Genetic Determinants of Tetracycline Resistance in Clinical Streptococcus pneumoniae Serotype 1 Isolates from Niger
Abstract
1. Introduction
2. Results
2.1. Patients and Isolates
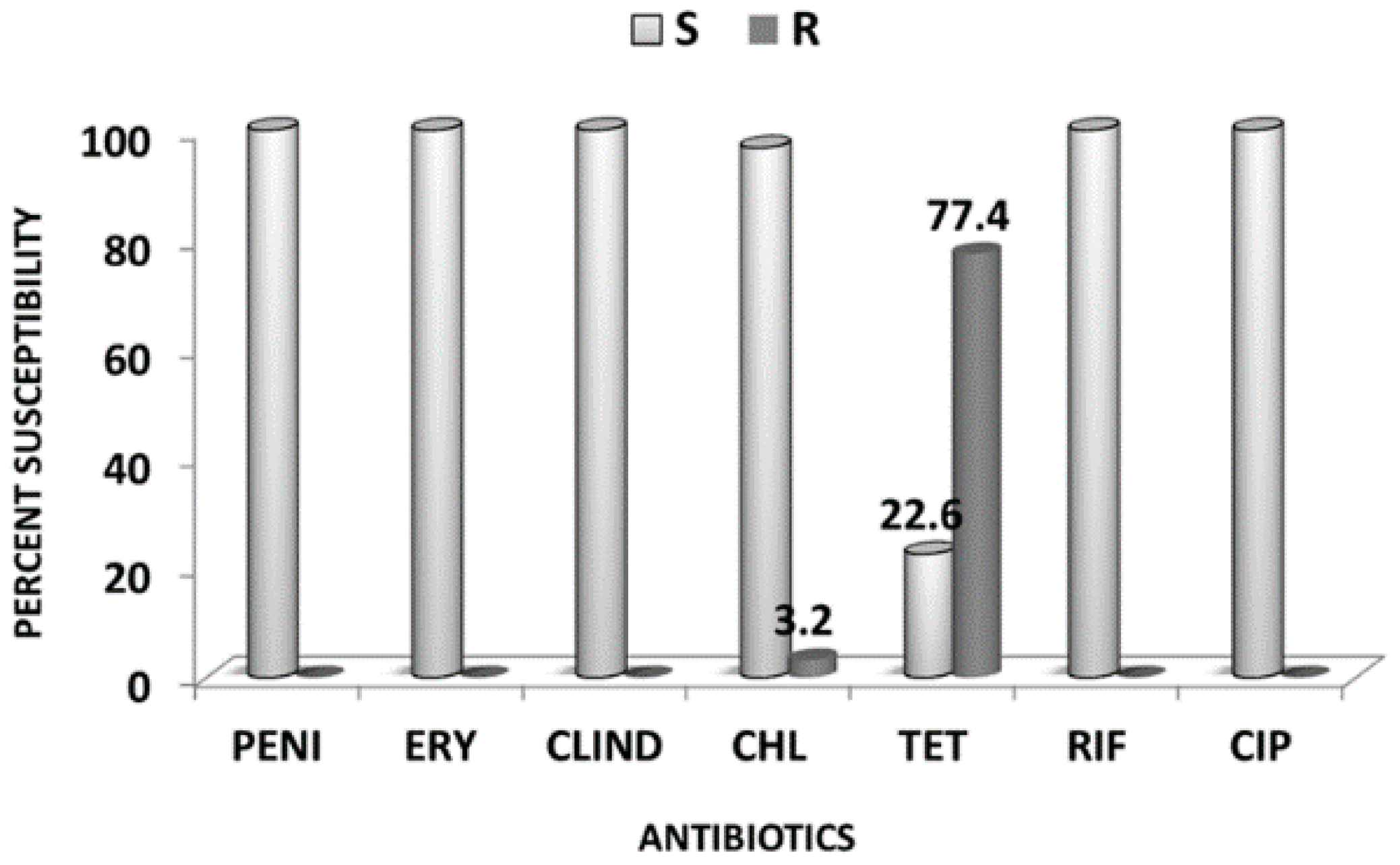
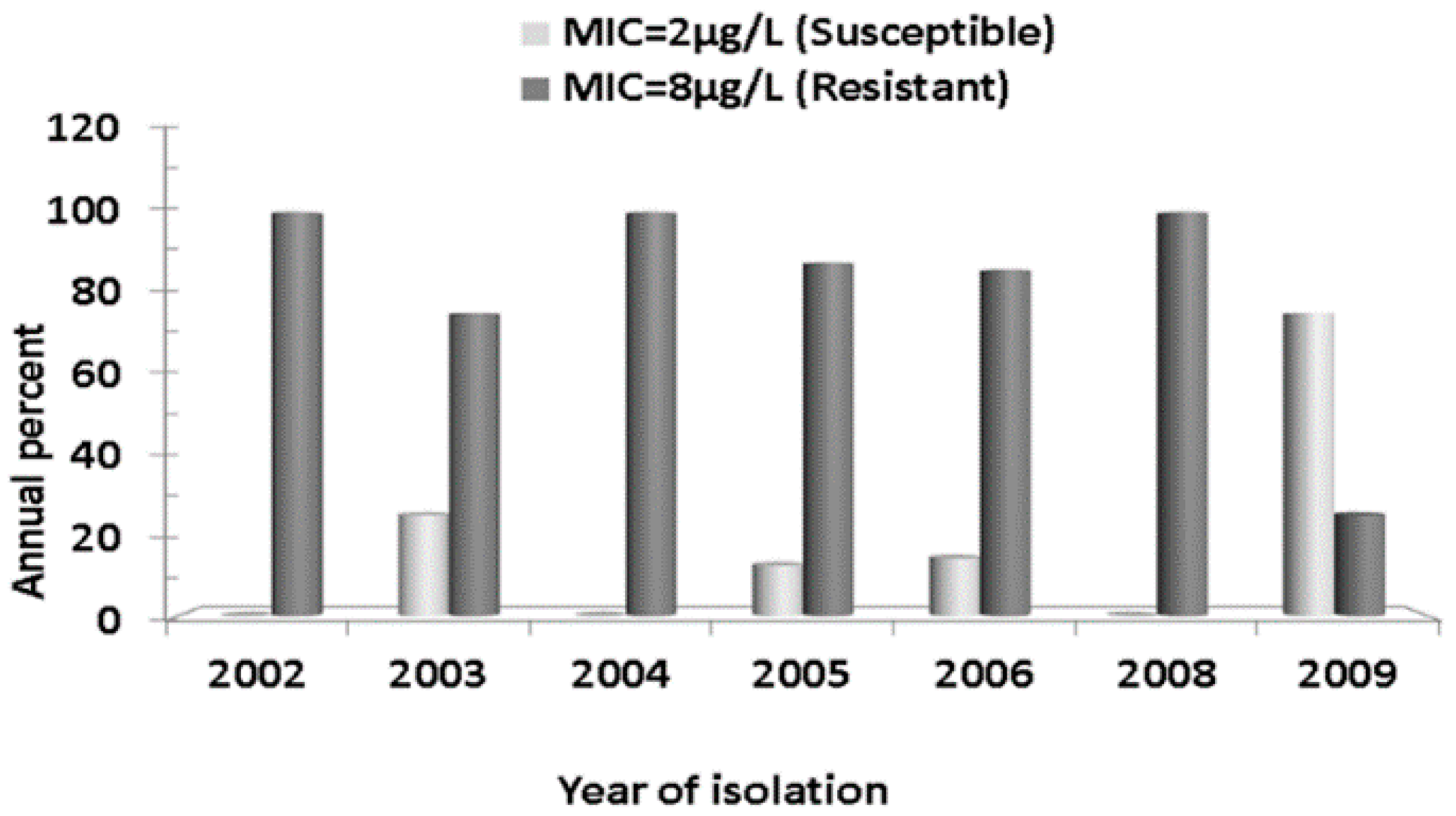
2.2. Antibiotics Susceptibility Testing
2.3. Multi Locus Sequence Typing
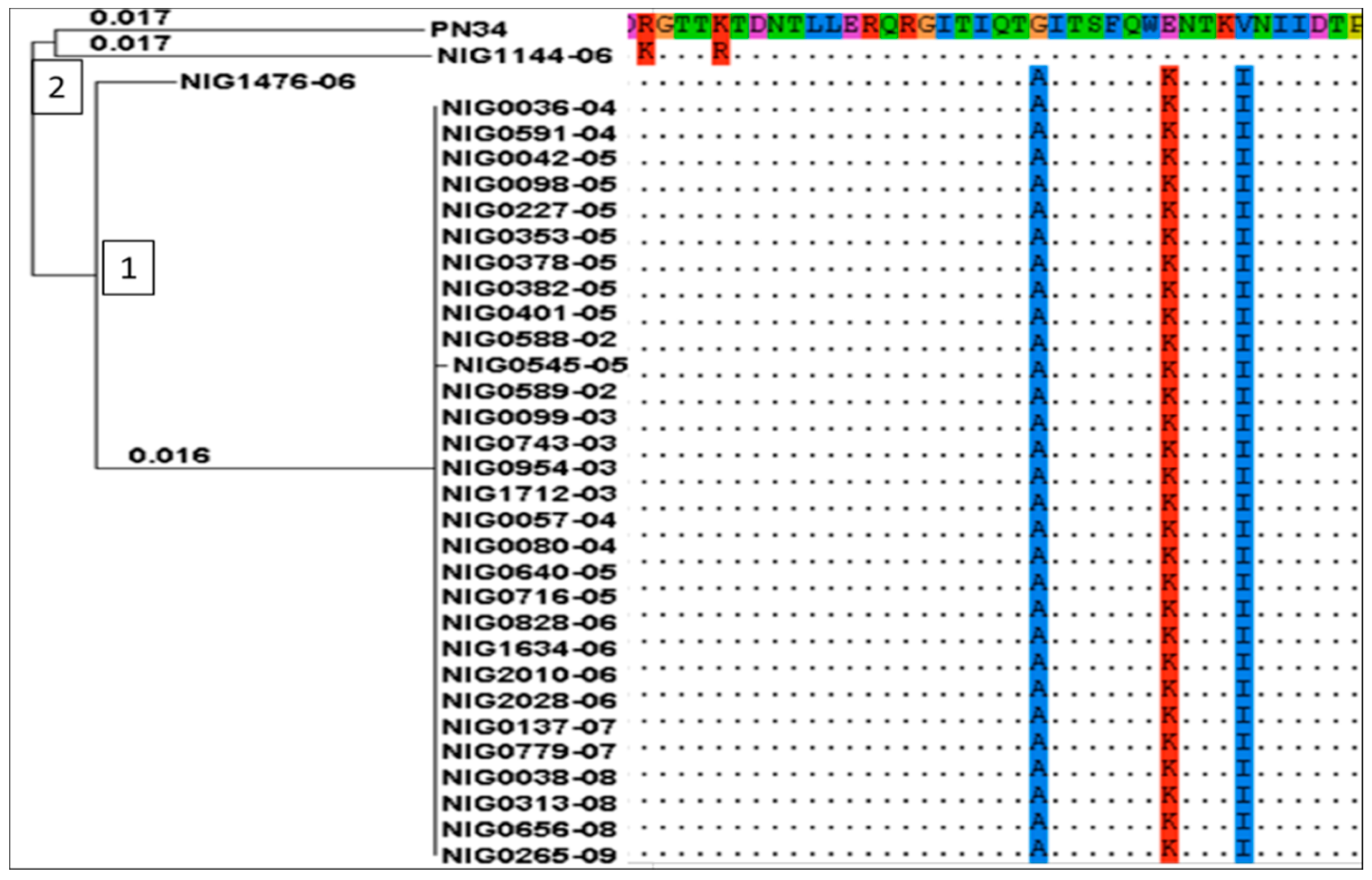
2.4. In Silico Detection of Tetracycline Resistance Determinants
Tetracycline and Chloramphenicol Resistance Genes
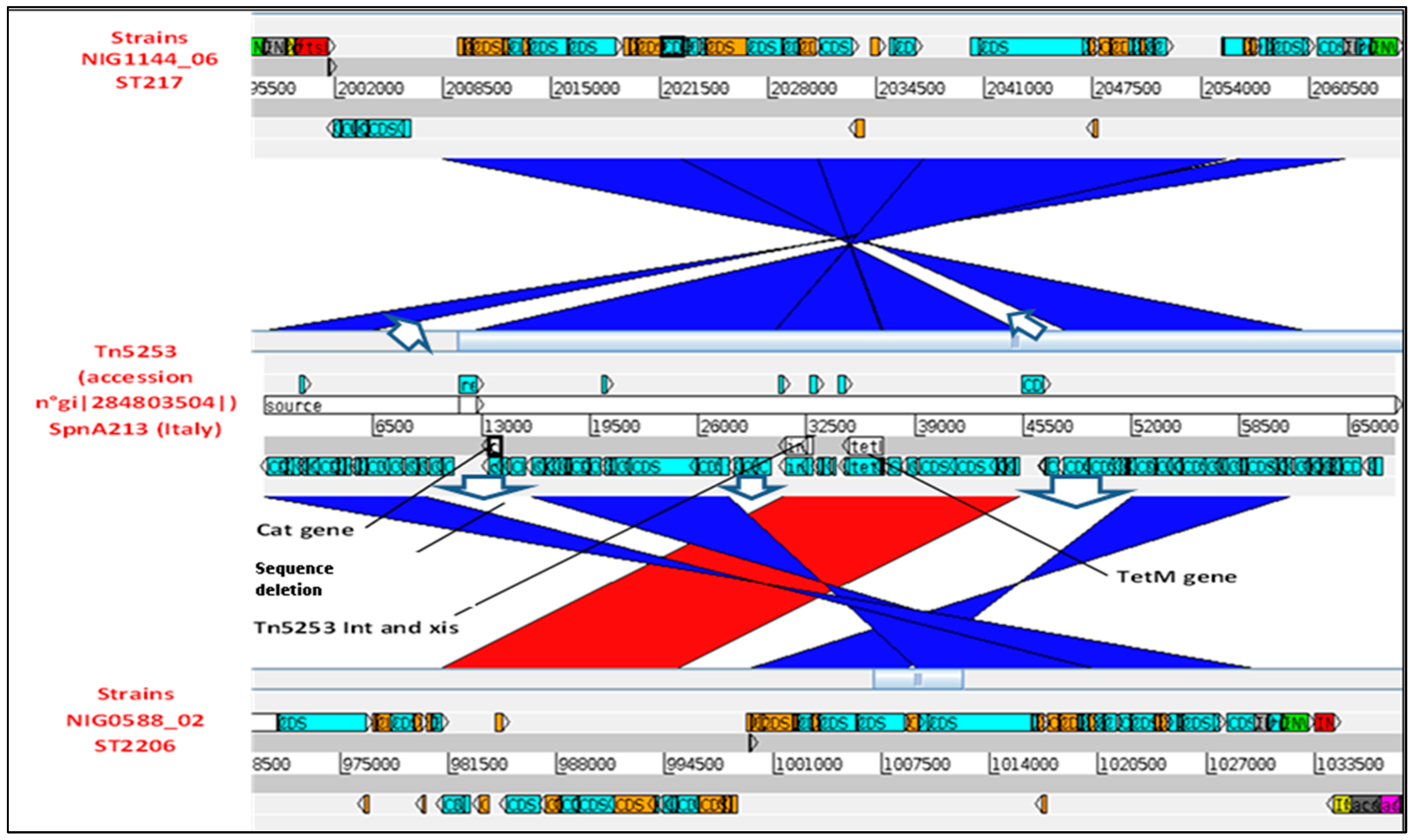
2.5. Carrier Transposons
3. Discussion
4. Materials and Methods
Ethics Statement
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Greenwood, B. The epidemiology of pneumococcal infection in children in the developing world. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1999, 354, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Cashat-Cruz, M.; Morales-Aguirre, J.J.; Mendoza-Azpiri, M. Respiratory tract infections in children in developing countries. Semin. Pediatr. Infect. Dis. 2005, 16, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Usen, S.; Adegbola, R.; Mulholland, K.; Jaffar, S.; Hilton, S.; Oparaugo, A.; Omosigho, C.; Lahai, G.; Corrah, T.; Palmer, A.; et al. Epidemiology of invasive pneumococcal disease in the Western Region, the Gambia. Pediatr. Infect. Dis. J. 1998, 17, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.M.; Lagos, R.; Levine, O.S.; Heitmann, I.; Enriquez, N.; Pinto, M.E.; Alvarez, A.M.; Wu, E.; Mayorga, C.; Reyes, A. Epidemiology of invasive pneumococcal infections in