Do Different Sutures with Triclosan Have Different Antimicrobial Activities? A Pharmacodynamic Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microbiology: Time-Kill Assays
2.2. Data Analysis
2.2.1. Plots of Time-Kill Assays
2.2.2. Statistical Analysis
2.2.3. Pharmacodynamic Fitting of Microbial Concentration
- (1)
- The targeted adjusted coefficient of determination R²adjusted (4) was within [0.9, 1] and
- (2)
- The predicted concentration at T24 was equal to the observed mean concentration at T24 among the six repetitions. n was the number of observations per combination (n = 4 measurements × 6 repetitions = 24 per combination), and k was the number of estimated explanatory parameters in the function using the n observations (k = 4, per combination).
3. Results
3.1. Time-Kill Assays
3.2. Statistical Analysis–Comprehensive Panel Model
3.3. Statistical Analysis–Focused Panel Model
3.4. Statistical Analysis–Post Hoc Paired t-Test
3.5. Pharmacodynamic Fitting of Microbial Concentration
4. Discussion
4.1. Protocol Specifications and Interpretation
4.2. Key Results
4.3. Interpretation of The Time-Kill Assays
4.4. Comparison with Other Preclinical Studies
4.5. Translational Interpretation to Live Operated Human Tissues
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hochberg, J.; Meyer, K.M.; Marion, M.D. Suture Choice and Other Methods of Skin Closure. Surg. Clin. N. Am. 2009, 89, 627–641. [Google Scholar] [CrossRef] [PubMed]
- Osunde, O.; Adebola, R.; Saheeb, B. A comparative study of the effect of suture-less and multiple suture techniques on inflammatory complications following third molar surgery. Int. J. Oral Maxillofac. Surg. 2012, 41, 1275–1279. [Google Scholar] [CrossRef]
- Lam, K.Y.; Reardon, M.J.; Yakubov, S.J.; Modine, T.; Fremes, S.; Tonino, P.A.; Tan, M.E.; Gleason, T.G.; Harrison, J.K.; Hughes, G.C.; et al. Surgical Sutureless and Sutured Aortic Valve Replacement in Low-risk Patients. Ann. Thorac. Surg. 2021, 113, 616–622. [Google Scholar] [CrossRef]
- Chisci, G.; Parrini, S.; Capuano, A. The use of suture-less technique following third molar surgery. Int. J. Oral Maxillofac. Surg. 2013, 42, 150–151. [Google Scholar] [CrossRef] [PubMed]
- Chisci, G.; Capuano, A.; Parrini, S. Alveolar Osteitis and Third Molar Pathologies. J. Oral Maxillofac. Surg. 2018, 76, 235–236. [Google Scholar] [CrossRef]
- Dhillon, G.S.; Kaur, S.; Pulicharla, R.; Brar, S.K.; Cledón, M.; Verma, M.; Surampalli, R.Y. Triclosan: Current Status, Occurrence, Environmental Risks and Bioaccumulation Potential. Int. J. Environ. Res. Public Health 2015, 12, 5657–5684. [Google Scholar] [CrossRef] [PubMed]
- Scientific Committee on Consumer Safety (SCCS). Opinion on Triclosan 2010. European Commission—Directorate General for Health and Consumers. Available online: https://op.europa.eu/en/publication-detail/-/publication/3b684b59-27f7-4d0e-853c-4a9efe0fb4cb/language-en (accessed on 3 December 2021).
- Villalaín, J.; Mateo, C.R.; Aranda, F.J.; Shapiro, S.; Micol, V. Membranotropic Effects of the Antibacterial Agent Triclosan. Arch. Biochem. Biophys. 2001, 390, 128–136. [Google Scholar] [CrossRef]
- Petersen, R.C. Triclosan Computational Conformational Chemistry Analysis for Antimicrobial Properties in Polymers. J. Nat. Appl. Sci. 2015, 1, e54. Available online: https://www.ncbi.nlm.nih.gov/pubmed/25879080 (accessed on 15 October 2021).
