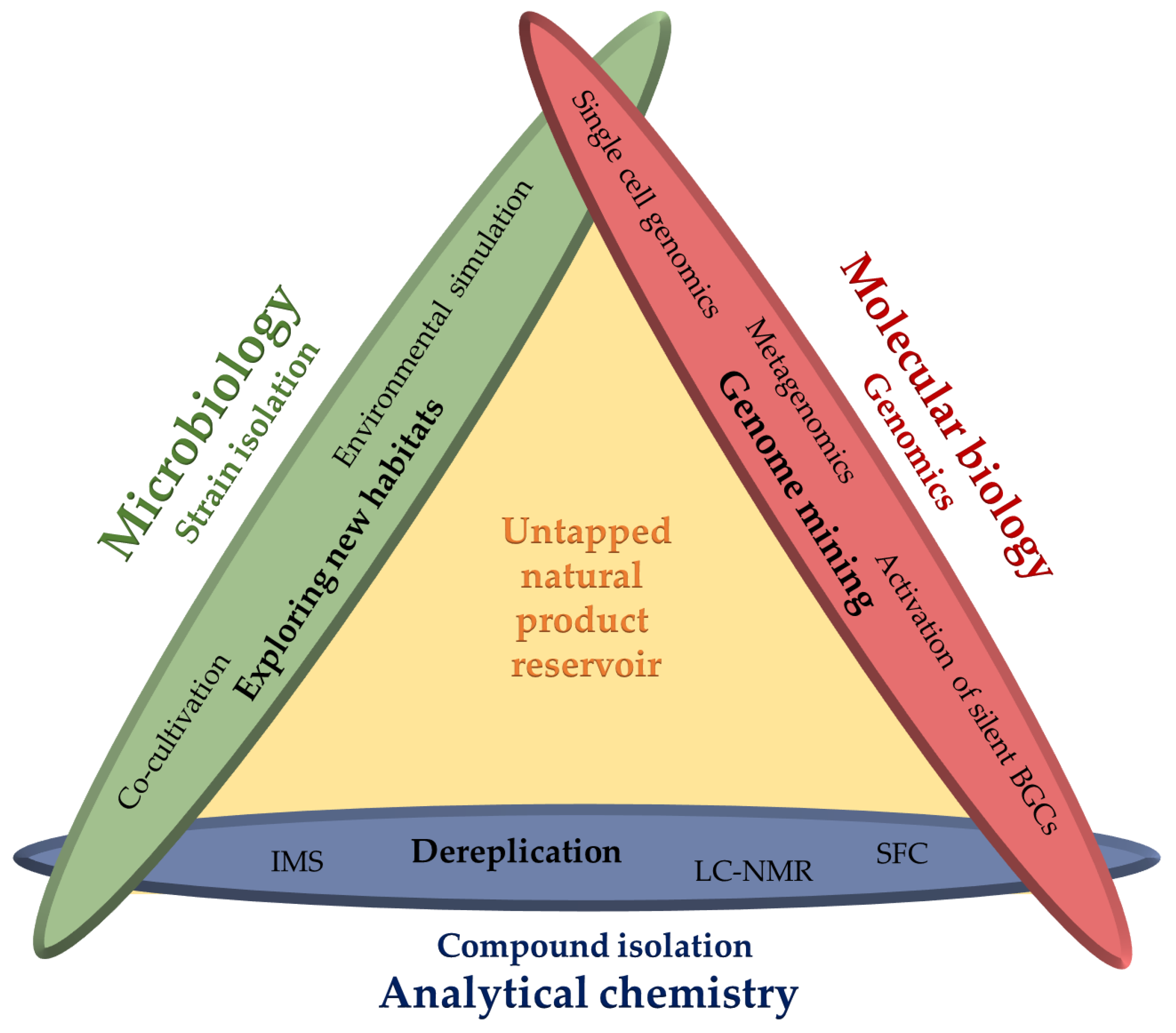
Outline of this Review
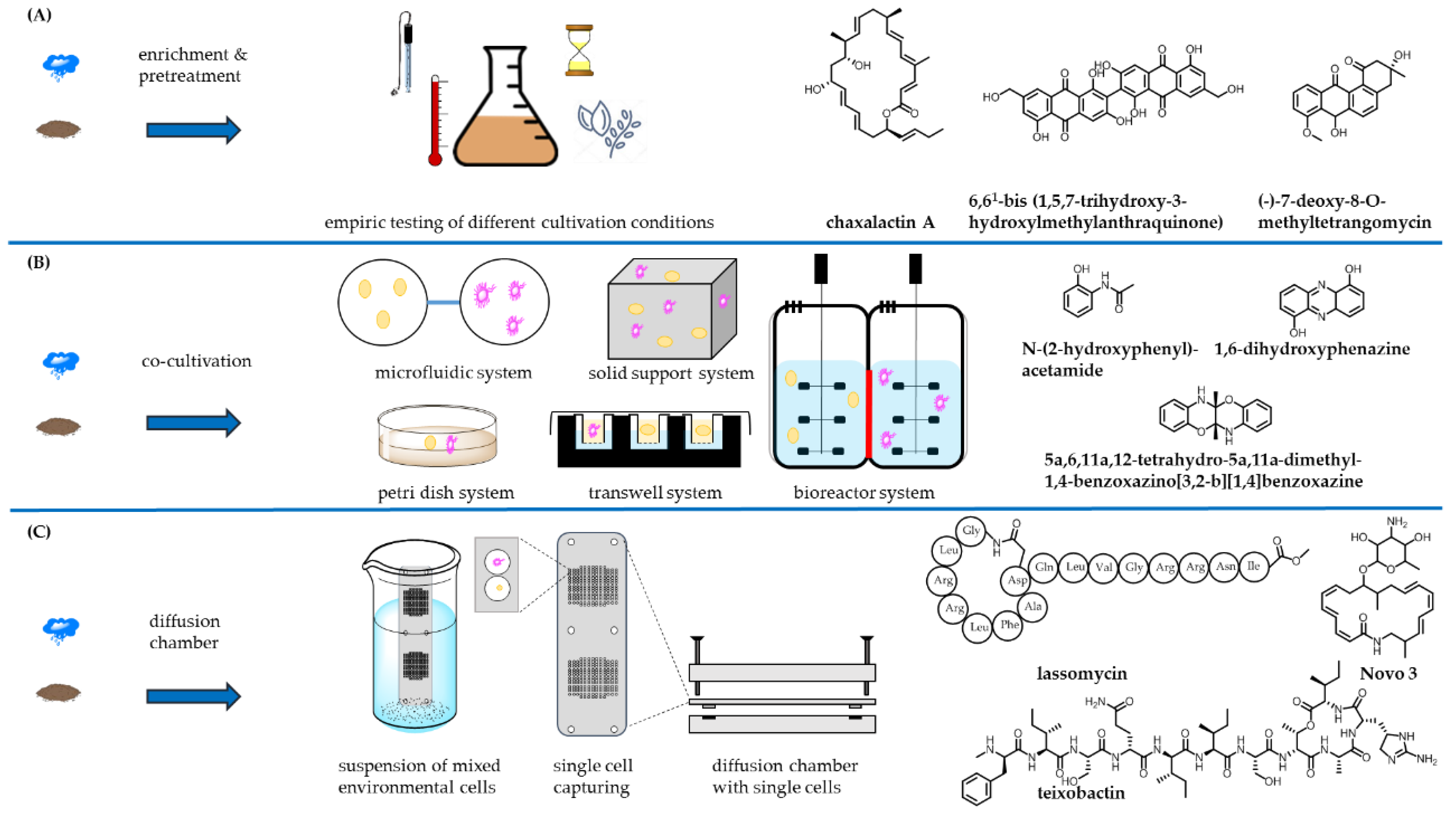
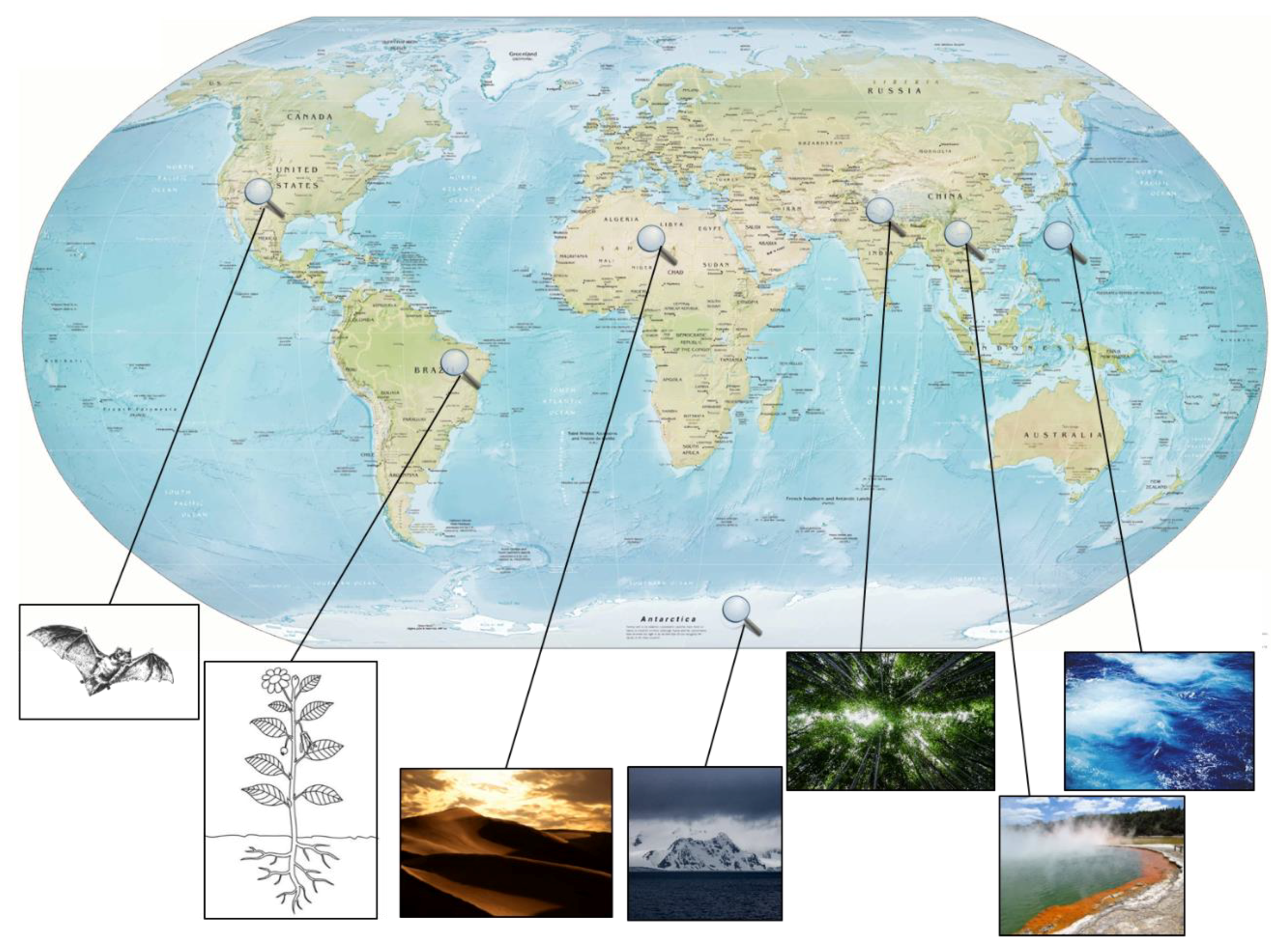
2. Exploring New Habitats
2.1. Extreme Environments as a Rich Source for Novel Strains
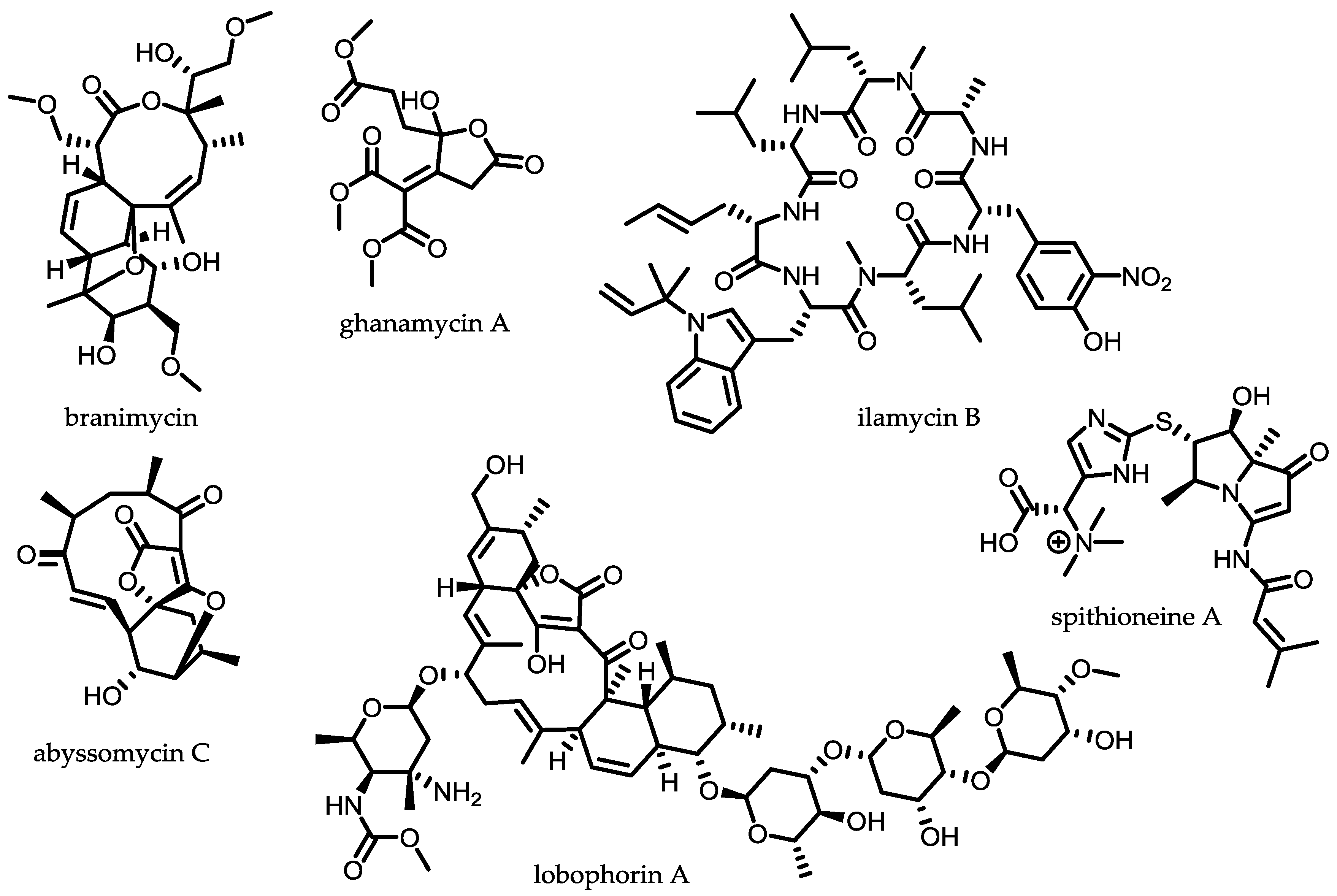
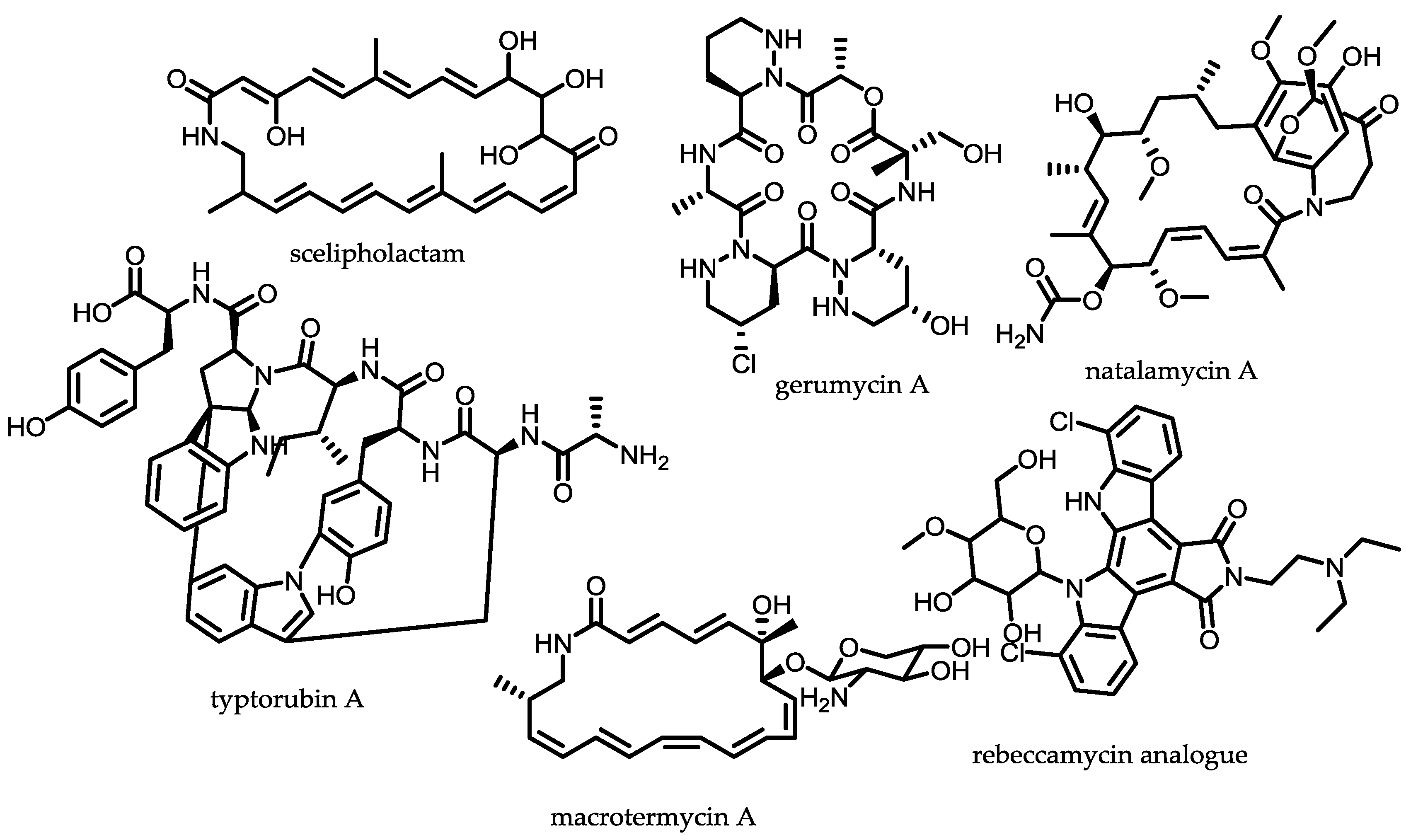
2.2. Endophytic Actinomycetes
2.3. Symbiotic Actinomycetes
2.4. Conclusions
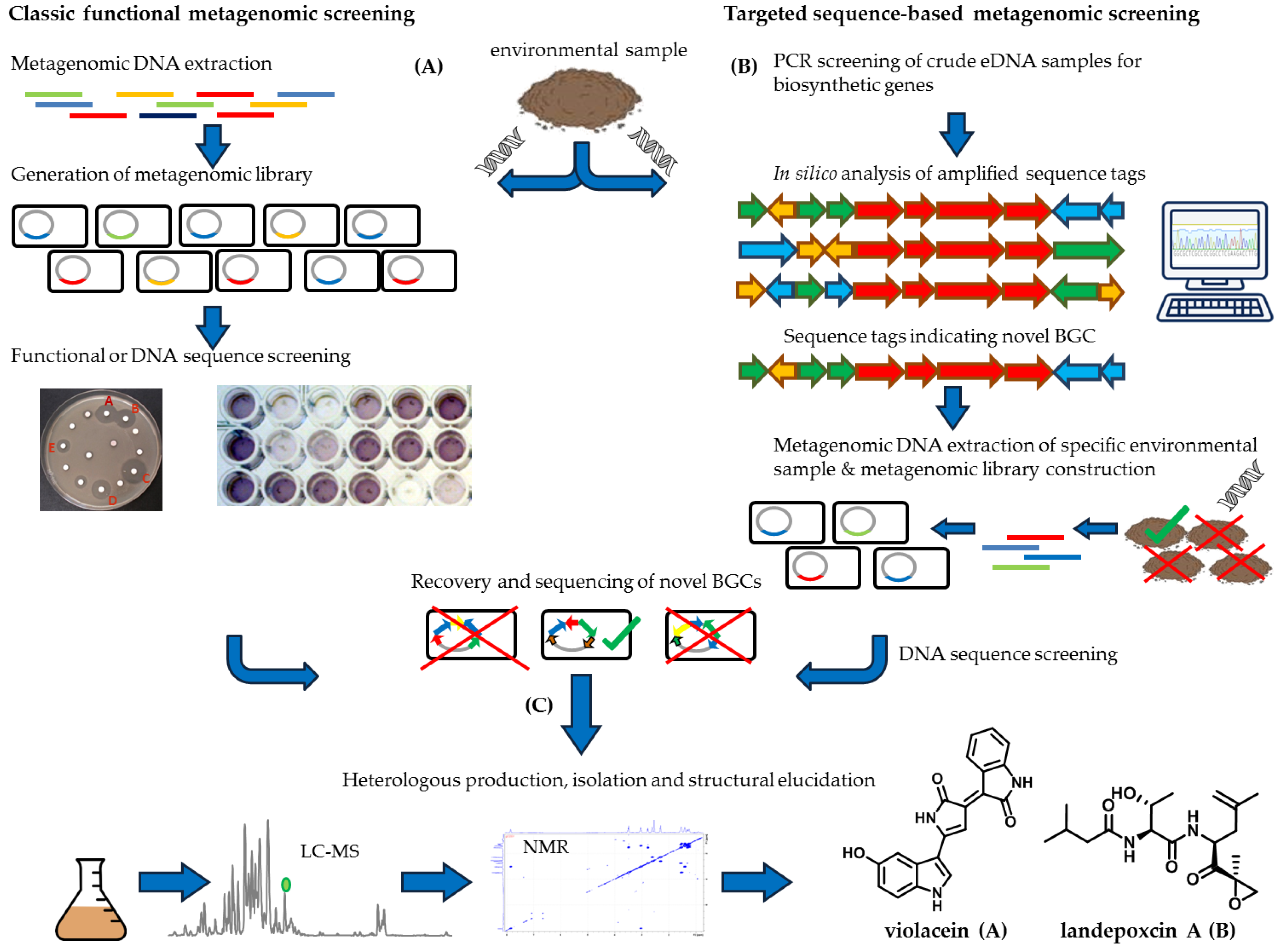
3. Metagenomic Approach to Exploit the Uncultured Bacterial Majority
3.1. The Metagenomic Screening Workflow
3.2. Direct Functional Metagenomic Screening
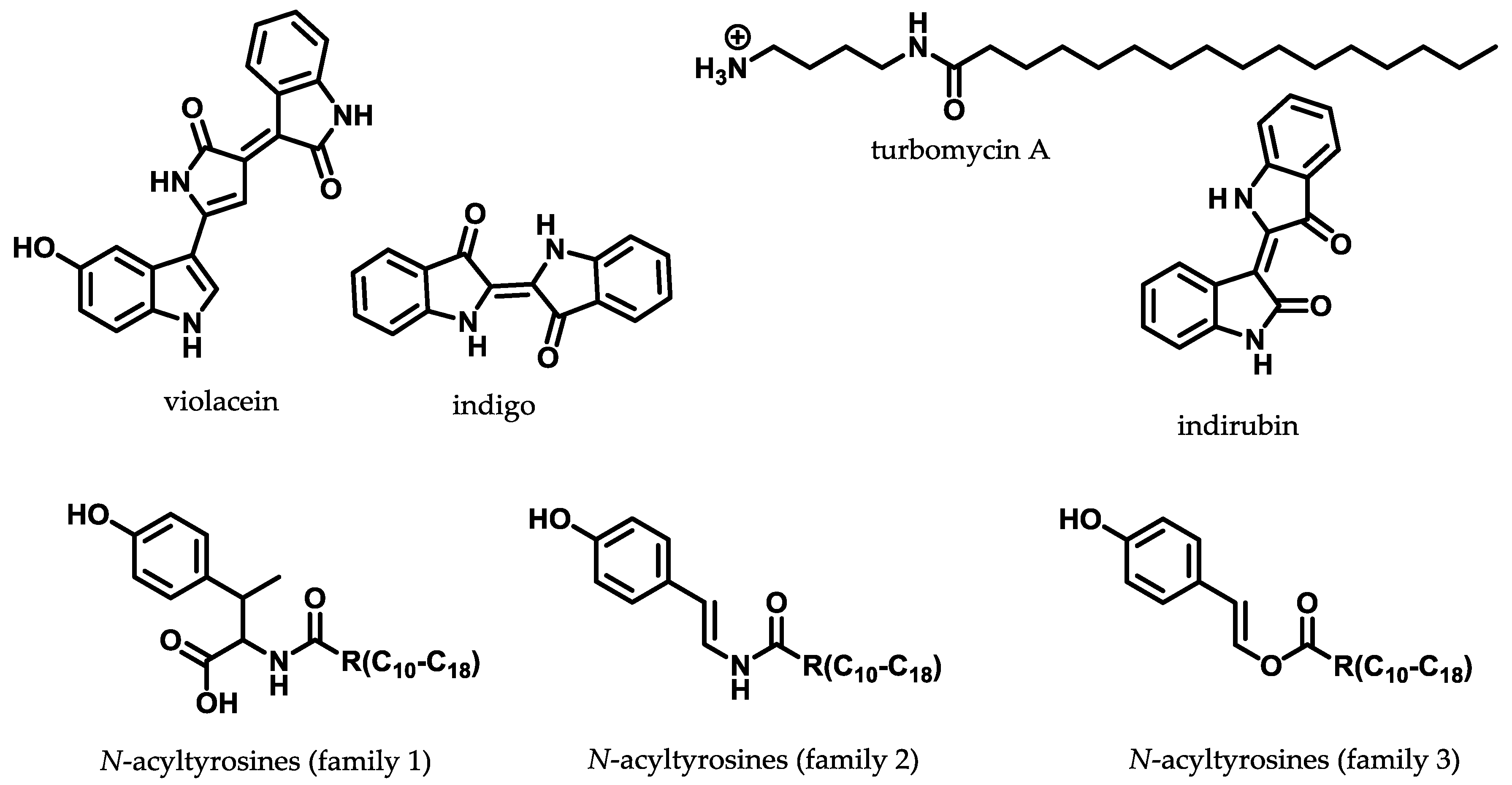
3.3. Sequence-Based Metagenomic Discovery Efforts
3.4. Metagenomics for the Assessment of Marine Endophytes
3.5. Sequence Boom: Potential of Next Generation Sequencing and Single-Cell Genomics
3.6. Conclusions and Future Considerations
4. Genome Mining: Current Reality and Future Promise of the Post-Genomic Era
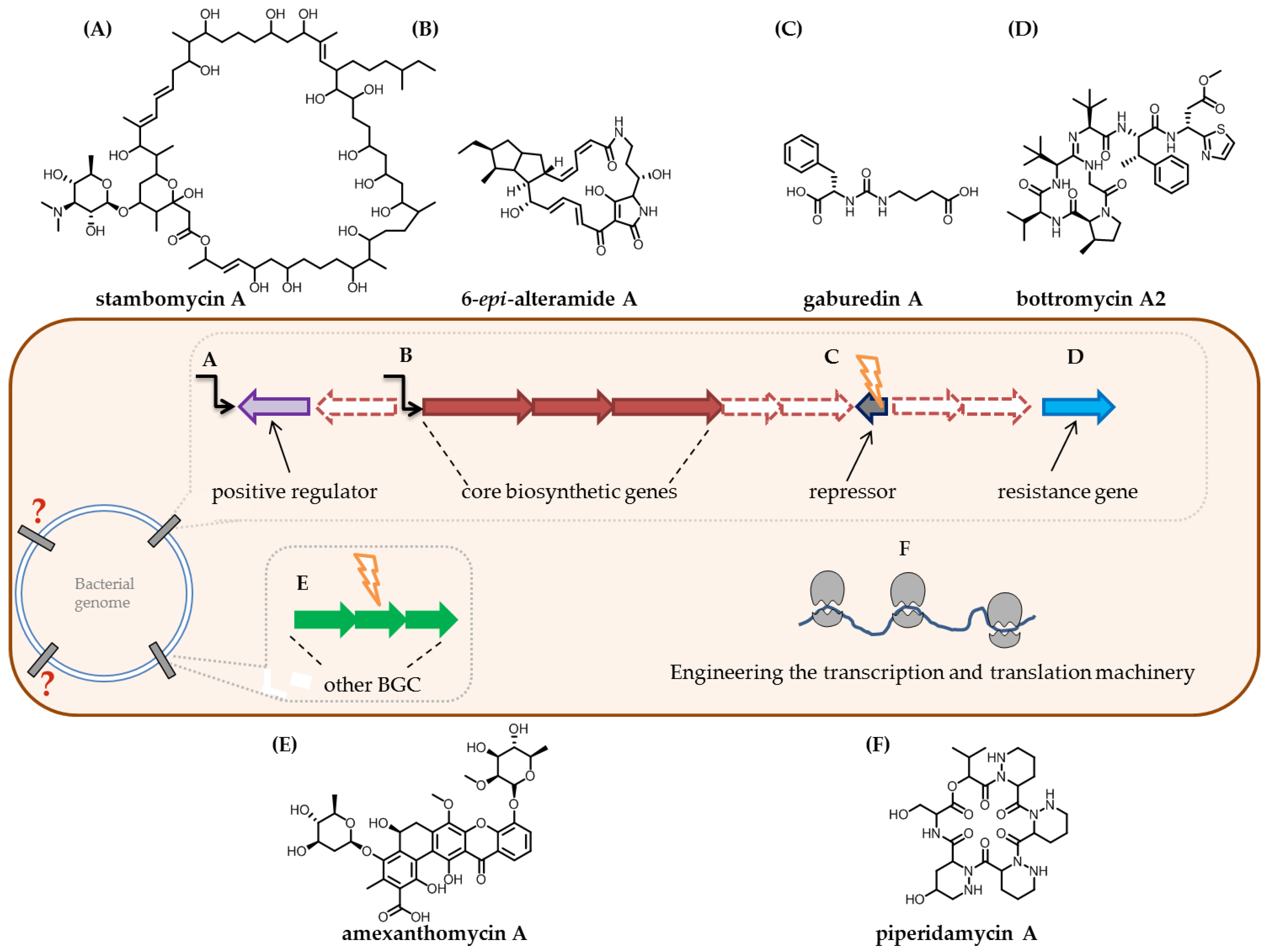
4.1. Biosynthetic Gene Cluster Prediction and Targeted Activation of BGCs
4.2. Utilising the Complexity of the Biosynthetic Machinery for the Discovery of Novel Natural Products
4.3. Silent BGC Activation by Chemical Elicitors, Ribosome Engineering and Chromatin Remodelling
4.4. Conclusions
5. Metabolomics for the Discovery of New Antibiotics Produced by Actinomycetes
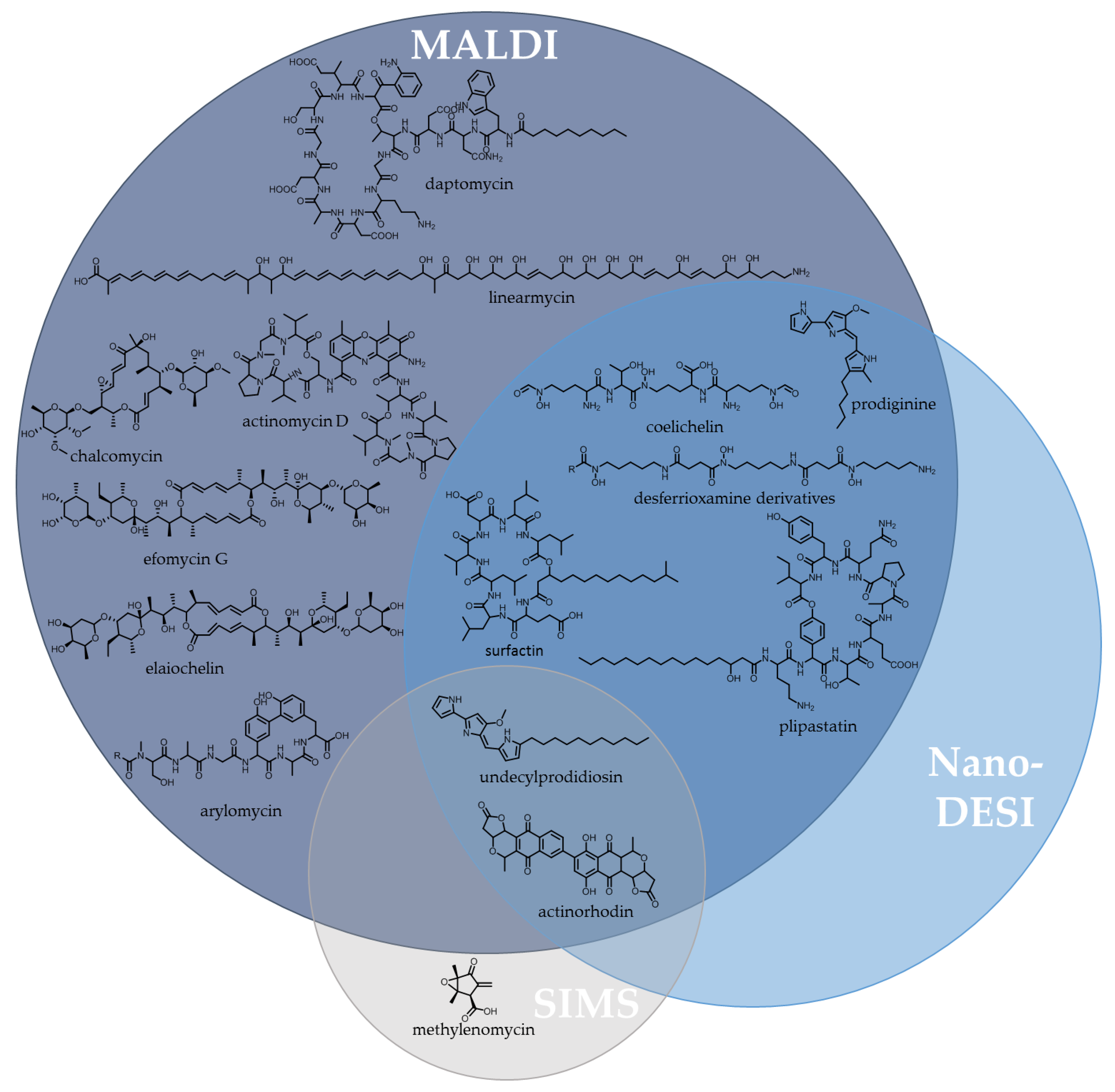
5.1. Innovations in Analytical Instrumentation for Natural Product Discovery
5.1.1. Imaging Mass Spectrometry (IMS)
5.1.2. Liquid Chromatography Coupled to Nuclear Magnetic Resonance Spectroscopy (LC-NMR)
5.1.3. Super Critical Fluid Chromatography (SFC)
5.2. Dereplication Using Metabolomic Data
5.3. Conclusions
6. Summary and Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation. Available online: http://apps.who.int/iris/bitstream/handle/10665/259744/9789241513449-eng.pdf;jsessionid=97E045EBFE3B29F286CBA3EE8360F0D1?sequence=1 (accessed on 31 January 2018).
- Aminov, R.I. A brief history of the antibiotic era: Lessons learned and challenges for the future. Front. Microbiol. 2010, 1. [Google Scholar] [CrossRef] [PubMed]
- De Lima Procópio, R.E.; da Silva, I.R.; Martins, M.K.; de Azevedo, J.L.; de Araújo, J.M. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 2012, 16, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Li, J.W.-H.; Vederas, J.C. Drug discovery and natural products: End of an era or an endless frontier? Science 2009, 325, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Luzhetskyy, A.; Pelzer, S.; Bechthold, A. The future of natural products as a source of new antibiotics. Curr. Opin. Investig. Drugs 2007, 8, 608–613. [Google Scholar] [PubMed]
- Thumar, J.T.; Dhulia, K.; Singh, S.P. Isolation and partial purification of an antimicrobial agent from halotolerant alkaliphilic Streptomyces aburaviensis strain Kut-8. World J. Microbiol. Biotechnol. 2010, 26, 2081–2087. [Google Scholar] [CrossRef]
- Hozzein, W.N.; Rabie, W.; Ali, M.I.A. Screening the Egyptian desert actinomycetes as candidates for new antimicrobial compounds and identification of a new desert Streptomyces strain. Afr. J. Biotechnol. 2011, 10, 2295–2301. [Google Scholar]
- Tiwari, K.; Gupta, R.K. Studies in Natural Products Chemistry. Bioactive Metabolites from Rare Actinomycetes; Elsevier: New York, NY, USA, 2014; Chapter 14. [Google Scholar]
- Bister, B.; Bischoff, D.; Strobele, M.; Riedlinger, J.; Reicke, A.; Wolter, F.; Bull, A.T.; Zahner, H.; Fiedler, H.P.; Sussmuth, R.D. Abyssomicin C—A polycyclic antibiotic from a marine Verrucosispora strain as an inhibitor of the p-aminobenzoic acid/tetrahydrofolate biosynthesis pathway. Angew. Chem. Int. Ed. Engl. 2004, 43, 2574–2576. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, H.-P.; Bruntner, C.; Riedlinger, J.; Bull, A.T.; Knutsen, G.; Goodfellow, M.; Jones, A.; Maldonado, L.; Pathom-Aree, W.; Beil, W.; et al. Proximicin A, B and C, novel aminofuran antibiotic and anticancer compounds isolated from marine strains of the actinomycete Verrucosispora. J. Antibiot. 2008, 61, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Genilloud, O.; Gonzalez, I.; Salazar, O.; Martin, J.; Tormo, J.R.; Vicente, F. Current approaches to exploit actinomycetes as a source of novel natural products. J. Ind. Microbiol. Biotechnol. 2011, 38, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Subramani, R.; Aalbersberg, W. Culturable rare Actinomycetes: Diversity, isolation and marine natural product discovery. Appl. Microbiol. Biotechnol. 2013, 97, 9291–9321. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Chater, K.F.; Cerdeño-Tárraga, A.-M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Vartoukian, S.R.; Palmer, R.M.; Wade, W.G. Strategies for culture of “unculturable” bacteria. FEMS Microbiol. Lett. 2010, 309, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, T.; Krug, D.; Bozkurt, N.; Duddela, S.; Jansen, R.; Garcia, R.; Gerth, K.; Steinmetz, H.; Müller, R. Correlating chemical diversity with taxonomic distance for discovery of natural products in myxobacteria. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Krug, D.; Müller, R. Secondary metabolomics: The impact of mass spectrometry-based approaches on the discovery and characterization of microbial natural products. Nat. Prod. Rep. 2014, 31, 768–783. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, S.C.; Müller, R. The impact of genomics on the exploitation of the myxobacterial secondary metabolome. Nat. Prod. Rep. 2009, 26, 1385–1407. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Müller, R. The impact of bacterial genomics on natural product research. Angew. Chem. Int. Ed. Engl. 2005, 44, 6828–6846. [Google Scholar] [CrossRef] [PubMed]
- Challinor, V.L.; Bode, H.B. Bioactive natural products from novel microbial sources. Ann. N. Y. Acad. Sci. 2015, 1354, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.S. Discovery of novel metabolites from marine actinomycetes. Curr. Opin. Microbiol. 2006, 9, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, G.B.; Balachandran, L. Sources of antibiotics: Hot springs. Biochem. Pharmacol. 2017, 134, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Passari, A.K.; Mishra, V.K.; Saikia, R.; Gupta, V.K.; Singh, B.P. Isolation, abundance and phylogenetic affiliation of endophytic actinomycetes associated with medicinal plants and screening for their in vitro antimicrobial biosynthetic potential. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Blunt, J.W.; Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, K.; Gupta, R.K. Diversity and isolation of rare actinomycetes: An overview. Crit. Rev. Microbiol. 2013, 39, 256–294. [Google Scholar] [CrossRef] [PubMed]
- Kurtboke, D.I.; French, J.R.J. Use of phage battery to investigate the actinofloral layers of termite gut microflora. J. Appl. Microbiol. 2007, 103, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Rateb, M.E.; Houssen, W.E.; Harrison, W.T.A.; Deng, H.; Okoro, C.K.; Asenjo, J.A.; Andrews, B.A.; Bull, A.T.; Goodfellow, M.; Ebel, R.; et al. Diverse metabolic profiles of a Streptomyces strain isolated from a hyper-arid environment. J. Nat. Prod. 2011, 74, 1965–1971. [Google Scholar] [CrossRef] [PubMed]
- Okoro, C.K.; Brown, R.; Jones, A.L.; Andrews, B.A.; Asenjo, J.A.; Goodfellow, M.; Bull, A.T. Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Anton. Leeuwenhoek 2009, 95, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Hadj Rabia-Boukhalfa, Y.; Eveno, Y.; Karama, S.; Selama, O.; Lauga, B.; Duran, R.; Hacène, H.; Eparvier, V. Isolation, purification and chemical characterization of a new angucyclinone compound produced by a new halotolerant Nocardiopsis sp. HR-4 strain. World J. Microbiol. Biotechnol. 2017, 33. [Google Scholar] [CrossRef] [PubMed]
- Duraipandiyan, V.; Al-Dhabi, N.A.; Ignacimuthu, S. New antimicrobial anthraquinone 6,61-bis (1,5,7-trihydroxy-3-hydroxymethylanthraquinone) isolated from Streptomyces sp. isolate ERI-26. Saudi J. Biol. Sci. 2016, 23, 731–735. [Google Scholar] [CrossRef] [PubMed]
- Arasu, M.V.; Duraipandiyan, V.; Agastian, P.; Ignacimuthu, S. Antimicrobial activity of Streptomyces spp. ERI-26 recovered from Western Ghats of Tamil Nadu. J. Mycol. Med. 2008, 18, 147–153. [Google Scholar] [CrossRef]
- Stewart, E.J. Growing Unculturable Bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef] [PubMed]
- Bollmann, A.; Lewis, K.; Epstein, S.S. Incubation of environmental samples in a diffusion chamber increases the diversity of recovered isolates. Appl. Environ. Microbiol. 2007, 73, 6386–6390. [Google Scholar] [CrossRef] [PubMed]
- Nichols, D.; Cahoon, N.; Trakhtenberg, E.M.; Pham, L.; Mehta, A.; Belanger, A.; Kanigan, T.; Lewis, K.; Epstein, S.S. Use of iChip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 2010, 76, 2445–2450. [Google Scholar] [CrossRef] [PubMed]
- Peoples, A.J.; Zhang, Q.; Millett, W.P.; Rothfeder, M.T.; Pescatore, B.C.; Madden, A.A.; Ling, L.L.; Moore, C.M. Neocitreamicins I and II, novel antibiotics with activity against methicillin resistant Staphylococcus aureus and vancomycin-resistant Enterococci. J. Antibiot. 2008, 61, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, T.; Lewis, K.; Epstein, S.S. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 2002, 296, 1127–1129. [Google Scholar] [CrossRef] [PubMed]