infants and young children in Metropolitan Santiago, Chile, a newly industrializing country. Pediatr. Infect. Dis. J. 1998, 17, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, R.; Togashi, T. Population-based incidence of invasive haemophilus influenzae and pneumococcal diseases before the introduction of vaccines in Japan. Pediatr. Infect. Dis. J. 2013, 32, 1394–1396. [Google Scholar] [CrossRef] [PubMed]
- Gessner, B.D.; Mueller, J.E.; Yaro, S. African meningitis belt pneumococcal disease epidemiology indicates a need for an effective serotype 1 containing vaccine, including for older children and adults. BMC Infect. Dis. 2010. [Google Scholar] [CrossRef] [PubMed]
- Boisier, P.; Maïnassara, H.B.; Sidikou, F.; Djibo, S.; Kairo, K.K.; Chanteau, S. Case-fatality ratio of bacterial meningitis in the African meningitis belt: We can do better. Vaccine 2007, 25, S24–S29. [Google Scholar] [CrossRef] [PubMed]
- Ihekweazu, C.; Basarab, M.; Wilson, D.; Oliver, I.; Dance, D.; George, R.; Pebody, R. Outbreaks of serious pneumococcal disease in closed settings in the post-antibiotic era: A systematic review. J. Infect. 2010, 61, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Gleich, S.; Morad, Y.; Echague, R.; Miller, J.R.; Kornblum, J.; Sampson, J.S.; Butler, J.C. Streptococcus pneumoniae serotype 4 outbreak in a home for the aged: Report and review of recent outbreaks. Infect. Control Hosp. Epidemiol. 2000, 21, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Leimkugel, J.; Adams Forgor, A.; Gagneux, S.; Pflüger, V.; Flierl, C.; Awine, E.; Naegeli, M.; Dangy, J.