- Bernabeu, A.; Shapiro, S.; Bernabeu-Sanz, A. Location and orientation of Triclosan in phospholipid model membranes. Eur. Biophys. J. 2004, 33, 448–453. [Google Scholar] [CrossRef]
- Hayami, M.; Okabe, A.; Kariyama, R.; Abe, M.; Kanemasa, Y. Lipid Composition of Staphylococcus aureus and Its Derived L-forms. Microbiol. Immunol. 1979, 23, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.D. Similarities and differences in the responses of microorganisms to biocides. J. Antimicrob. Chemother. 2003, 52, 750–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heath, R.J.; Su, N.; Murphy, C.K.; Rock, C.O. The Enoyl-[acyl-carrier-protein] Reductases FabI and FabL from Bacillus subtilis. J. Biol. Chem. 2000, 275, 40128–40133. [Google Scholar] [CrossRef]
- Schweizer, H.P. Triclosan: A widely used biocide and its link to antibiotics. FEMS Microbiol. Lett. 2001, 202, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chuanchuen, R.; Karkhoff-Schweizer, R.R.; Schweizer, H.P. High-level triclosan resistance in Pseudomonas aeruginosa is solely a result of efflux. Am. J. Infect. Control 2003, 31, 124–127. [Google Scholar] [CrossRef]
- Zhu, L.; Lin, J.; Ma, J.; Cronan, J.E.; Wang, H. Triclosan Resistance of Pseudomonas aeruginosa PAO1 Is Due to FabV, a Triclosan-Resistant Enoyl-Acyl Carrier Protein Reductase. Antimicrob. Agents Chemother. 2010, 54, 689–698. [Google Scholar] [CrossRef]
- BASF Names West Coast Distributor for Care Chemicals Business. Available online: https://www.basf.com/us/en/media/news-releases/2013/10/p-13-441.html (accessed on 8 September 2021).
- Heath, R.J.; Li, J.; Roland, G.E.; Rock, C.O. Inhibition of the Staphylococcus aureus NADPH-dependent Enoyl-Acyl Carrier Protein Reductase by Triclosan and Hexachlorophene. J. Biol. Chem. 2000, 275, 4654–4659. [Google Scholar] [CrossRef] [PubMed]
- Heath, R.J.; Rubin, J.R.; Holland, D.R.; Zhang, E.; Snow, M.E.; Rock, C.O. Mechanism of Triclosan Inhibition of Bacterial Fatty Acid Synthesis. J. Biol. Chem. 1999, 274, 11110–11114. [Google Scholar] [CrossRef] [PubMed]
- Levy, C.W.; Roujeinikova, A.; Sedelnikova, S.; Baker, P.J.; Stuitje, A.R.; Slabas, A.R.; Rice, D.W.; Rafferty, J.B. Molecular basis of triclosan activity. Nature 1999, 398, 383–384. [Google Scholar] [CrossRef] [PubMed]
- Escalada, M.G.; Harwood, J.L.; Maillard, J.-Y.; Ochs, D. Triclosan inhibition of fatty acid synthesis and its effect on growth of Escherichia coli and Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2005, 55, 879–882. [Google Scholar] [CrossRef] [PubMed]
- Giuliana, G.; Pizzo, G.; Milici, M.E.; Musotto, G.C.; Giangreco, R. In Vitro Antifungal Properties of Mouthrinses Containing Antimicrobial Agents. J. Periodontol. 1997, 68, 729–733. [Google Scholar] [CrossRef]
- VICRYL Plus—389595.R04; Ethicon Inc.: Raritan, NJ, USA, 2002.
- PDS Plus—389688.R02; Ethicon Inc.: Raritan, NJ, USA, 2006.
- MONOCRYL Plus—389680.R02; Ethicon Inc.: Raritan, NJ, USA, 2005.
- Daoud, F.C.; Goncalves, R.; Moore, N. How Long Do Implanted Triclosan Sutures Inhibit Staphylococcus aureus in Surgical Conditions? A Pharmacological Model. Pharmaceutics 2022, 14, 539. [Google Scholar] [CrossRef]
- Rothenburger, S.; Spangler, D.; Bhende, S.; Burkley, D. In Vitro Antimicrobial Evaluation of Coated VICRYL* Plus Antibacterial Suture (Coated Polyglactin 910 with Triclosan) using Zone of Inhibition Assays. Surg. Infect. 2002, 3 (Suppl. 1), s79–s87. [Google Scholar] [CrossRef]
- Ming, X.; Rothenburger, S.; Nichols, M.M. In Vivo and In Vitro Antibacterial Efficacy of PDS Plus (Polidioxanone with Triclosan) Suture. Surg. Infect. 2008, 9, 451–457. [Google Scholar] [CrossRef]
- Ming, X.; Rothenburger, S.; Yang, D. In Vitro Antibacterial Efficacy of MONOCRYL Plus Antibacterial Suture (Poliglecaprone 25 with Triclosan). Surg. Infect. 2007, 8, 201–208. [Google Scholar] [CrossRef]
- Storch, M.L.; Rothenburger, S.J.; Jacinto, G. Experimental Efficacy Study of Coated VICRYL plus Antibacterial Suture in Guinea Pigs Challenged with Staphylococcus aureus. Surg. Infect. 2004, 5, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.; Boulton, A.J.; Rizvi, S.; Carlos, W.; Dickenson, E.; A Smith, N.; Reed, M. The use of triclosan-coated sutures to prevent surgical site infections: A systematic review and meta-analysis of the literature. BMJ Open 2019, 9, e029727. [Google Scholar] [CrossRef]
- WHO. World Health Organization Global Guidelines for the Prevention of Surgical Site Infection, 2nd ed.; Licence: CC BY-NC-SA 3.0 IGO. 2018; WHO: Geneva, Switzerland, 2018; Available online: https://apps.who.int›handle›9789241550475-eng.pdf (accessed on 15 October 2021).