- Peoples, A.; Zhang, Q.; Moore, C.; Ling, L.; Rothfeder, M.; Lewis, K. Macrolactam Compounds. Patent WO2009026527A1, 17 January 2012. [Google Scholar]
- Gavrish, E.; Sit, C.S.; Cao, S.; Kandror, O.; Spoering, A.; Peoples, A.; Ling, L.; Fetterman, A.; Hughes, D.; Bissell, A.; et al. Lassomycin, a ribosomally synthesized cyclic peptide, kills Mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem. Biol. 2014, 21, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Goers, L.; Freemont, P.; Polizzi, K.M. Co-culture systems and technologies: Taking synthetic biology to the next level. J. R. Soc. Interface 2014, 11. [Google Scholar] [CrossRef] [PubMed]
- Dashti, Y.; Grkovic, T.; Abdelmohsen, U.R.; Hentschel, U.; Quinn, R.J. Production of induced secondary metabolites by a co-culture of sponge-associated actinomycetes, Actinokineospora sp. EG49 and Nocardiopsis sp. RV163. Mar. Drugs 2014, 12, 3046–3059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zengler, K.; Toledo, G.; Rappe, M.; Elkins, J.; Mathur, E.J.; Short, J.M.; Keller, M. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 2002, 99, 15681–15686. [Google Scholar] [CrossRef] [PubMed]
- Zengler, K.; Walcher, M.; Clark, G.; Haller, I.; Toledo, G.; Holland, T.; Mathur, E.J.; Woodnutt, G.; Short, J.M.; Keller, M. High-throughput cultivation of microorganisms using microcapsules. In Environmental Microbiology; Leadbetter, J.R., Ed.; Elsevier Academic Press: Amsterdam, The Netherlands; Boston, MA, USA, 2005; pp. 124–130. [Google Scholar]
- Ben-Dov, E.; Kramarsky-Winter, E.; Kushmaro, A. An in situ method for cultivating microorganisms using a double encapsulation technique. FEMS Microbiol. Ecol. 2009, 68, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Bomar, L.; Maltz, M.; Colston, S.; Graf, J. Directed culturing of microorganisms using metatranscriptomics. mBio 2011, 2. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.J.; Kirkegaard, R.; Szul, M.J.; Johnson, Z.I.; Zinser, E.R. Facilitation of robust growth of prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl. Environ. Microbiol. 2008, 74, 4530–4534. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Hanada, S.; Manome, A.; Tsuchida, T.; Kurane, R.; Nakamura, K.; Kamagata, Y. Catellibacterium nectariphilum gen. nov., sp. nov., which requires a diffusible compound from a strain related to the genus Sphingomonas for vigorous growth. Int. J. Syst. Evol. Microbiol. 2004, 54, 955–959. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, J.-J.; Masui, R.; Kuramitsu, S.; Sung, M.-H. A commensal symbiotic interrelationship for the growth of Symbiobacterium toebii with its partner bacterium, Geobacillus toebii. BMC Res. Notes 2011, 4. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.J.; Lenski, R.E.; Zinser, E.R. The Black Queen Hypothesis: Evolution of dependencies through adaptive gene loss. mBio 2012, 3, e00036-12. [Google Scholar] [CrossRef] [PubMed]
- Schofield, M.M.; Jain, S.; Porat, D.; Dick, G.J.; Sherman, D.H. Identification and analysis of the bacterial endosymbiont specialized for production of the chemotherapeutic natural product ET-743. Environ. Microbiol. 2015, 17, 3964–3975. [Google Scholar] [CrossRef] [PubMed]
- Rashad, F.M.; Fathy, H.M.; El-Zayat, A.S.; Elghonaimy, A.M. Isolation and characterization of multifunctional Streptomyces species with antimicrobial, nematicidal and phytohormone activities from marine environments in Egypt. Microbiol. Res. 2015, 175, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. ChemBioChem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Engelhardt, K.; Degnes, K.F.; Kemmler, M.; Bredholt, H.; Fjaervik, E.; Klinkenberg, G.; Sletta, H.; Ellingsen, T.E.; Zotchev, S.B. Production of a new thiopeptide antibiotic, TP-1161, by a marine Nocardiopsis species. Appl. Environ. Microbiol. 2010, 76, 4969–4976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dufour, C.; Wink, J.; Kurz, M.; Kogler, H.; Olivan, H.; Sablé, S.; Heyse, W.; Gerlitz, M.; Toti, L.; Nußer, A.; et al. Isolation and structural elucidation of armeniaspirols A–C: Potent antibiotics against Gram-positive pathogens. Chem. Eur. J. 2012, 18, 16123–16128. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Huang, H.; Xie, Y.; Liu, Z.; Zhao, J.; Zhang, C.; Jia, Y.; Zhang, Y.; Zhang, H.; Zhang, T.; et al. Biosynthesis of ilamycins featuring unusual building blocks and engineered production of enhanced anti-tuberculosis agents. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Boubetra, D.; Sabaou, N.; Zitouni, A.; Bijani, C.; Lebrihi, A.; Mathieu, F. Taxonomy and chemical characterization of new antibiotics produced by Saccharothrix SA198 isolated from a Saharan soil. Microbiol. Res. 2013, 168, 223–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, L.-H.; Cheah, Y.-K.; Mohd Sidik, S.; Ab Mutalib, N.-S.; Tang, Y.-L.; Lin, H.-P.; Hong, K. Molecular characterization of Antarctic actinobacteria and screening for antimicrobial metabolite production. World J. Microbiol. Biotechnol. 2012, 28, 2125–2137. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Salam, N.; Jiao, J.-Y.; Jiang, H.-C.; Zhou, E.-M.; Yin, Y.-R.; Ming, H.; Li, W.-J. Diversity of culturable thermophilic Actinobacteria in hot springs in Tengchong, china and studies of their biosynthetic gene profiles. Microb. Ecol. 2016, 72, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Silva-Lacerda, G.R.; Santana, R.C.F.; Vicalvi-Costa, M.C.V.; Solidônio, E.G.; Sena, K.X.F.R.; Lima, G.M.S.; Araújo, J.M. Antimicrobial potential of actinobacteria isolated from the rhizosphere of the Caatinga biome plant Caesalpinia pyramidalis Tul. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Hamm, P.S.; Caimi, N.A.; Northup, D.E.; Valdez, E.W.; Buecher, D.C.; Dunlap, C.A.; Labeda, D.P.; Lueschow, S.; Porras-Alfaro, A. Western bats as a reservoir of novel Streptomyces species with antifungal activity. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Braña, A.F.; Sarmiento-Vizcaíno, A.; Pérez-Victoria, I.; Otero, L.; Fernández, J.; Palacios, J.J.; Martín, J.; De La Cruz, M.; Díaz, C.; Vicente, F.; et al. Branimycins B and C, antibiotics produced by the abyssal actinobacterium Pseudonocardia carboxydivorans M-227. J. Nat. Prod. 2017, 80, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.-H.; Gu, K.-B.; Zhang, D.-J.; Li, Y.-G.; Tian, L. Ghanamycins A and B, two novel γ-butyrolactones from marine-derived Streptomyces ghanaensis TXC6-16. J. Antibiot. 2017, 70, 733–736. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Li, S.; Chen, Y.; Tian, X.; Zhang, H.; Zhang, G.; Zhang, W.; Yang, X.; Zhang, S.; Ju, J.; et al. Lobophorins E and F, new spirotetronate antibiotics from a South China Sea-derived Streptomyces sp. SCSIO 01127. J. Antibiot. 2011, 64, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.; MacMillan, J.B. Spithioneines A and B, two new bohemamine derivatives possessing ergothioneine moiety from a marine-derived Streptomyces spinoverrucosus. Org. Lett. 2015, 17, 3046–3049. [Google Scholar] [CrossRef] [PubMed]
- Keller, S.; Nicholson, G.; Drahl, C.; Sorensen, E.; Fiedler, H.-P.; Sussmuth, R.D. Abyssomicins G and H and atrop-abyssomicin C from the marine Verrucosispora strain AB-18-032. J. Antibiot. 2007, 60, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, Y.; Yu, L.; Miyanaga, S.; Fukuda, T.; Saitoh, N.; Sakurai, H.; Saiki, I.; Alonso-Vega, P.; Trujillo, M.E. Abyssomicin I, a modified polycyclic polyketide from Streptomyces sp. CHI39. J. Nat. Prod. 2010, 73, 1943–1946. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.-M.; Li, S.-H.; Gorls, H.; Schollmeyer, D.; Hilliger, M.; Grabley, S.; Sattler, I. Abyssomicin E, a highly functionalized polycyclic metabolite from Streptomyces species. Org. Lett. 2007, 9, 2437–2440. [Google Scholar] [CrossRef] [PubMed]
- Feling, R.H.; Buchanan, G.O.; Mincer, T.J.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Salinosporamide A: A highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. Engl. 2003, 42, 355–357. [Google Scholar] [CrossRef] [PubMed]
- Stadler, M.; Bitzer, J.; Mayer-Bartschmid, A.; Muller, H.; et-Buchholz, J.; Gantner, F.; Tichy, H.V.; Reinemer, P.; Bacon, K.B. Cinnabaramides A–G: Analogues of lactacystin and salinosporamide from a terrestrial streptomycete. J. Nat. Prod. 2007, 70, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Messaoudi, O.; Bendahou, M.; Benamar, I.; Abdelwouhid, D.-E. Identification and preliminary characterization of non-polyene antibiotics secreted by new strain of actinomycete isolated from sebkha of Kenadsa, Algeria. Asian Pac. J. Trop. Biomed. 2015, 5, 438–445. [Google Scholar] [CrossRef]
- Shin, B.; Kim, B.-Y.; Cho, E.; Oh, K.-B.; Shin, J.; Goodfellow, M.; Oh, D.-C. Actinomadurol, an antibacterial norditerpenoid from a rare Actinomycete, Actinomadura sp. KC 191. J. Nat. Prod. 2016, 79, 1886–1890. [Google Scholar] [CrossRef] [PubMed]
- Al-Dhabi, N.A.; Esmail, G.A.; Duraipandiyan, V.; Valan Arasu, M.; Salem-Bekhit, M.M. Isolation, identification and screening of antimicrobial thermophilic Streptomyces sp. Al-Dhabi-1 isolated from Tharban hot spring, Saudi Arabia. Extremophiles 2016, 20, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Strobel, G.; Daisy, B. Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. Rev. 2003, 67, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, A.; Takahashi, Y. Endophytic actinomycetes: Promising source of novel bioactive compounds. J. Antibiot. 2017, 70, 514–519. [Google Scholar] [CrossRef] [PubMed]