-P.; Smith, T.; Hodgson, A.; et al. An outbreak of serotype 1 Streptococcus pneumoniae meningitis in northern Ghana with features that are characteristic of Neisseria meningitidis meningitis epidemics. J. Infect. Dis. 2005, 192, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Yaro, S.; Lourd, M.; Traoré, Y.; Njanpop-Lafourcade, B.-M.; Sawadogo, A.; Sangare, L.; Hien, A.; Ouedraogo Macaire, S.; Sanou, O.; Parent du Châtelet, I.; et al. Epidemiological and molecular characteristics of a highly lethal pneumococcal meningitis epidemic in Burkina Faso. Clin. Infect. Dis. 2006, 43, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Chaguza, C.; Cornick, J.E.; Everett, D.B. Mechanisms and impact of genetic recombination in the evolution of Streptococcus pneumoniae. Comput. Struct. Biotechnol. J. 2015, 13, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Klugman, K.P. The successful clone: The vector of dissemination of resistance in Streptococcus pneumoniae. J. Antimicrob. Chemother. 2002, 50, 1–6. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liñares, J.; Ardanuy, C.; Pallares, R.; Fenoll, A. Changes in antimicrobial resistance, serotypes and genotypes in Streptococcus pneumoniae over a 30-year period. Clin. Microbiol. Infect. 2010, 16, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Widdowson, C.A.; Klugman, K.P. The molecular mechanisms of tetracycline resistance in the pneumococcus. Microbiol. Drug Resist. 1998, 4, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Mingoia, M.; Tili, E.; Manso, E.; Varaldo, P.E.; Montanari, M.P. Heterogeneity of Tn5253-like composite elements in clinical Streptococcus pneumoniae isolates. Antimicrob. Agents Chemother. 2011, 55, 1453–1459. [Google Scholar] [CrossRef] [PubMed]