- Horan, T.C.; Gaynes, R.P.; Martone, W.J.; Jarvis, W.R.; Emori, T.G. CDC definitions of nosocomial surgical site infections, 1992: A modification of CDC definitions of surgical wound infections. Am. J. Infect. Control. 1992, 20, 271–274. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. M26-A Methods for Determining Bactericidal Activity of Antimicrobial Agents; Approved Guideline. September 1999. Available online: https://clsi.org/standards/products/microbiology/documents/m26/ (accessed on 15 October 2021).
- Halaby, C.N. Panel Models in Sociological Research: Theory into Practice. Annu. Rev. Sociol. 2004, 30, 507–544. [Google Scholar] [CrossRef]
- Smith, B.T. Solubility and Dissolution. Remington Education: Physical Pharmacy; Pharmaceutical Press: London, UK, 2016; pp. 31–50. Available online: https://www.pharmpress.com/product/9780857112521/remington-education-physical-pharmacy-ebook (accessed on 1 August 2020).
- Mircioiu, C.; Voicu, V.; Anuta, V.; Tudose, A.; Celia, C.; Paolino, D.; Fresta, M.; Sandulovici, R.; Mircioiu, I. Mathematical Modeling of Release Kinetics from Supramolecular Drug Delivery Systems. Pharmaceutics 2019, 11, 140. [Google Scholar] [CrossRef] [PubMed]
- The Merck Index—An Encyclopedia of Chemicals, Drugs, and Biologicals, 15th ed.; O’Neil, M.J.; Maryadele, J. (Eds.) The Royal Society of Chemistry: Cambridge, UK, 2013; 1789p. [Google Scholar]