- Chagas, F.O.; Ruzzini, A.C.; Bacha, L.V.; Samborskyy, M.; Conti, R.; Pessotti, R.C.; de Oliveira, L.G.; Clardy, J.; Pupo, M.T. Genome sequence of Streptomyces sp. strain RTd22, an endophyte of the Mexican sunflower. Genome Announc. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Barke, J.; Seipke, R.F.; Grüschow, S.; Heavens, D.; Drou, N.; Bibb, M.J.; Goss, R.J.M.; Yu, D.W.; Hutchings, M.I. A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol. 2010, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, D.-C.; Poulsen, M.; Currie, C.R.; Clardy, J. Sceliphrolactam, a polyene macrocyclic lactam from a wasp-associated Streptomyces sp. Org. Lett. 2011, 13, 752–755. [Google Scholar] [CrossRef] [PubMed]
- Beemelmanns, C.; Ramadhar, T.R.; Kim, K.H.; Klassen, J.L.; Cao, S.; Wyche, T.P.; Hou, Y.; Poulsen, M.; Bugni, T.S.; Currie, C.R.; et al. Macrotermycins A–D, glycosylated macrolactams from a termite-associated Amycolatopsis sp. M39. Org. Lett. 2017, 19, 1000–1003. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Ramadhar, T.R.; Beemelmanns, C.; Cao, S.; Poulsen, M.; Currie, C.R.; Clardy, J. Natalamycin A, an ansamycin from a termite-associated Streptomyces sp. Chem. Sci. 2014, 5, 4333–4338. [Google Scholar] [CrossRef] [PubMed]
- Sit, C.S.; Ruzzini, A.C.; van Arnam, E.B.; Ramadhar, T.R.; Currie, C.R.; Clardy, J. Variable genetic architectures produce virtually identical molecules in bacterial symbionts of fungus-growing ants. Proc. Natl. Acad. Sci. USA 2015, 112, 13150–13154. [Google Scholar] [CrossRef] [PubMed]
- Nettleton, D.E.; Doyle, T.W.; Krishnan, B.; Matsumoto, G.K.; Clardy, J. Isolation and structure of rebeccamycin—A new antitumor antibiotic from Nocardia aerocoligenes. Tetrahedron Lett. 1985, 26, 4011–4014. [Google Scholar] [CrossRef]
- Van Arnam, E.B.; Ruzzini, A.C.; Sit, C.S.; Currie, C.R.; Clardy, J. A rebeccamycin analog provides plasmid-encoded niche defense. J. Am. Chem. Soc. 2015, 137, 14272–14274. [Google Scholar] [CrossRef] [PubMed]
- Wyche, T.P.; Ruzzini, A.C.; Schwab, L.; Currie, C.R.; Clardy, J. Tryptorubin A: A polycyclic peptide from a fungus-derived streptomycete. J. Am. Chem. Soc. 2017, 139, 12899–12902. [Google Scholar] [CrossRef] [PubMed]
- Pettit, R.K. Culturability and secondary metabolite diversity of extreme microbes: Expanding contribution of deep sea and deep-sea vent microbes to natural product discovery. Mar. Biotechnol. 2011, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wilson, Z.E.; Brimble, M.A. Molecules derived from the extremes of life. Nat. Prod. Rep. 2009, 26, 44–71. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.F.; Simmons, L.; Kim, J.H.; Schmidt, E.W. Metagenomic approaches to natural products from free-living and symbiotic organisms. Nat. Prod. Rep. 2009, 26, 1488–1503. [Google Scholar] [CrossRef] [PubMed]
- Techtmann, S.M.; Hazen, T.C. Metagenomic applications in environmental monitoring and bioremediation. J. Ind. Microbiol. Biotechnol. 2016, 43, 1345–1354. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, S.; Selvakumar, R.; Sathishkumar, M.; Swaminathan, K.; Lakshmanaperumalsamy, P.; Singh, A.; Jain, S.K. Nitrate removal using Brevundimonas diminuta MTCC 8486 from ground water. Water Sci. Technol. 2009, 60, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Green, S.J.; Prakash, O.; Jasrotia, P.; Overholt, W.A.; Cardenas, E.; Hubbard, D.; Tiedje, J.M.; Watson, D.B.; Schadt, C.W.; Brooks, S.C.; et al. Denitrifying bacteria from the genus Rhodanobacter dominate bacterial communities in the highly contaminated subsurface of a nuclear legacy waste site. Appl. Environ. Microbiol. 2012, 78, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
- Dubinsky, E.A.; Conrad, M.E.; Chakraborty, R.; Bill, M.; Borglin, S.E.; Hollibaugh, J.T.; Mason, O.U.; Piceno, Y.M.; Reid, F.C.; Stringfellow, W.T.; et al. Succession of hydrocarbon-degrading bacteria in the aftermath of the deepwater horizon oil spill in the gulf of Mexico. Environ. Sci. Technol. 2013, 47, 10860–10867. [Google Scholar] [CrossRef] [PubMed]
- Lok, C. Mining the microbial dark matter. Nature 2015, 522, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.F.; Chao, C.J.; Handelsman, J.; Clardy, J. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org. Lett. 2001, 3, 1981–1984. [Google Scholar] [CrossRef] [PubMed]
- Owen, J.G.; Charlop-Powers, Z.; Smith, A.G.; Ternei, M.A.; Calle, P.Y.; Reddy, B.V.B.; Montiel, D.; Brady, S.F. Multiplexed metagenome mining using short DNA sequence tags facilitates targeted discovery of epoxyketone proteasome inhibitors. Proc. Natl. Acad. Sci. USA 2015, 112, 4221–4226. [Google Scholar] [CrossRef] [PubMed]
- Katz, M.; Hover, B.M.; Brady, S.F. Culture-independent discovery of natural products from soil metagenomes. J. Ind. Microbiol. Biotechnol. 2016, 43, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Altaee, N.; Jawad, M.; Hameed, I. Characterization of metabolites produced by E. coli and analysis of its chemical compounds using GC-MS. IJCPR 2017, 7, 393–399. [Google Scholar]
- Wexler, M.; Johnston, A.W.B. Wide host-range cloning for functional metagenomics. In Metagenomics: Methods and Protocols; Streit, W.R., Daniel, R., Eds.; Humana: London, UK; Springer: New York, NY, USA, 2010; pp. 77–96. [Google Scholar]
- Gabor, E.M.; Alkema, W.B.; Janssen, D.B. Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ. Microbiol. 2004, 6, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Stevens, D.C.; Conway, K.R.; Pearce, N.; Villegas-Penaranda, L.R.; Garza, A.G.; Boddy, C.N. Alternative sigma factor over-expression enables heterologous expression of a type II polyketide biosynthetic pathway in Escherichia coli. PLoS ONE 2013, 8, e64858. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, B.A.; Admiraal, S.J.; Gramajo, H.; Cane, D.E.; Khosla, C. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 2001, 291, 1790–1792. [Google Scholar] [CrossRef] [PubMed]
- Boddy, C.N.; Hotta, K.; Tse, M.L.; Watts, R.E.; Khosla, C. Precursor-directed biosynthesis of epothilone in Escherichia coli. J. Am. Chem. Soc. 2004, 126, 7436–7437. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Hotta, K.; Praseuth, A.P.; Koketsu, K.; Migita, A.; Boddy, C.N.; Wang, C.C.; Oguri, H.; Oikawa, H. Total biosynthesis of antitumor nonribosomal peptides in Escherichia coli. Nat. Chem. Biol. 2006, 2, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.A.; Van Der Donk, W.A. New insights into the biosynthetic logic of ribosomally synthesized and post-translationally modified peptide natural products. Cell Chem. Biol. 2016, 23, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Ongley, S.; Bian, X.; Neilan, B.A.; Müller, R. Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat. Prod. Rep. 2013, 30, 1121–1138. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, S.C. Biosynthesis and Heterologous Production of Myxobacterial Secondary Metabolites. Ph.D. Thesis, Fachbereich Biowissenschaftern und Psychologie, Technische Carolo-Wilhemina-Universität, Braunschweig, Germany, 2005. [Google Scholar]
- Ferrari, B.C.; Binnerup, S.J.; Gillings, M. Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl. Environ. Microbiol. 2005, 71, 8714–8720. [Google Scholar] [CrossRef] [PubMed]
- Cueto, M.; Jensen, P.R.; Kauffman, C.; Fenical, W.; Lobkovsky, E.; Clardy, J. Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J. Nat. Prod. 2001, 64, 1444–1446. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.M.; Bradshaw, E.; Seipke, R.F.; Hutchings, M.I.; McArthur, M. Use and discovery of chemical elicitors that stimulate biosynthetic gene clusters in Streptomyces bacteria. Methods Enzymol. 2012, 517, 367–385. [Google Scholar] [PubMed]
- Montiel, D.; Kang, H.-S.; Chang, F.-Y.; Charlop-Powers, Z.; Brady, S.F. Yeast homologous recombination-based promoter engineering for the activation of silent natural product biosynthetic gene clusters. Proc. Natl. Acad. Sci. USA 2015, 112, 8953–8958. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.-Y.; Ternei, M.A.; Calle, P.Y.; Brady, S.F. Discovery and synthetic refactoring of tryptophan dimer gene clusters from the environment. J. Am. Chem. Soc. 2013, 135, 17906–17912. [Google Scholar] [CrossRef] [PubMed]