- Franke, A.E.; Clewell, D.B. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 1981, 145, 494–502. [Google Scholar] [PubMed]
- Izdebski, R.; Sadowy, E.; Fiett, J.; Grzesiowski, P.; Gniadkowski, M.; Hryniewicz, W. Clonal diversity and resistance mechanisms in tetracycline-nonsusceptible Streptococcus pneumoniae isolates in Poland. Antimicrob. Agents Chemother. 2007, 51, 1155–1163. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Katarzyna, D.-F.; Katarzyna, S.; Paulina, G.; Stefania, G.-K.; Maria, K.; Alfred, S.; Stefan, T.; Danuta, D.; Krzysztof, T. Evidence for tetracycline resistance determinant tet(M) allele replacement in a Streptococcus pneumoniae population of limited geographical origin. Int. J. Antimicrob. Agents 2006, 27, 159–164. [Google Scholar]
- Widdowson, C.A.; Adrian, P.V.; Klugman, K.P. Acquisition of chloramphenicol resistance by the linearization and integration of the entire staphylococcal plasmid pC194 into the chromosome of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2000, 44, 393–395. [Google Scholar] [CrossRef] [PubMed]
- Hansman, D. Chloramphenicol-resistant pneumococci in West Africa. Lancet 1978, 311, 1102–1103. [Google Scholar] [CrossRef]