- Yalkowsky, S.H.; Parijat, H.Y.; Triclosan, J. Handbook of Aqueous Solubility Data; CRC Press: Boca Raton, FL, USA, 2010; p. 844. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Triclosan#section=Melting-Point (accessed on 1 August 2020).
- Ciusa, M.L.; Furi, L.; Knight, D.; Decorosi, F.; Fondi, M.; Raggi, C.; Coelho, J.R.; Aragones, L.; Moce, L.; Visa, P.; et al. A novel resistance mechanism to triclosan that suggests horizontal gene transfer and demonstrates a potential selective pressure for reduced biocide susceptibility in clinical strains of Staphylococcus aureus. Int. J. Antimicrob. Agents 2012, 40, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, I.; Oggioni, M.R.; Knight, D.R.; Curiao, T.; Coque, T.; Kalkanci, A.; Martinez, J.L. The BIOHYPO Consortium Evaluation of Epidemiological Cut-Off Values Indicates that Biocide Resistant Subpopulations Are Uncommon in Natural Isolates of Clinically-Relevant Microorganisms. PLoS ONE 2014, 9, e86669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCagherty, J.; Yool, D.A.; Paterson, G.K.; Mitchell, S.R.; Woods, S.; Marques, A.I.; Hall, J.L.; Mosley, J.R.; Nuttall, T.J. Investigation of the in vitro antimicrobial activity of triclosan-coated suture material on bacteria commonly isolated from wounds in dogs. Am. J. Veter Res. 2020, 81, 84–90. [Google Scholar] [CrossRef] [PubMed]

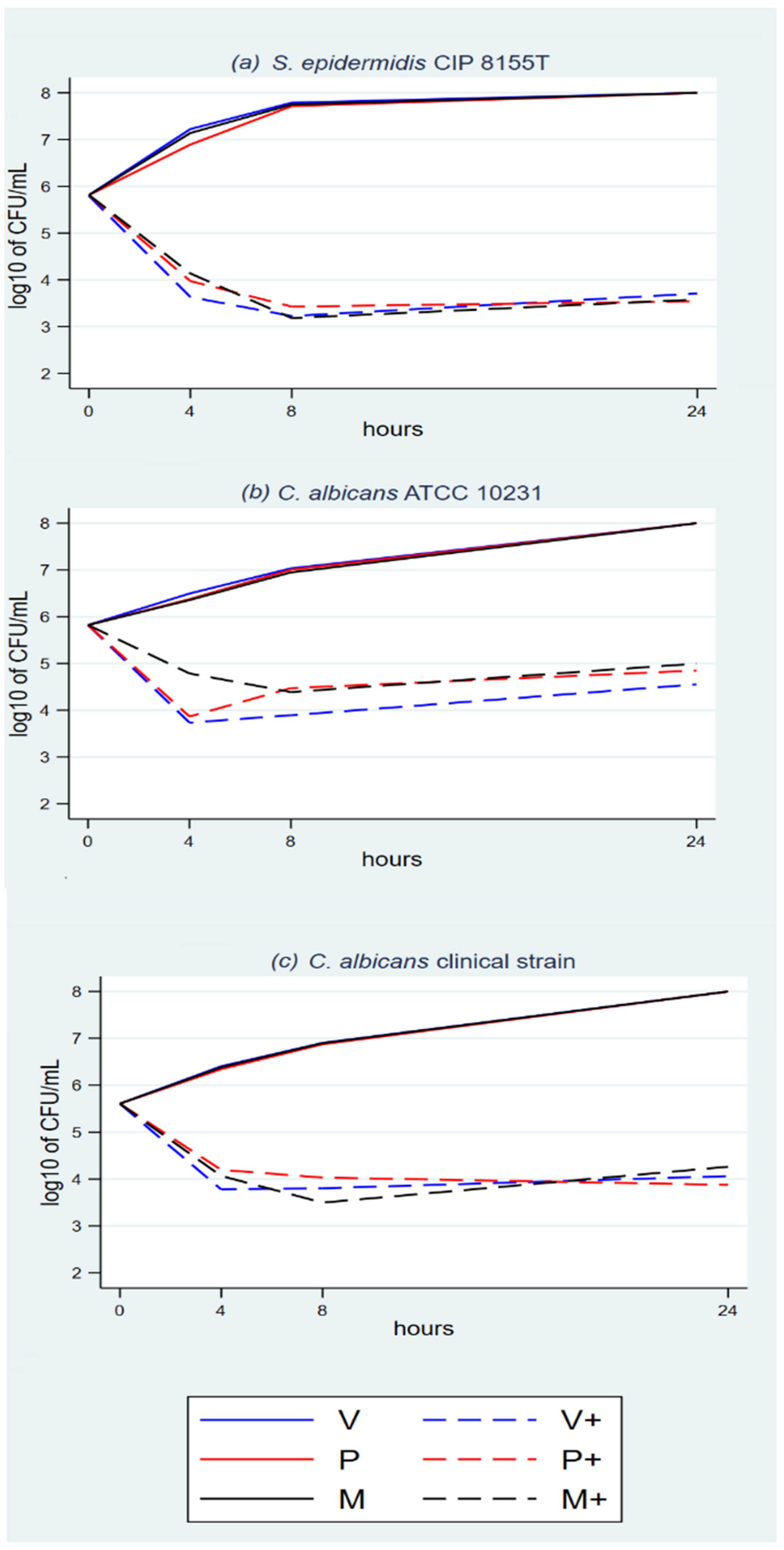
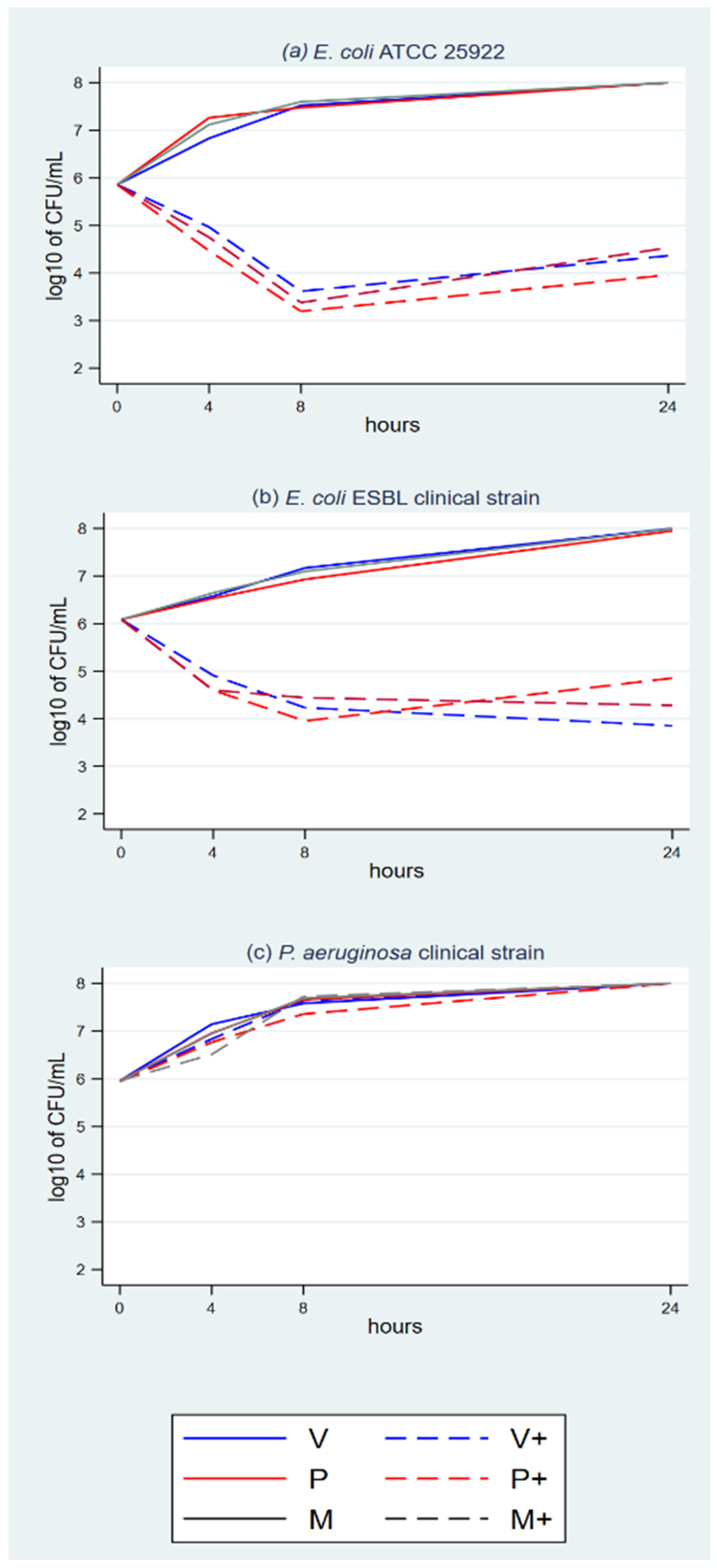


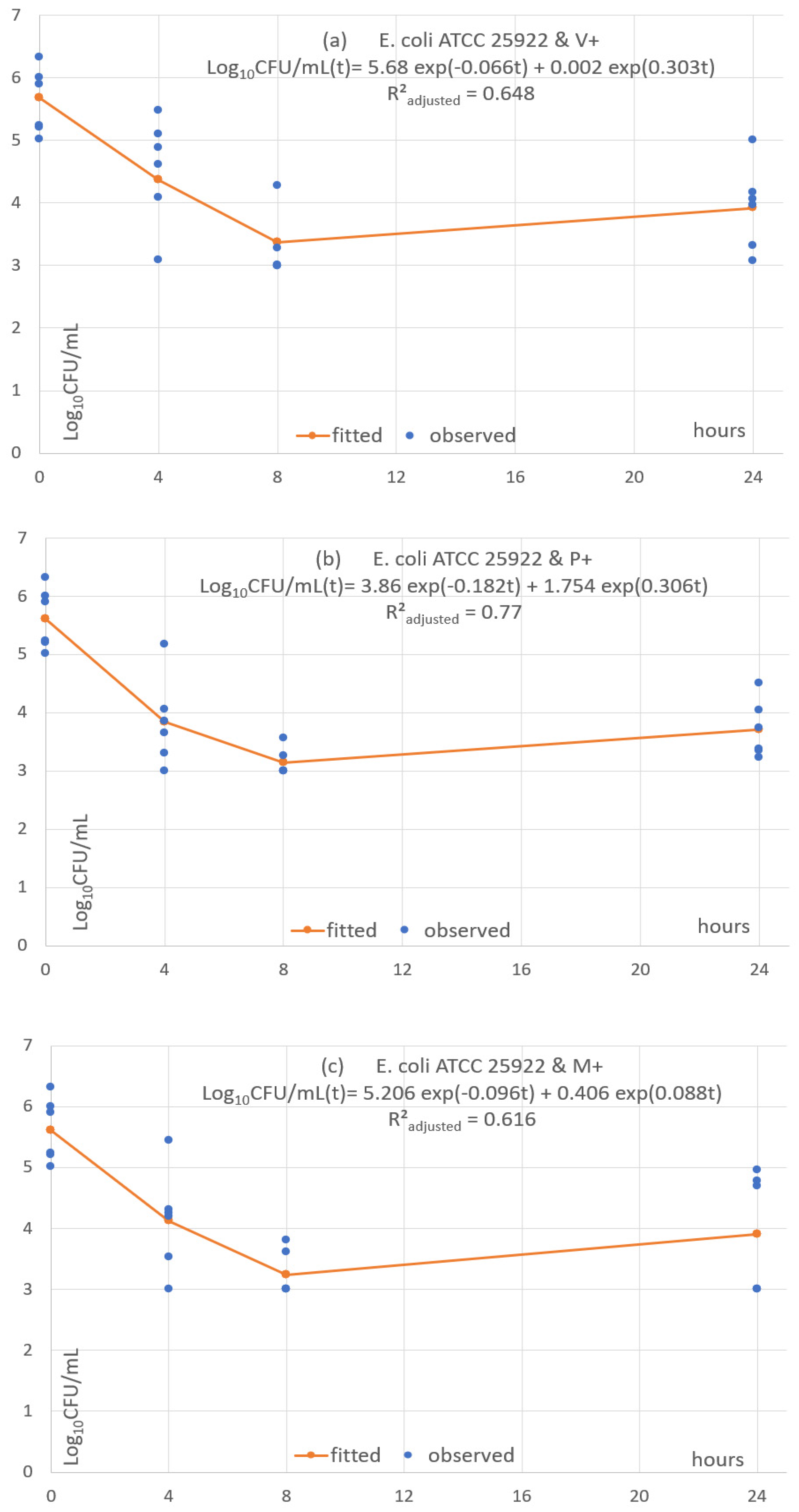
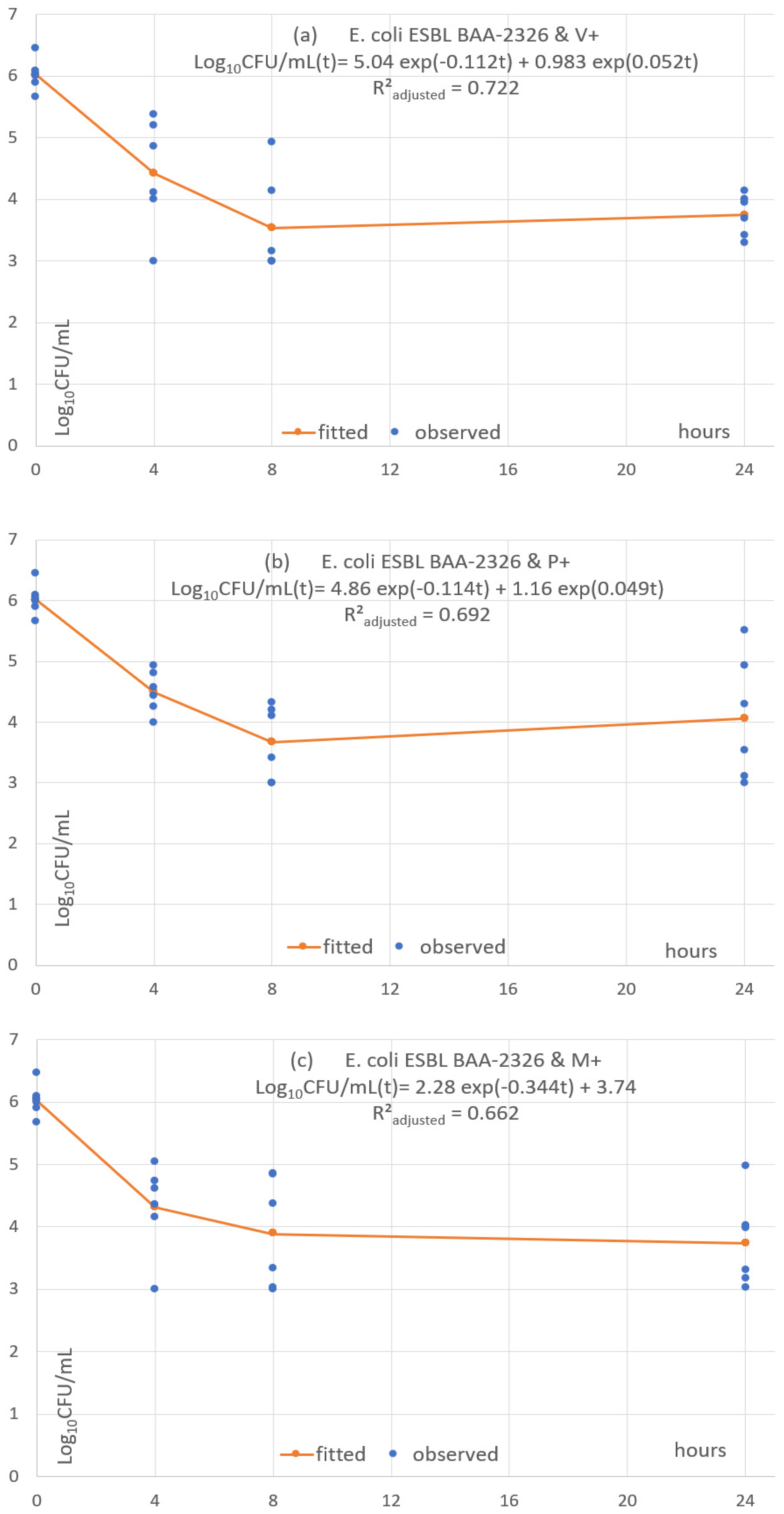
| log(CFU) | Coef. | St.Err. | t-Value | p-Value | 95% Conf | Interval | Sig. |