- Trindade, M.; van Zyl, L.J.; Navarro-Fernández, J.; Abd Elrazak, A. Targeted metagenomics as a tool to tap into marine natural product diversity for the discovery and production of drug candidates. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Williamson, L.L.; Borlee, B.R.; Schloss, P.D.; Guan, C.; Allen, H.K.; Handelsman, J. Intracellular screen to identify metagenomic clones that induce or inhibit a quorum-sensing biosensor. Appl. Environ. Microbiol. 2005, 71, 6335–6344. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, T.; Abe, T.; Ikemura, T.; Watanabe, K. Substrate-induced gene-expression screening of environmental metagenome libraries for isolation of catabolic genes. Nat. Biotechnol. 2005, 23, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.K.; Chung, E.J.; Kim, J.-C.; Choi, G.J.; Jang, K.S.; Chung, Y.R.; Cho, K.Y.; Lee, S.-W. Characterization of a forest soil metagenome clone that confers indirubin and indigo production on Escherichia coli. Appl. Environ. Microbiol. 2005, 71, 7768–7777. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.F.; Chao, C.J.; Clardy, J. New natural product families from an environmental DNA (eDNA) gene cluster. J. Am. Chem. Soc. 2002, 124, 9968–9969. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, D.E.; Brady, S.F.; Bettermann, A.D.; Cianciotto, N.P.; Liles, M.R.; Rondon, M.R.; Clardy, J.; Goodman, R.M.; Handelsman, J. Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl. Environ. Microbiol. 2002, 68, 4301–4306. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.M.; Qiao, Y.; Ang, E.L.; Zhao, H. Using natural products for drug discovery: The impact of the genomics era. Expert Opin. Drug Discov. 2017, 12, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Beloqui, A.; Vieites, J.M.; Guazzaroni, M.E.; Berger, I.; Aharoni, A. Interplay of metagenomics and in vitro compartmentalization. Microb. Biotechnol. 2009, 2, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Owen, J.G.; Robins, K.J.; Parachin, N.S.; Ackerley, D.F. A functional screen for recovery of 4’-phosphopantetheinyl transferase and associated natural product biosynthesis genes from metagenome libraries. Environ. Microbiol. 2012, 14, 1198–1209. [Google Scholar] [CrossRef] [PubMed]
- Beld, J.; Sonnenschein, E.C.; Vickery, C.R.; Noel, J.P.; Burkart, M.D. The phosphopantetheinyl transferases: Catalysis of a post-translational modification crucial for life. Nat. Prod. Rep. 2013, 31, 61–108. [Google Scholar] [CrossRef] [PubMed]
- Bitok, J.K.; Lemetre, C.; Ternei, M.A.; Brady, S.F. Identification of biosynthetic gene clusters from metagenomic libraries using PPTase complementation in a Streptomyces host. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed]
- Gaitatzis, N.; Kunze, B.; Müller, R. In vitro reconstitution of the myxochelin biosynthetic machinery of Stigmatella aurantiaca Sg a15: Biochemical characterization of a reductive release mechanism from nonribosomal peptide synthetases. Proc. Natl. Acad. Sci. USA 2001, 98, 11136–11141. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; de Los Santos, E.L.C.; Kim, H.U.; Nave, M.; et al. antiSMASH 4.0—Improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 2017, 45, W36–W41. [Google Scholar] [CrossRef] [PubMed]
- Reddy, B.V.B.; Milshteyn, A.; Charlop-Powers, Z.; Brady, S.F. eSNaPD: A versatile, web-based bioinformatics platform for surveying and mining natural product biosynthetic diversity from metagenomes. Chem. Biol. 2014, 21, 1023–1033. [Google Scholar] [CrossRef] [PubMed]
- Ziemert, N.; Podell, S.; Penn, K.; Badger, J.H.; Allen, E.; Jensen, P.R. The natural product domain seeker NaPDoS: A phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS ONE 2012, 7, e34064. [Google Scholar] [CrossRef] [PubMed]
- Owen, J.G.; Reddy, B.V.; Ternei, M.A.; Charlop-Powers, Z.; Calle, P.Y.; Kim, J.H.; Brady, S.F. Mapping gene clusters within arrayed metagenomic libraries to expand the structural diversity of biomedically relevant natural products. Proc. Natl. Acad. Sci. USA 2013, 110, 11797–11802. [Google Scholar] [CrossRef] [PubMed]
- Charlop-Powers, Z.; Milshteyn, A.; Brady, S.F. Metagenomic small molecule discovery methods. Curr. Opin. Microbiol. 2014, 0, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Woodhouse, J.N.; Fan, L.; Brown, M.V.; Thomas, T.; Neilan, B.A. Deep sequencing of non-ribosomal peptide synthetases and polyketide synthases from the microbiomes of Australian marine sponges. ISME J. 2013, 7, 1842–1851. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.; Tan, H. Nucleoside antibiotics: Biosynthesis, regulation, and biotechnology. Trends Microbiol. 2015, 23, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.Y.; Kallifidas, D.; Brady, S.F. Functional analysis of environmental DNA-derived type II polyketide synthases reveals structurally diverse secondary metabolites. Proc. Natl. Acad. Sci. USA 2011, 108, 12629–12634. [Google Scholar] [CrossRef] [PubMed]
- Banik, J.J.; Craig, J.W.; Calle, P.Y.; Brady, S.F. Tailoring enzyme-rich environmental DNA clones: A source of enzymes for generating libraries of unnatural natural products. J. Am. Chem. Soc. 2010, 132, 15661–15670. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Fan, X.J.; Lu, Y.; Liu, Y.H. Isolation and characterization of a novel tannase from a metagenomic library. J. Agric. Food Chem. 2011, 59, 3812–3818. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wu, H.; Wang, A.; Du, P.; Pei, X.; Li, H.; Yin, X.; Huang, L.; Xiong, X. Prospecting metagenomic enzyme subfamily genes for DNA family shuffling by a novel PCR-based approach. J. Biol. Chem. 2010, 285, 41509–41516. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, G.; Yu, S.Q.; Zhang, C.T.; Liu, Y.H. A novel metagenome-derived β-galactosidase: Gene cloning, overexpression, purification and characterization. Appl. Microbiol. Biotechnol. 2010, 88, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.F.; Bauer, J.D.; Clarke-Pearson, M.F.; Daniels, R. Natural products from isnA-containing biosynthetic gene clusters recovered from the genomes of cultured and uncultured bacteria. J. Am. Chem. Soc. 2007, 129, 12102–12103. [Google Scholar] [CrossRef] [PubMed]
- Chang, F.-Y.; Ternei, M.A.; Calle, P.Y.; Brady, S.F. Targeted metagenomics: Finding rare tryptophan dimer natural products in the environment. J. Am. Chem. Soc. 2015, 137, 6044–6052. [Google Scholar] [CrossRef] [PubMed]
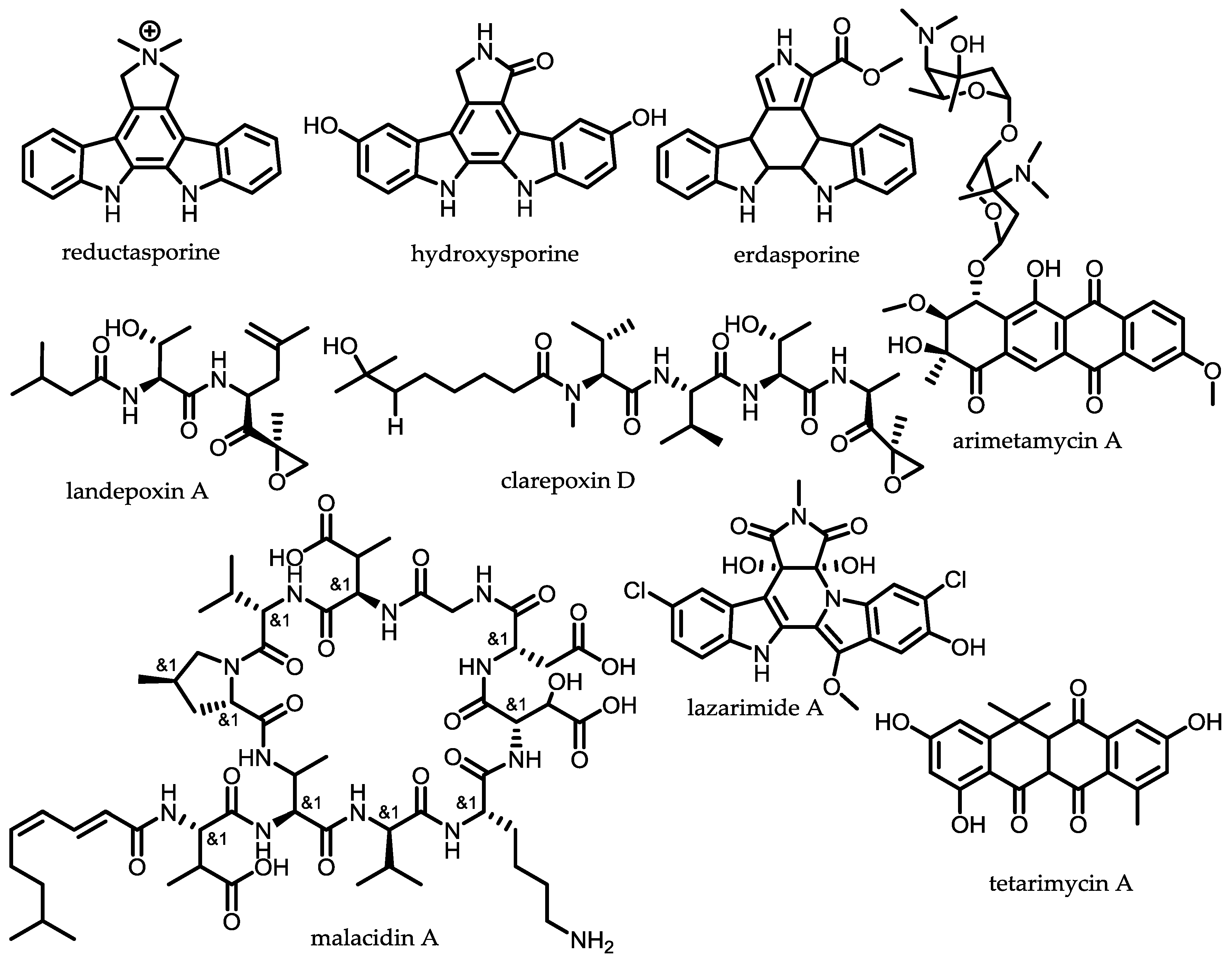
- Kang, H.-S.; Brady, S.F. Arimetamycin A: Improving clinically relevant families of natural products through sequence-guided screening of soil metagenomes. Angew. Chem. Int. Ed. 2013, 52, 11063–11067. [Google Scholar] [CrossRef] [PubMed]
- Kallifidas, D.; Kang, H.-S.; Brady, S.F. Tetarimycin A, an MRSA-active antibiotic identified through induced expression of environmental DNA gene clusters. J. Am. Chem. Soc. 2012, 134, 19552–19555. [Google Scholar] [CrossRef] [PubMed]
- Hover, B.M.; Kim, S.-H.; Katz, M.; Charlop-Powers, Z.; Owen, J.G.; Ternei, M.A.; Maniko, J.; Estrela, A.B.; Molina, H.; Park, S.; et al. Culture-independent discovery of the malacidins as calcium-dependent antibiotics with activity against multidrug-resistant Gram-positive pathogens. Nat. Microbiol. 2018, 3, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Kallifidas, D.; Jiang, G.; Ding, Y.; Luesch, H. Rational engineering of Streptomyces albus J1074 for the overexpression of secondary metabolite gene clusters. Microb. Cell Fact. 2018, 17. [Google Scholar] [CrossRef] [PubMed]
- Leis, B.; Angelov, A.; Liebl, W. Screening and expression of genes from metagenomes. Adv. Appl. Microbiol. 2013, 83, 1–68. [Google Scholar] [PubMed]
- Montaser, R.; Luesch, H. Marine natural products: A new wave of drugs? Future Med. Chem. 2011, 3, 1475–1489. [Google Scholar] [CrossRef] [PubMed]
- Towbin, H.; Bair, K.W.; DeCaprio, J.A.; Eck, M.J.; Kim, S.; Kinder, F.R.; Morollo, A.; Mueller, D.R.; Schindler, P.; Song, H.K.; et al. Proteomics-based target identification—Bengamides as a new class of methionine aminopeptidase inhibitors. J. Biol. Chem. 2003, 278, 52964–52971. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, S.C.; Hoffmann, H.; Zhang, J.; Debussche, L.; Haag-Richter, S.; Kurz, M.; Nardi, F.; Lukat, P.; Kochems, I.; Tietgen, H.; et al. Production of the bengamide class of marine natural products in myxobacteria: Biosynthesis and structure-activity relationships. Angew. Chem. Int. Ed. Engl. 2015, 54, 15560–15564. [Google Scholar] [CrossRef] [PubMed]
- Quinoa, E.; Adamczeski, M.; Crews, P.; Bakus, G.J. Bengamides, heterocyclic anthelmintics from a Jaspidae marine sponge. J. Org. Chem. 1986, 51, 4494–4497. [Google Scholar] [CrossRef]
- Piel, J. Metabolites from symbiotic bacteria. Nat. Prod. Rep. 2009, 26, 338–362. [Google Scholar] [CrossRef] [PubMed]
- Bewley, C.A.; Debitus, C.; Faulkner, D.J. Microsclerodermins A and B. Antifungal cyclic peptides from the lithistid sponge Microscleroderma sp. J. Am. Chem. Soc. 1994, 116, 7631–7636. [Google Scholar] [CrossRef]
- Schmidt, E.W.; Faulkner, D.J. Microsclerodermins C–E, antifungal cyclic peptides from the lithistid marine sponges Theonella sp. and Microscleroderma sp. Tetrahedron 1998, 54, 3043–3056. [Google Scholar] [CrossRef]
- Hoffmann, T.; Müller, S.; Nadmid, S.; Garcia, R.; Müller, R. Microsclerodermins from terrestrial myxobacteria: An intriguing biosynthesis likely connected to a sponge symbiont. J. Am. Chem. Soc. 2013, 45, 16904–16911. [Google Scholar] [CrossRef] [PubMed]
- Simister, R.L.; Deines, P.; Botté, E.S.; Webster, N.S.; Taylor, M.W. Sponge-specific clusters revisited: A comprehensive phylogeny of sponge-associated microorganisms. Environ. Microbiol. 2012, 14, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [PubMed]
- Hamada, T.; Matsunaga, S.; Yano, G.; Fusetani, N. Polytheonamides A and B, highly cytotoxic, linear polypeptides with unprecedented structural features, from the marine sponge, Theonella swinhoei. J. Am. Chem. Soc. 2005, 127, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Shinohara, N.; Tanabe, S.; Takahashi, T.; Okura, K.; Itoh, H.; Mizoguchi, Y.; Iida, M.; Lee, N.; Matsuoka, S. Total synthesis of the large non-ribosomal peptide polytheonamide B. Nat. Chem. 2010, 2, 280–285. [Google Scholar] [CrossRef] [PubMed]
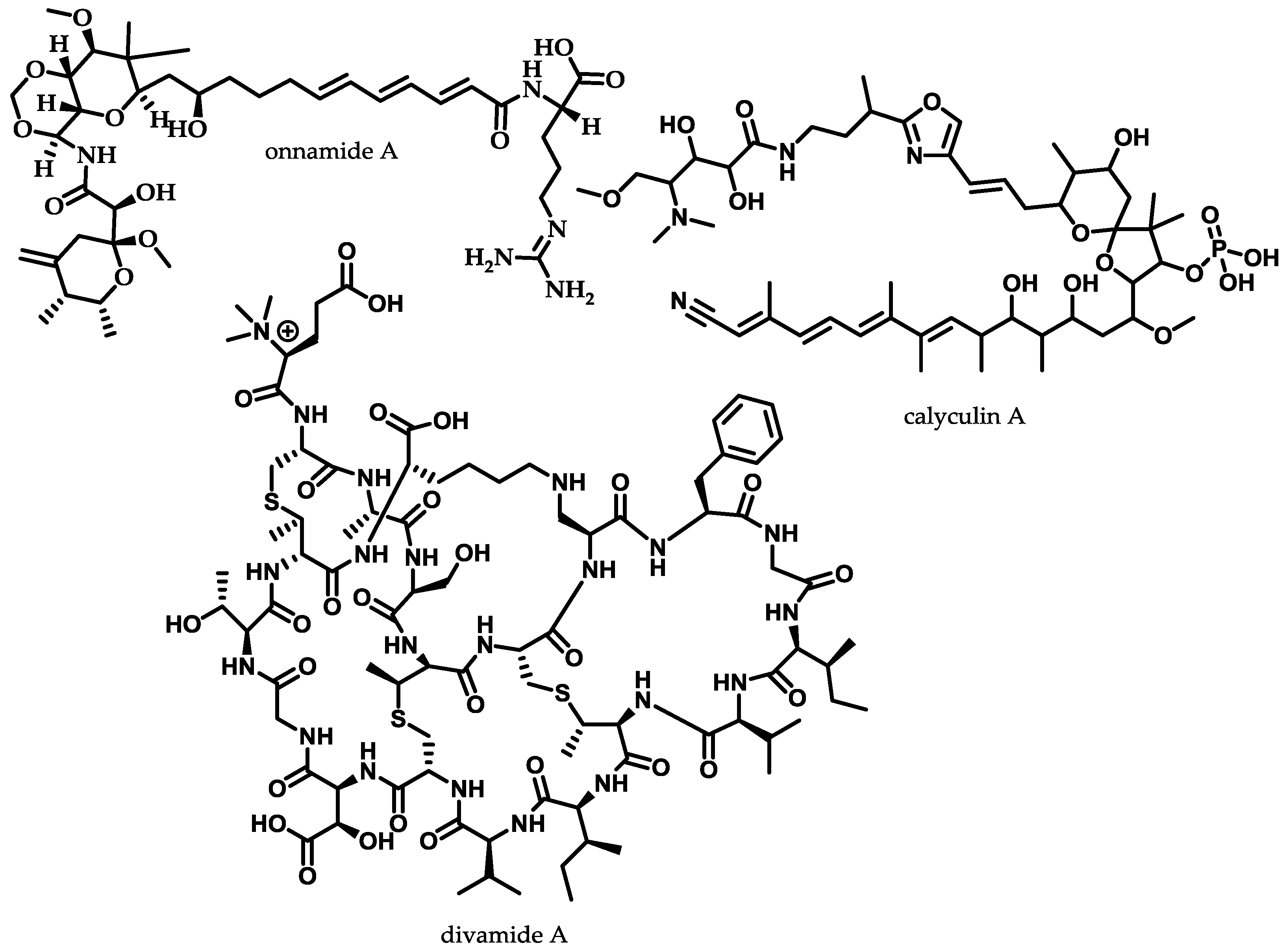