- Cornick, J.E.; Chaguza, C.; Harris, S.R.; Yalcin, F.; Senghore, M.; Kiran, A.M.; Govindpershad, S.; Ousmane, S.; Plessis, M.D.; Pluschke, G.; et al. Region-specific diversification of the highly virulent serotype 1 Streptococcus pneumoniae. Microb. Genomics 2015. [Google Scholar] [CrossRef] [PubMed]
- Byarugaba, D.K. Antimicrobial resistance in developing countries and responsible risk factors. Int. J. Antimicrob. Agents 2004, 24, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Okeke, I.N.; Klugman, K.P.; Bhutta, Z.A.; Duse, A.G.; Jenkins, P.; O’Brien, T.F.; Pablos-Mendez, A.; Laxminarayan, R. Antimicrobial resistance in developing countries. Part II: Strategies for containment. Lancet Infect. Dis. 2005, 5, 568–580. [Google Scholar] [CrossRef]
- Collard, J.M.; Alio Sanda, A.K.; Jusot, J.F. Determination of pneumococcal serotypes in meningitis cases in Niger, 2003–2011. PLoS ONE 2013, 8, e60432. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Greenwood, B. Pneumococcal meningitis epidemics in Africa. Clin. Infect. Dis. 2006, 43, 701–703. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Antonio, M.; Hakeem, I.; Awine, T.; Secka, O.; Sankareh, K.; Nsekpong, D.; Lahai, G.; Akisanya, A.; Egere, U.; Enwere, G.; et al. Seasonality and outbreak of a predominant Streptococcus pneumoniae serotype 1 clone from the Gambia: Expansion of ST217 hypervirulent clonal complex in West Africa. BMC Microbiol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Chaguza, C.; Cornick, J.E.; Harris, S.R.; Andam, C.P.; Bricio-Moreno, L.; Yang, M.; Yalcin, F.; Ousmane, S.; Govindpersad, S.; Senghore, M.; et al. Understanding pneumococcal serotype 1 biology through population genomic analysis. BMC Infect. Dis. 2016. [Google Scholar] [CrossRef] [PubMed]