|---|---|---|---|---|---|---|---|
| S. epidermidis CIP 8155T | 0 | Reference level for microorganisms | |||||
| C. albicans ATCC 10231 | −0.097 | 0.124 | −0.78 | 0.436 | −0.339 | 0.147 | |
| C. albicans clinical | −0.205 | 0.124 | −1.66 | 0.098 | −0.448 | 0.038 | |
| E. coli ATCC 25929 | 0.052 | 0.124 | 0.42 | 0.674 | −0.191 | 0.295 | |
| E. coli ESBL clinical | 0.183 | 0.124 | 1.48 | 0.140 | −0.060 | 0.426 | |
| MRSA ATCC 33592 | 0.152 | 0.124 | 1.23 | 0.221 | −0.195 | 0.395 | |
| MRSA clinical strain | 0.048 | 0.124 | 0.38 | 0.700 | −0.205 | 0.291 | |
| P. aeruginosa clinical | 1.306 | 0.124 | 10.55 | 0.000 | 1.063 | 1.549 | * |
| S. aureus ATCC 29213 | 0.170 | 0.124 | 1.37 | 0.170 | −0.073 | 0.413 | |
| Polyglecaprone 25 | 0 | Reference level for suture materials | |||||
| Polyglactin 910 | 0.035 | 0.072 | 0.47 | 0.637 | −0.105 | 0.175 | |
| Polydioxanone | −0.015 | 0.072 | −0.21 | 0.835 | −0.156 | 0.125 | |
| No triclosan | 0 | Reference level for use of triclosan | |||||
| Triclosan | −2.256 | 0.058 | −38.64 | 0.000 | −2.370 | −2.141 | * |
| 0 h | 0 | Reference level for hours | |||||
| 4 | −0.379 | 0.083 | −4.66 | 0.000 | −0.541 | −0.217 | * |
| 8 | −0.237 | 0.083 | −2.90 | 0.005 | −0.399 | −0.075 | ** |
| 24 | 0.416 | 0.083 | 5.11 | 0.000 | 0.254 | 0.578 | * |
| Overall model intercept | 4.400 | 0.113 | 38.92 | 0.000 | 4.179 | 4.622 | * |
| Number of obs | 1296 | Number of groups | 324 | ||||