- Freeman, M.F.; Gurgui, C.; Helf, M.J.; Morinaka, B.I.; Uria, A.R.; Oldham, N.J.; Sahl, H.-G.; Matsunaga, S.; Piel, J. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science 2012, 338, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Freeman, M.F.; Helf, M.J.; Bhushan, A.; Morinaka, B.I.; Piel, J. Seven enzymes create extraordinary molecular complexity in an uncultivated bacterium. Nat. Chem. 2017, 9, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.C.; Mori, T.; Rückert, C.; Uria, A.R.; Helf, M.J.; Takada, K.; Gernert, C.; Steffens, U.A.E.; Heycke, N.; Schmitt, S.; et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature 2014, 506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, Y.; Fusetani, N.; Matsunaga, S.; Hashimoto, K.; Fujita, S.; Furuya, T. Bioactive marine metabolites. Part 16. Calyculin A. A novel antitumor metabolite from the marine sponge Discodermia calyx. J. Am. Chem. Soc. 1986, 108, 2780–2781. [Google Scholar] [CrossRef]
- Edrada, R.A.; Ebel, R.; Supriyono, A.; Wray, V.; Schupp, P.; Steube, K.; van Soest, R.; Proksch, P. Swinhoeiamide A, a new highly active calyculin derivative from the marine sponge Theonella swinhoei. J. Nat. Prod. 2002, 65, 1168–1172. [Google Scholar] [CrossRef] [PubMed]
- Hrvatin, S.; Piel, J. Rapid isolation of rare clones from highly complex DNA libraries by PCR analysis of liquid gel pools. J. Microbiol. Methods 2007, 68, 434–436. [Google Scholar] [CrossRef] [PubMed]
- Wakimoto, T.; Egami, Y.; Nakashima, Y.; Wakimoto, Y.; Mori, T.; Awakawa, T.; Ito, T.; Kenmoku, H.; Asakawa, Y.; Piel, J.; et al. Calyculin biogenesis from a pyrophosphate protoxin produced by a sponge symbiont. Nat. Chem. Biol. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Repka, L.M.; Chekan, J.R.; Nair, S.K.; Van Der Donk, W.A. Mechanistic understanding of lanthipeptide biosynthetic enzymes. Chem. Rev. 2017, 117, 5457–5520. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.E.; Pond, C.D.; Pierce, E.; Harmer, Z.P.; Kwan, J.; Zachariah, M.M.; Harper, M.K.; Wyche, T.P.; Matainaho, T.K.; Bugni, T.S.; et al. Accessing chemical diversity from the uncultivated symbionts of small marine animals. Nat. Chem. Biol. 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Minowa, Y.; Araki, M.; Kanehisa, M. Comprehensive analysis of distinctive polyketide and nonribosomal peptide structural motifs encoded in microbial genomes. J. Mol. Biol. 2007, 368, 1500–1517. [Google Scholar] [CrossRef] [PubMed]
- Gawad, C.; Koh, W.; Quake, S.R. Single-cell genome sequencing: Current state of the science. Nat. Rev. Genet. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Genilloud, O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017, 34, 1203–1232. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, J.; Fayad, A.A.; Müller, R. Natural products from myxobacteria: Novel metabolites and bioactivities. Nat. Prod. Rep. 2017, 34, 135–160. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Escribano, J.P.; Alt, S.; Bibb, M.J. Next generation sequencing of Actinobacteria for the discovery of novel natural products. Mar. Drugs 2016, 14. [Google Scholar] [CrossRef] [PubMed]
- Grindberg, R.V.; Ishoey, T.; Brinza, D.; Esquenazi, E.; Coates, R.C.; Liu, W.-T.; Gerwick, L.; Dorrestein, P.C.; Pevzner, P.; Lasken, R.; et al. Single cell genome amplification accelerates identification of the apratoxin biosynthetic pathway from a complex microbial assemblage. PLoS ONE 2011, 6, e18565. [Google Scholar] [CrossRef] [PubMed]
- Mason, O.U.; Hazen, T.C.; Borglin, S.; Chain, P.S.G.; Dubinsky, E.A.; Fortney, J.L.; Han, J.; Holman, H.-Y.N.; Hultman, J.; Lamendella, R.; et al. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 2012, 6, 1715–1727. [Google Scholar] [CrossRef] [PubMed]
- Engel, K.; Pinnell, L.; Cheng, J.; Charles, T.C.; Neufeld, J.D. Nonlinear electrophoresis for purification of soil DNA for metagenomics. J. Microbiol. Methods 2012, 88, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Pel, J.; Broemeling, D.; Mai, L.; Poon, H.-L.; Tropini, G.; Warren, R.L.; Holt, R.A.; Marziali, A. Nonlinear electrophoretic response yields a unique parameter for separation of biomolecules. Proc. Natl. Acad. Sci. USA 2009, 106, 14796–14801. [Google Scholar] [CrossRef] [PubMed]
- Liles, M.R.; Williamson, L.L.; Rodbumrer, J.; Torsvik, V.; Goodman, R.M.; Handelsman, J. Recovery, purification, and cloning of high-molecular-weight DNA from soil microorganisms. Appl. Environ. Microbiol. 2008, 74, 3302–3305. [Google Scholar] [CrossRef] [PubMed]
- Teeling, H.; Glöckner, F.O. Current opportunities and challenges in microbial metagenome analysis—A bioinformatic perspective. Brief. Bioinform. 2012, 13, 728–742. [Google Scholar] [CrossRef] [PubMed]
- Lakey, J.H.; Lea, E.J.; Rudd, B.A.; Wright, H.M.; Hopwood, D.A. A new channel-forming antibiotic from Streptomyces coelicolor A3(2) which requires calcium for its activity. J. Gen. Microbiol. 1983, 129, 3565–3573. [Google Scholar] [CrossRef] [PubMed]
- Rudd, B.A.; Hopwood, D.A. A pigmented mycelial antibiotic in Streptomyces coelicolor: Control by a chromosomal gene cluster. J. Gen. Microbiol. 1980, 119, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Tsao, S.-W.; Rudd, B.A.M.; He, X.-G.; Chang, C.-J.; Floss, H.G. Identification of a red pigment from Streptomyces coelicolor A3(2) as a mixture of prodigiosin derivatives. J. Antibiot. 1985, 38, 128–131. [Google Scholar] [CrossRef] [PubMed]
- Wright, L.F.; Hopwood, D.A. Actinorhodin is a chromosomally-determined antibiotic in Streptomyces coelicolor A3(2). J. Gen. Microbiol. 1976, 96, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Kazempour, D.; Wohlleben, W.; Weber, T. Improved lanthipeptide detection and prediction for antiSMASH. PLoS ONE 2014, 9, e89420. [Google Scholar] [CrossRef] [PubMed]
- Röttig, M.; Medema, M.H.; Blin, K.; Weber, T.; Rausch, C.; Kohlbacher, O. NRPSpredictor2—A web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011, 39, W362–W367. [Google Scholar] [CrossRef] [PubMed]
- Stachelhaus, T.; Mootz, H.D.; Marahiel, M.A. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 1999, 6, 493–505. [Google Scholar] [CrossRef]
- Yadav, G.; Gokhale, R.S.; Mohanty, D. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 2003, 328, 335–363. [Google Scholar] [CrossRef]
- Mohimani, H.; Pevzner, P.A. Dereplication, sequencing and identification of peptidic natural products: From genome mining to peptidogenomics to spectral networks. Nat. Prod. Rep. 2016, 33, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Mohimani, H.; Liu, W.-T.; Kersten, R.D.; Moore, B.S.; Dorrestein, P.C.; Pevzner, P.A. NRPquest: Coupling mass spectrometry and genome mining for nonribosomal peptide discovery. J. Nat. Prod. 2014, 77, 1902–1909. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, M.A.; Merwin, N.J.; Johnston, C.W.; Magarvey, N.A. PRISM 3: Expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 2017, 45, W49–W54. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, M.A.; Dejong, C.A.; Rees, P.N.; Johnston, C.W.; Li, H.; Webster, A.L.H.; Wyatt, M.A.; Magarvey, N.A. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res. 2015, 43, 9645–9662. [Google Scholar] [CrossRef] [PubMed]
- Alanjary, M.; Kronmiller, B.; Adamek, M.; Blin, K.; Weber, T.; Huson, D.; Philmus, B.; Ziemert, N. The Antibiotic Resistant Target Seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery. Nucleic Acids Res. 2017, 45, W42–W48. [Google Scholar] [CrossRef] [PubMed]
- Doyle, D.; McDowall, K.J.; Butler, M.J.; Hunter, I.S. Characterization of an oxytetracycline-resistance gene, otrA, of Streptomyces rimosus. Mol. Microbiol. 1991, 5, 2923–2933. [Google Scholar] [CrossRef] [PubMed]
- Kling, A.; Lukat, P.; Almeida, D.V.; Bauer, A.; Fontaine, E.; Sordello, S.; Zaburannyi, N.; Herrmann, J.; Wenzel, S.C.; König, C.; et al. Targeting DnaN for tuberculosis therapy using novel griselimycins. Science 2015, 348, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Huo, L. Synthetic Biotechnology to Study and Engineer Natural Product Biosynthesis in Actinomycetes. Ph.D. Thesis, Saarland University, Saarbrücken, Germany, 2014. [Google Scholar]
- Li, X.; Wu, X.; Zhu, J.; Shen, Y. Amexanthomycins A–J, pentangular polyphenols produced by Amycolatopsis mediterranei S699∆rifA. Appl. Microbiol. Biotechnol. 2018, 102, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Rebets, Y.; Brötz, E.; Tokovenko, B.; Luzhetskyy, A. Actinomycetes biosynthetic potential: How to bridge in silico and in vivo? J. Ind. Microbiol. Biotechnol. 2014, 41, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Zerikly, M.; Challis, G.L. Strategies for the discovery of new natural products by genome mining. ChemBioChem 2009, 10, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Rigali, S.; Anderssen, S.; Naômé, A.; van Wezel, G.P. Cracking the regulatory code of biosynthetic gene clusters as a strategy for natural product discovery. Biochem. Pharmacol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Metsä-Ketelä, M.; Ylihonko, K.; Mäntsälä, P. Partial activation of a silent angucycline-type gene cluster from a rubromycin β producing Streptomyces sp. PGA64. J. Antibiot. 2004, 57, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Bunet, R.; Song, L.; Mendes, M.V.; Corre, C.; Hotel, L.; Rouhier, N.; Framboisier, X.; Leblond, P.; Challis, G.L.; Aigle, B. Characterization and manipulation of the pathway-specific late regulator AlpW reveals Streptomyces ambofaciens as a new producer of kinamycins. J. Bacteriol. 2011, 193, 1142–1153. [Google Scholar] [CrossRef] [PubMed]