- Ousmane, S.; Diallo, B.A.; Ouedraogo, R.; Sanda, A.K.A.; Soussou, A.M.; Collard, J.M. Serotype distribution and antimicrobial sensitivity profile of Streptococcus pneumoniae carried in healthy toddlers before PCV13 introduction in Niamey, Niger. PLoS ONE 2017, 12, e0169547. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance standards for antimicrobial susceptibility testing: Twenty-first informational supplement. Clin. Lab. Stand. Inst. 2011, 31, 1–172. [Google Scholar]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ousmane, S.; Diallo, B.A.; Ouedraogo, R. Genetic Determinants of Tetracycline Resistance in Clinical Streptococcus pneumoniae Serotype 1 Isolates from Niger. Antibiotics 2018, 7, 19. https://doi.org/10.3390/antibiotics7010019
Ousmane S, Diallo BA, Ouedraogo R. Genetic Determinants of Tetracycline Resistance in Clinical Streptococcus pneumoniae Serotype 1 Isolates from Niger. Antibiotics. 2018; 7(1):19. https://doi.org/10.3390/antibiotics7010019
Chicago/Turabian StyleOusmane, Sani, Bouli A. Diallo, and Rasmata Ouedraogo. 2018. "Genetic Determinants of Tetracycline Resistance in Clinical Streptococcus pneumoniae Serotype 1 Isolates from Niger" Antibiotics 7, no. 1: 19. https://doi.org/10.3390/antibiotics7010019
APA StyleOusmane, S., Diallo, B. A., & Ouedraogo, R. (2018). Genetic Determinants of Tetracycline Resistance in Clinical Streptococcus pneumoniae Serotype 1 Isolates from Niger. Antibiotics, 7(1), 19. https://doi.org/10.3390/antibiotics7010019