| F-test | df = 141,281 | p-value | <0.0001 | ||||
| R-squared | 0.5847 | Adjusted R-squared | 0.5802 | ||||
| log(CFU) | Coef. | St.Err. | t-Value | p-Value | 95% Conf | Interval | Sig. |
|---|---|---|---|---|---|---|---|
| S. epidermidis CIP 8155T | 0 | Reference level for microorganisms | |||||
| C. albicans ATCC 10231 | 0.341 | 0.128 | 2.65 | 0.008 | 0.089 | 0.592 | |
| C. albicans clinical | 0.098 | 0.128 | 0.77 | 0.444 | −0.153 | 0.350 | |
| E. coli ATCC 25929 | 0.265 | 0.128 | 2.07 | 0.039 | 0.014 | 0.517 | |
| E. coli ESBL clinical | 0.551 | 0.128 | 4.30 | 0.000 | 0.210 | 0.803 | * |
| MRSA ATCC 33592 | 0.537 | 0.128 | 4.19 | 0.000 | 0.286 | 0.789 | * |
| MRSA clinical strain | 0.221 | 0.128 | 1.72 | 0.085 | −0.030 | 0.472 | |
| S. aureus ATCC 29213 | 0.545 | 0.128 | 4.25 | 0.000 | 0.294 | 0.796 | * |
| Polyglecaprone 25 | 0 | Reference level for suture materials | |||||
| Polyglactin 910 | 0.071 | 0.079 | 0.90 | 0.369 | −0.083 | 0.225 | |
| Polydioxanone | −0.008 | 0.079 | −0.11 | 0.915 | −0.162 | 0.146 | |
| 0 h | 0 | Reference level for hours | |||||
| 4 | −1.795 | 0.063 | −28.28 | 0.000 | −1.919 | −1.670 | * |
| 8 | −2.148 | 0.063 | −33.85 | 0.000 | −2.273 | −2.023 | * |
| 24 | −1.936 | 0.063 | −30.50 | 0.000 | −2.060 | −1.811 | * |
| Overall model intercept | 5.392 | 0.109 | 49.64 | 0.000 | 5.179 | 5.605 | ** |
| Number of obs | 576 | Number of groups | 144 | ||||
| Wald Chi-squared | df = 12 | p-value | <0.0001 | ||||
| R-squared | 0.687 | ||||||