- Sidda, J.D.; Song, L.; Poon, V.; Al-Bassam, M.; Lazos, O.; Buttner, M.J.; Challis, G.L.; Corre, C. Discovery of a family of γ-aminobutyrate ureas via rational derepression of a silent bacterial gene cluster. Chem. Sci. 2013, 5, 86–89. [Google Scholar] [CrossRef]
- Laureti, L.; Song, L.J.; Huang, S.; Corre, C.; Leblond, P.; Challis, G.L.; Aigle, B. Identification of a bioactive 51-membered macrolide complex by activation of a silent polyketide synthase in Streptomyces ambofaciens. Proc. Natl. Acad. Sci. USA 2011, 108, 6258–6263. [Google Scholar] [CrossRef] [PubMed]
- Olano, C.; García, I.; González, A.; Rodriguez, M.; Rozas, D.; Rubio, J.; Sánchez-Hidalgo, M.; Braña, A.F.; Méndez, C.; Salas, J.A. Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microb. Biotechnol. 2014, 7, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Bode, E.; Brachmann, A.O.; Kegler, C.; Simsek, R.; Dauth, C.; Zhou, Q.; Kaiser, M.; Klemmt, P.; Bode, H.B. Simple “on-demand” production of bioactive natural products. ChemBioChem 2015, 16, 1115–1119. [Google Scholar] [CrossRef] [PubMed]
- Siegl, T.; Tokovenko, B.; Myronovskyi, M.; Luzhetskyy, A. Design, construction and characterisation of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metab. Eng. 2013, 19, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Seghezzi, N.; Amar, P.; Koebmann, B.; Jensen, P.R.; Virolle, M.-J. The construction of a library of synthetic promoters revealed some specific features of strong Streptomyces promoters. Appl. Microbiol. Biotechnol. 2011, 90, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J. Ind. Microbiol. Biotechnol. 2016, 43, 343–370. [Google Scholar] [CrossRef] [PubMed]
- Zaburannyi, N.; Rabyk, M.; Ostash, B.; Fedorenko, V.; Luzhetskyy, A. Insights into naturally minimised Streptomyces albus J1074 genome. BMC Genom. 2014, 15. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Escribano, J.P.; Bibb, M.J. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 2011, 4, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Okada, B.K.; Seyedsayamdost, M.R. Antibiotic dialogues: Induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol. Rev. 2017, 41, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, P.J.; Challis, G.L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Seyedsayamdost, M.R. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc. Natl. Acad. Sci. USA 2014, 111, 7266–7271. [Google Scholar] [CrossRef] [PubMed]
- Locatelli, F.M.; Goo, K.-S.; Ulanova, D. Effects of trace metal ions on secondary metabolism and the morphological development of streptomycetes. Metallomics 2016, 8, 469–480. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, T.; Ohnishi-Kameyama, M.; Muramatsu, H.; Murakami, K.; Tsurumi, Y.; Kodani, S.; Yoshida, M.; Fujie, A.; Ochi, K. Antibacterial discovery in actinomycetes strains with mutations in RNA polymerase or ribosomal protein S12. Nat. Biotechnol. 2009, 27, 462–464. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Kasahara, K.; Hirose, Y.; Murakami, K.; Kugimiya, R.; Ochi, K. Activation and products of the cryptic secondary metabolite biosynthetic gene clusters by rifampin resistance (rpoB) mutations in actinomycetes. J. Bacteriol. 2013, 195, 2959–2970. [Google Scholar] [CrossRef] [PubMed]
- Ochi, K.; Hosaka, T. New strategies for drug discovery: Activation of silent or weakly expressed microbial gene clusters. Appl. Microbiol. Biotechnol. 2013, 97, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Onaka, H.; Tabata, H.; Igarashi, Y.; Sato, Y.; Furumai, T. Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in streptomycetes. I. Purification and Characterization. J. Antibiot. 2001, 54, 1036–1044. [Google Scholar] [CrossRef] [PubMed]
- Truong, T.T.; Seyedsayamdost, M.; Greenberg, E.P.; Chandler, J.R. A Burkholderia thailandensis acyl-homoserine lactone-independent orphan luxr homolog that activates production of the cytotoxin malleilactone. J. Bacteriol. 2015, 197, 3456–3462. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K.; Oikawa, H.; Ogawa, H.O.; Hosono, K.; Shinmachi, F.; Takano, H.; Sakuda, S.; Beppu, T.; Ueda, K. Desferrioxamine E produced by Streptomyces griseus stimulates growth and development of Streptomyces tanashiensis. Microbiology 2005, 151, 2899–2905. [Google Scholar] [CrossRef] [PubMed]
- Craney, A.; Ozimok, C.; Pimentel-Elardo, S.M.; Capretta, A.; Nodwell, J.R. Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem. Biol. 2012, 19, 1020–1027. [Google Scholar] [CrossRef] [PubMed]
- Pimentel-Elardo, S.M.; Sørensen, D.; Ho, L.; Ziko, M.; Bueler, S.A.; Lu, S.; Tao, J.; Moser, A.; Lee, R.; Agard, D.; et al. Activity-independent discovery of secondary metabolites using chemical elicitation and cheminformatic inference. ACS Chem. Biol. 2015, 10, 2616–2623. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, P.M.; Angell, H.D.; Whittington, D.A.; Flynn, E.F.; Rajashankar, K.R.; Christianson, D.W. Structure of prokaryotic polyamine deacetylase reveals evolutionary functional relationships with eukaryotic histone deacetylases. Biochemistry 2011, 50, 1808–1817. [Google Scholar] [CrossRef] [PubMed]
- Gacek, A.; Strauss, J. The chromatin code of fungal secondary metabolite gene clusters. Appl. Microbiol. Biotechnol. 2012, 95, 1389–1404. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.; Baltz, R.H. Natural product discovery: Past, present, and future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Penesyan, A.; Kjelleberg, S.; Egan, S. Development of novel drugs from marine surface associated microorganisms. Mar. Drugs 2010, 8, 438–459. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zhang, Q.; Tan, B.; Zheng, L.; Li, H.; Zhu, Y.; Zhang, C. Genome mining and activation of a silent PKS/NRPS gene cluster direct the production of totopotensamides. Org. Lett. 2017, 19, 5697–5700. [Google Scholar] [CrossRef] [PubMed]
- Reen, F.J.; Romano, S.; Dobson, A.D.W.; O’Gara, F. The Sound of silence: Activating silent biosynthetic gene clusters in marine microorganisms. Mar. Drugs 2015, 13, 4754–4783. [Google Scholar] [CrossRef] [PubMed]
- Thanapipatsiri, A.; Gomez-Escribano, J.P.; Song, L.; Bibb, M.J.; Al-Bassam, M.; Chandra, G.; Thamchaipenet, A.; Challis, G.L.; Bibb, M.J. Discovery of unusual biaryl polyketides by activation of a silent Streptomyces venezuelae biosynthetic gene cluster. ChemBioChem 2016, 17, 2189–2198. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.M.; Wong, F.T.; Wang, Y.; Luo, S.; Lim, Y.H.; Heng, E.; Yeo, W.L.; Cobb, R.E.; Enghiad, B.; Ang, E.L.; et al. CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat. Chem. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Challis, G.L. Genome mining for novel natural product discovery. J. Med. Chem. 2008, 51, 2618–2628. [Google Scholar] [CrossRef] [PubMed]
- Hubert, J.; Nuzillard, J.-M.; Renault, J.-H. Dereplication strategies in natural product research: How many tools and methodologies behind the same concept? Phytochem. Rev. 2017, 16, 55–95. [Google Scholar] [CrossRef]
- Breitling, R.; Ceniceros, A.; Jankevics, A.; Takano, E. Metabolomics for secondary metabolite research. Metabolites 2013, 2013, 1076–1083. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.T.; Merlo, M.E.; Medema, M.H.; Jankevics, A.; Breitling, R.; Takano, E. Metabolomics methods for the synthetic biology of secondary metabolism. FEBS Lett. 2012, 586, 2177–2183. [Google Scholar] [CrossRef] [PubMed]
- Wolfender, J.-L.; Marti, G.; Ferreira Queiroz, E. Advances in techniques for profiling crude extracts and for the rapid identification of natural products: Dereplication, quality control and metabolomics. Curr. Org. Chem. 2010, 14, 1808–1832. [Google Scholar] [CrossRef]
- Hou, Y.; Braun, D.R.; Michel, C.R.; Klassen, J.L.; Adnani, N.; Wyche, T.P.; Bugni, T.S. Microbial strain prioritization using metabolomics tools for the discovery of natural products. Anal. Chem. 2012, 84, 4277–4283. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-T.; Kersten, R.D.; Yang, Y.-L.; Moore, B.S.; Dorrestein, P.C. Imaging mass spectrometry and genome mining via short sequence tagging identified the anti-infective agent arylomycin in Streptomyces roseosporus. J. Am. Chem. Soc. 2011, 133, 18010–18013. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, X.; Jiang, Y.; Jiang, C. Morphological identification of actinobacteria. In Actinobacteria—Basics and Biotechnological Applications; Dhanasekaran, D., Jiang, Y., Eds.; InTech: Rijeka, Croatia, 2016. [Google Scholar]
- Onaka, H.; Mori, Y.; Igarashi, Y.; Furumai, T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 2011, 77, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.L.; Xu, Y.; Straight, P.; Dorrestein, P.C. Translating metabolic exchange with imaging mass spectrometry. Nat. Chem. Biol. 2009, 5, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Esquenazi, E.; Yang, Y.L.; Watrous, J.; Gerwick, W.H.; Dorrestein, P.C. Imaging mass spectrometry of natural products. Nat. Prod. Rep. 2009, 26, 1521–1534. [Google Scholar] [CrossRef] [PubMed]