| Microbial Strain | Suture | R²adjusted | C1 µg/mL | HF1 Hours | C2 µg/mL | HL2 Hours | Plot Shape |
|---|---|---|---|---|---|---|---|
| S. aureus ATCC 29213 | V+ | 0.631 | 1.600 | 2.820 | 4.129 | NA. | T0–T8: approx. −1.5log10 T8–T24: plateau & mild decrease |
| S. aureus ATCC 29213 | P+ | 0.787 | 2.279 | 1.852 | 3.451 | 451 | T0–T8: approx. −2log10 T8–T24: steady plateau |
| S. aureus ATCC 29213 | M+ | 0.700 | 5.382 | 10.558 | 0.359 | 8.171 | T0–T8: approx. −2log10 T8–T24: plateau & mild decrease |
| MRSA ATCC 33592 | V+ | 0.676 | 2.542 | 2.633 | 3.445 | 98.943 | T0–T8: approx. −2log10 T8–T24: plateau & mild increase |
| MRSA ATCC 33592 | P+ | 0.710 | 2.696 | 1.974 | 3.691 | 210.724 | T0–T8: approx. T8–T24: plateau & mild increase |
| MRSA ATCC 33592 | M+ | 0.858 | 3.017 | 2.000 | 2.9700 | 64.903 | T0–T8: approx. −2.5log10 T8–T24: plateau & mild increase |
| MRSA clinical | V+ | 0.700 | 2.070 | 0.633 | 3.776 | NA. | T0–T4: approx. −2log10 T4–T24: steady plateau |
| MRSA clinical | P+ | 0.836 | 2.680 | 1.613 | 3.166 | 152.646 | T0–T8: approx. −2.5log10 T8–T24: plateau & mild increase |
| MRSA clinical | M+ | 0.818 | 2.547 | 1.677 | 3.297 | NA. | T0–T8: approx. −2.5log10 T8–T24: plateau & mild decrease |
| S. epidermidis CIP 8155T | V+ | 0.889 | 2.891 | 0.971 | 2.834 | 64.203 | T0–T8: approx. −2.5log10 T8–T24: plateau & mild increase |
| S. epidermidis CIP 8155T | P+ | 0.833 | 2.960 | 1.778 | 2.790 | 83.821 | T0–T8: approx. −2.5log10 T8–T24: plateau & mild increase |