- Watrous, J.D.; Dorrestein, P.C. Imaging mass spectrometry in microbiology. Nat. Rev. Microbiol. 2011, 9, 683–694. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.B.; Cooks, R.G. Simulation of atmospheric transport and droplet-thin film collisions in desorption electrospray ionization. Chem. Commun. 2007, 3915–3917. [Google Scholar] [CrossRef] [PubMed]
- Kersten, R.D.; Yang, Y.L.; Xu, Y.; Cimermancic, P.; Nam, S.J.; Fenical, W.; Fischbach, M.A.; Moore, B.S.; Dorrestein, P.C. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat. Chem. Biol. 2011, 7, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, S.; Fletcher, J.S.; Goodacre, R.; Lockyer, N.P.; Micklefield, J.; Vickerman, J.C. Subsurface biomolecular imaging of Streptomyces coelicolor using secondary ion mass spectrometry. Anal. Chem. 2008, 80, 1942–1951. [Google Scholar] [CrossRef] [PubMed]
- Traxler, M.F.; Watrous, J.D.; Alexandrov, T.; Dorrestein, P.C.; Kolter, R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. mBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.N.; Liyu, A.V.; Chu, R.K.; Anderton, C.R.; Laskin, J. Constant-distance mode nanospray desorption electrospray ionization mass spectrometry imaging of biological samples with complex topography. Anal. Chem. 2017, 89, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, D.J.; Haste, N.M.; Hollands, A.; Fleming, T.C.; Hamby, M.; Pogliano, K.; Nizet, V.; Dorrestein, P.C. Microbial competition between Bacillus subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiology 2011, 157, 2485–2492. [Google Scholar] [CrossRef] [PubMed]
- Barger, S.R.; Hoefler, B.C.; Cubillos-Ruiz, A.; Russell, W.K.; Russell, D.H.; Straight, P.D. Imaging secondary metabolism of Streptomyces sp. Mg1 during cellular lysis and colony degradation of competing Bacillus subtilis. Anton. Leeuwenhoek 2012, 102, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Boya, P.C.A.; Fernández-Marín, H.; Mejía, L.C.; Spadafora, C.; Dorrestein, P.C.; Gutiérrez, M. Imaging mass spectrometry and MS/MS molecular networking reveals chemical interactions among cuticular bacteria and pathogenic fungi associated with fungus-growing ants. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Hoefler, B.C.; Stubbendieck, R.M.; Josyula, N.K.; Moisan, S.M.; Schulze, E.M.; Straight, P.D. A link between linearmycin biosynthesis and extracellular vesicle genesis connects specialized metabolism and bacterial membrane physiology. Cell Chem. Biol. 2017, 24, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, E.K.; Moore, C.M.; Krastel, P.; Petersen, F. Natural products as catalysts for innovation: A pharmaceutical industry perspective. Curr. Opin. Chem. Biol. 2011, 15, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Exarchou, V.; Krucker, M.; van Beek, T.A.; Vervoort, J.; Gerothanassis, I.P.; Albert, K. LC-NMR coupling technology: Recent advancements and applications in natural products analysis. Magn. Reson. Chem. 2005, 43, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, F.C.; Gronquist, M. Extending the scope of NMR spectroscopy with microcoil probes. Angew. Chem. Int. Ed. Engl. 2006, 45, 7122–7131. [Google Scholar] [CrossRef] [PubMed]
- Iwasa, K.; Takahashi, T.; Nishiyama, Y.; Moriyasu, M.; Sugiura, M.; Takeuchi, A.; Tode, C.; Tokuda, H.; Takeda, K. Online structural elucidation of alkaloids and other constituents in crude extracts and cultured cells of Nandina domestica by combination of LC-MS/MS, LC-NMR, and LC-CD analyses. J. Nat. Prod. 2008, 71, 1376–1385. [Google Scholar] [CrossRef] [PubMed]
- Iwasa, K.; Cui, W.; Takahashi, T.; Nishiyama, Y.; Kamigauchi, M.; Koyama, J.; Takeuchi, A.; Moriyasu, M.; Takeda, K. Biotransformation of phenolic tetrahydroprotoberberines in plant cell cultures followed by LC-NMR, LC-MS, and LC-CD. J. Nat. Prod. 2010, 73, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Iwasa, K.; Sugiura, M.; Takeuchi, A.; Tode, C.; Nishiyama, Y.; Moriyasu, M.; Tokuda, H.; Takeda, K. Biotransformation of phenolic 1-benzyl-N-methyltetrahydroisoquinolines in plant cell cultures followed by LC/NMR, LC/MS, and LC/CD. J. Nat. Prod. 2007, 70, 1771–1778. [Google Scholar] [CrossRef] [PubMed]
- Exarchou, V.; Fiamegos, Y.C.; van Beek, T.A.; Nanos, C.; Vervoort, J. Hyphenated chromatographic techniques for the rapid screening and identification of antioxidants in methanolic extracts of pharmaceutically used plants. J. Chromatogr. A 2006, 1112, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Krucker, M.; Lienau, A.; Putzbach, K.; Grynbaum, M.D.; Schuler, P.; Albert, K. Hyphenation of capillary HPLC to microcoil 1H NMR spectroscopy for the determination of tocopherol homologues. Anal. Chem. 2004, 76, 2623–2628. [Google Scholar] [CrossRef] [PubMed]
- Zehl, M.; Braunberger, C.; Conrad, J.; Crnogorac, M.; Krasteva, S.; Vogler, B.; Beifuss, U.; Krenn, L. Identification and quantification of flavonoids and ellagic acid derivatives in therapeutically important Drosera species by LC-DAD, LC-NMR, NMR, and LC-MS. Anal. Bioanal. Chem. 2011, 400, 2565–2576. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.-F.; Garo, E.; Yoo, H.-D.; Cremin, P.A.; Goering, M.G.; O’Neil-Johnson, M.; Eldridge, G.R. Cyclolignans from Scyphocephalium ochocoa via high-throughput natural product chemistry methods. Phytochemistry 2005, 66, 1077–1082. [Google Scholar] [CrossRef] [PubMed]
- Johansen, K.T.; Wubshet, S.G.; Nyberg, N.T.; Jaroszewski, J.W. From retrospective assessment to prospective decisions in natural product isolation: HPLC-SPE-NMR analysis of Carthamus oxyacantha. J. Nat. Prod. 2011, 74, 2454–2461. [Google Scholar] [CrossRef] [PubMed]
- Lang, G.; Mayhudin, N.A.; Mitova, M.I.; Sun, L.; van der, S.S.; Blunt, J.W.; Cole, A.L.; Ellis, G.; Laatsch, H.; Munro, M.H. Evolving trends in the dereplication of natural product extracts: New methodology for rapid, small-scale investigation of natural product extracts. J. Nat. Prod. 2008, 71, 1595–1599. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Plaza, A.; Dubiella, C.; Groll, M.; Kaiser, M.; Muller, R. Macyranones: Structure, biosynthesis, and binding mode of an unprecedented epoxyketone that targets the 20S proteasome. J. Am. Chem. Soc. 2015, 137, 8121–8130. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Schiavo, S.; Orjala, J.; Vouros, P.; Kautz, R. Microscale LC-MS-NMR platform applied to the identification of active cyanobacterial metabolites. Anal. Chem. 2008, 80, 8045–8054. [Google Scholar] [CrossRef] [PubMed]
- Pham, L.H.; Vater, J.; Rotard, W.; Mügge, C. Identification of secondary metabolites from Streptomyces violaceoruber TU22 by means of on-flow LC-NMR and LC-DAD-MS. Magn. Reson. Chem. 2005, 43, 710–723. [Google Scholar] [CrossRef] [PubMed]
- Alshaibani, M.; Zin, N.; Jalil, J.; Sidik, N.; Ahmad, S.J.; Kamal, N.; Edrada-Ebel, R. Isolation, purification, and characterization of five active diketopiperazine derivatives from endophytic Streptomyces SUK 25 with antimicrobial and cytotoxic activities. J. Microbiol. Biotechnol. 2017, 27, 1249–1256. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.; Rather, M.A.; Hassan, Q.P.; Aga, M.A.; Mushtaq, S.; Shah, A.M.; Hussain, A.; Baba, S.A.; Ahmad, Z. Discovery of anti-microbial and anti-tubercular molecules from Fusarium solani: An endophyte of Glycyrrhiza glabra. J. Appl. Microbiol. 2017, 122, 1168–1176. [Google Scholar] [CrossRef] [PubMed]
- Greule, A.; Zhang, S.; Paululat, T.; Bechthold, A. From a natural product to its biosynthetic gene cluster: a demonstration using polyketomycin from Streptomyces diastatochromogenes Tü6028. J. Vis. Exp. 2017. [Google Scholar] [CrossRef] [PubMed]
- Gibitz Eisath, N.; Sturm, S.; Stuppner, H. Supercritical fluid chromatography in natural product analysis—An update. Planta Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- Khater, S.; Lozac’h, M.-A.; Adam, I.; Francotte, E.; West, C. Comparison of liquid and supercritical fluid chromatography mobile phases for enantioselective separations on polysaccharide stationary phases. J. Chromatogr. A 2016, 1467, 463–472. [Google Scholar] [CrossRef] [PubMed]
- De Klerck, K.; Mangelings, D.; Vander Heyden, Y. Supercritical fluid chromatography for the enantioseparation of pharmaceuticals. J. Pharm. Biomed. Anal. 2012, 69, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Novakova, L.; Perrenoud, A.G.-G.; Francois, I.; West, C.; Lesellier, E.; Guillarme, D. Modern analytical supercritical fluid chromatography using columns packed with sub-2 µm particles: A tutorial. Anal. Chim. Acta 2014, 824, 18–35. [Google Scholar] [CrossRef] [PubMed]
- Lesellier, E.; West, C. The many faces of packed column supercritical fluid chromatography—A critical review. J. Chromatogr. A 2015, 1382, 2–46. [Google Scholar] [CrossRef] [PubMed]
- West, C.; Lemasson, E.; Bertin, S.; Hennig, P.; Lesellier, E. An improved classification of stationary phases for ultra-high performance supercritical fluid chromatography. J. Chromatogr. A 2016, 1440, 212–228. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carrell, E.J.; Chittiboyina, A.G.; Avula, B.; Wang, Y.-H.; Zhao, J.; Parcher, J.F.; Khan, I.A. Concurrent supercritical fluid chromatographic analysis of terpene lactones and ginkgolic acids in Ginkgo biloba extracts and dietary supplements. Anal. Bioanal. Chem. 2016, 408, 4649–4660. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Feng, Y.; Tang, G.; Li, M.; Zhang, T.; Fillet, M.; Crommen, J.; Jiang, Z. Development and validation of a fast SFC method for the analysis of flavonoids in plant extracts. J. Pharm. Biomed. Anal. 2017, 140, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Vicente, G.; García-Risco, M.R.; Fornari, T.; Reglero, G. Isolation of carnosic acid from rosemary extracts using semi-preparative supercritical fluid chromatography. J. Chromatogr. A 2013, 1286, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Salimi, A.; Fatemi, S.; Zakizadeh Nei Nei, H.; Safaralie, A. Mathematical modeling of supercritical extraction of valerenic acid from Valeriana officinalis L. Chem. Eng. Technol. 2008, 31, 1470–1480. [Google Scholar] [CrossRef]
- Ramsden, J.J. Metabolomics and metabonomics. In Bioinformatics: An Introduction, 2nd ed.; Ramsden, J., Ed.; Springer: New York, NY, USA, 2009; pp. 1–6. [Google Scholar]
- Mohamed, A.; Nguyen, C.H.; Mamitsuka, H. Current status and prospects of computational resources for natural product dereplication: A review. Brief. Bioinform. 2016, 17, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Beutler, J.A.; Alvarado, A.B.; Schaufelberger, D.E.; Andrews, P.; McCloud, T.G. Dereplication of phorbol bioactives: Lyngbya majuscula and Croton cuneatus. J. Nat. Prod. 1990, 53, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Chanana, S.; Thomas, C.S.; Braun, D.R.; Hou, Y.; Wyche, T.P.; Bugni, T.S. Natural product discovery using planes of principal component analysis in R (PoPCAR). Metabolites 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Bitzer, J.; Köpcke, B.; Stadler, M.; Hellwig, V.; Ju, Y.-M.; Seip, S.; Henkel, T. Accelerated dereplication of natural products, supported by reference libraries. CHIMIA 2007, 61, 332–338. [Google Scholar] [CrossRef]
- Dictionary of Natural Products. Available online: http://dnp.chemnetbase.com/faces/chemical/ChemicalSearch.xhtml (accessed on 31 January 2018).
- Laatsch, H. AntiBase 2014: The Natural Compound Identifier; Wiley-VCH: Weinheim, Germany, 2014. [Google Scholar]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.G.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2017, 34, 235–294. [Google Scholar] [CrossRef] [PubMed]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. METLIN: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef] [PubMed]
- mzCloud: Advanced Mass Spectral Database. Available online: https://www.mzcloud.org/ (accessed on 10 May 2018).
- Sawada, Y.; Nakabayashi, R.; Yamada, Y.; Suzuki, M.; Sato, M.; Sakata, A.; Akiyama, K.; Sakurai, T.; Matsuda, F.; Aoki, T.; et al. RIKEN tandem mass spectral database (ReSpect) for phytochemicals: A plant-specific MS/MS-based data resource and database. Phytochemistry 2012, 82, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Johnston, C.W.; Skinnider, M.A.; Wyatt, M.A.; Li, X.; Ranieri, M.R.M.; Yang, L.; Zechel, D.L.; Ma, B.; Magarvey, N.A. An automated Genomes-to-Natural Products platform (GNP) for the discovery of modular natural products. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Crüsemann, M.; O’Neill, E.C.; Larson, C.B.; Melnik, A.V.; Floros, D.J.; da Silva, R.R.; Jensen, P.R.; Dorrestein, P.C.; Moore, B.S. Prioritizing natural product diversity in a collection of 146 bacterial strains based on growth and extraction protocols. J. Nat. Prod. 2016. [Google Scholar] [CrossRef] [PubMed]
- Mohimani, H.; Gurevich, A.; Mikheenko, A.; Garg, N.; Nothias, L.-F.; Ninomiya, A.; Takada, K.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of peptidic natural products through database search of mass spectra. Nat. Chem. Biol. 2017, 13, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Mikheenko, A.; Shlemov, A.; Korobeynikov, A.; Mohimani, H.; Pevzner, P.A. Increased diversity of peptidic natural products revealed by modification-tolerant database search of mass spectra. Nat. Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zierep, P.F.; Padilla, N.; Yonchev, D.G.; Telukunta, K.K.; Klementz, D.; Günther, S. SeMPI: A genome-based secondary metabolite prediction and identification web server. Nucleic Acids Res. 2017, 45, W64–W71. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Medema, M.H.; Kazempour, D.; Fischbach, M.A.; Breitling, R.; Takano, E.; Weber, T. antiSMASH 2.0—A versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013, 41, W204–W212. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Paalvast, Y.; Nguyen, D.D.; Melnik, A.; Dorrestein, P.C.; Takano, E.; Breitling, R. Pep2Path: Automated mass spectrometry-guided genome mining of peptidic natural products. PLoS Comput. Biol. 2014, 10, e1003822. [Google Scholar] [CrossRef] [PubMed]
- Mohimani, H.; Kersten, R.D.; Liu, W.-T.; Wang, M.; Purvine, S.O.; Wu, S.; Brewer, H.M.; Pasa-Tolic, L.; Bandeira, N.; Moore, B.S.; et al. Automated genome mining of ribosomal peptide natural products. ACS Chem. Biol. 2014, 9, 1545–1551. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Medema, M.H.; Kottmann, R.; Lee, S.Y.; Weber, T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2017, 45, D555–D559. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.T.; Lamsa, A.; Wong, W.R.; Boudreau, P.D.; Kersten, R.; Peng, Y.; Moree, W.J.; Duggan, B.M.; Moore, B.S.; Gerwick, W.H.; et al. MS/MS-based networking and peptidogenomics guided genome mining revealed the stenothricin gene cluster in Streptomyces roseosporus. J. Antibiot. 2014, 67, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Udwary, D.W.; Zeigler, L.; Asolkar, R.N.; Singan, V.; Lapidus, A.; Fenical, W.; Jensen, P.R.; Moore, B.S. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. USA 2007, 104, 10376–10381. [Google Scholar] [CrossRef] [PubMed]
- Kjaerulff, L.; Sikandar, A.; Zaburannyi, N.; Adam, S.; Herrmann, J.; Koehnke, J.; Müller, R. Thioholgamides: Thioamide-containing cytotoxic RiPPs natural products. ACS Chem. Biol. 2017, 12, 2837–2841. [Google Scholar] [CrossRef] [PubMed]
- Salvador-Reyes, L.A.; Luesch, H. Biological targets and mechanisms of action of natural products from marine cyanobacteria. Nat. Prod. Rep. 2015, 32, 478–503. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, S.C.; Müller, R. Myxobacteria—Unique microbial secondary metabolite factories. In Comprehensive Natural Products Chemistry II, Vol 2: Structural Diversity II—Secondary Metabolite Sources, Evolution and Selected Molecular Structures; Moore, B.S., Ed.; Elsevier: Oxford, UK, 2010; pp. 189–222. [Google Scholar]
- Tiwari, K.; Gupta, R.K. Rare actinomycetes: A potential storehouse for novel antibiotics. Crit. Rev. Biotechnol. 2012, 32, 108–132. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.R. Natural products and the gene cluster revolution. Trends Microbiol. 2016, 24, 968–977. [Google Scholar] [CrossRef] [PubMed]
- Goss, R.J.M.; Shankar, S.; Fayad, A.A. The generation of “unnatural” products: Synthetic biology meets synthetic chemistry. Nat. Prod. Rep. 2012, 29, 870–889. [Google Scholar]
- Smanski, M.J.; Zhou, H.; Claesen, J.; Shen, B.; Fischbach, M.A.; Voigt, C.A. Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 2016, 14, 135–149. [Google Scholar] [CrossRef] [PubMed]
| Acronym | Input | Output | Type of BGC | Highlights & Limitations | Accessibility | Source |
|---|---|---|---|---|---|---|
| PRISM 3 | Genome sequence | Gene clusters | NRPS, PKS | Tailoring reactions implemented 1 | Open-source web application | [184,185] |
| SeMPI | Genome sequence (also raw DNA code) | Domains and 10 best matching compounds | Type I PKS | Generation of the non-modified PKS products 1 | Open-source web application | [291] |
| antiSMASH 4.0 | Genome sequence | NRPS/PKS domains, chemical structure prediction, Cluster Blast | NRPS, PKS, RiPPs terpenes | Terpene prediction Trans AT PKS domain alignments 1 | Open-source webserver | [121,122,292] |
| Pep2Path | Genome sequence mass shifts or amino acid-sequence | Tag-BGC alignment and scoring | NRPS, RiPPs | Automatic identification of BGC corresponding to mass shift sequence or amino acid-sequence tags 2 | Open-source application | [293] |
| RiPPquest | Genome sequence MS/MS dataset | Peptide-spectrum match (p-value) | RiPPs | Molecular network analysis using input from various MS/MS datasets possible 3 | Open-source application, (implemented in GNPS) | [294] |
| NRPquest | Genome sequence MS/MS dataset | Spectral network of matching peptides with p-values | NRPs | Molecular network analysis of various MS/MS datasets possible 4 | Open-source application | [183] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hug, J.J.; Bader, C.D.; Remškar, M.; Cirnski, K.; Müller, R. Concepts and Methods to Access Novel Antibiotics from Actinomycetes. Antibiotics 2018, 7, 44. https://doi.org/10.3390/antibiotics7020044
Hug JJ, Bader CD, Remškar M, Cirnski K, Müller R. Concepts and Methods to Access Novel Antibiotics from Actinomycetes. Antibiotics. 2018; 7(2):44. https://doi.org/10.3390/antibiotics7020044
Chicago/Turabian StyleHug, Joachim J., Chantal D. Bader, Maja Remškar, Katarina Cirnski, and Rolf Müller. 2018. "Concepts and Methods to Access Novel Antibiotics from Actinomycetes" Antibiotics 7, no. 2: 44. https://doi.org/10.3390/antibiotics7020044
APA StyleHug, J. J., Bader, C. D., Remškar, M., Cirnski, K., & Müller, R. (2018). Concepts and Methods to Access Novel Antibiotics from Actinomycetes. Antibiotics, 7(2), 44. https://doi.org/10.3390/antibiotics7020044