| S. epidermidis CIP 8155T | M+ | 0.857 | 3.364 | 2.714 | 2.389 | 50.196 | T0–T8: approx. −2.5log10 T8–T24: plateau & mild increase |
| E. coli ATCC 25922 | V+ | 0.648 | 5.682 | 10.528 | 0.0019 | 2.287 | T0–T8: approx. −2log10 T8–T24: plateau & mild increase |
| E. coli ATCC 25922 | P+ | 0.771 | 3.858 | 3.791 | 1.754 | 22.728 | T0–T8: approx. −2.5log10 T8–T24: plateau & mild increase |
| E. coli ATCC 25922 | M+ | 0.616 | 5.206 | 7.196 | 0.406 | 7.884 | T0–T8: approx. −2.5log10 T8–T24: plateau & mild increase |
| E. coli ESBL BAA-2326 | V+ | 0.721 | 5.037 | 6.162 | 0.983 | 13.36 | T0–T8: approx. −2.5log10 T8–T24: plateau & mild increase |
| E. coli ESBL BAA-2326 | P+ | 0.692 | 4.857 | 6.091 | 1.162 | 14.216 | T0–T8: approx. −2.5log10 T8–T24: plateau & mild increase |
| E. coli ESBL BAA-2326 | M+ | 0.662 | 2.78 | 2.014 | 2.015 | NA. | T0–T8: approx. −2log10 T8–T24: plateau & mild decrease |
| C. albicans ATCC 10231 | V+ | 0.629 | 2.104 | 0.185 | 3.371 | 93.158 | T0–T4: approx. −2log10 T4–T24: plateau & mild increase |
| C. albicans ATCC 10231 | P+ | 0.494 | 1.864 | 0.166 | 3.611 | 86.005 | T0–T4: approx. −1.7log10 T4–T24: plateau & mild increase |
| C. albicans ATCC 10231 | M+ | 0.381 | 1.824 | 0.184 | 3.651 | 98.992 | T0–T4: approx. −1.7log10 T4–T24: plateau & mild increase |
| C. albicans clinical | V+ | 0.647 | 5.673 | 10.400 | 0.006 | 2.723 | T0–T8: approx. −2log10 T8–T24: plateau & mild increase |
| C. albicans clinical | P+ | 0.648 | 5.675 | 10.568 | 0.001 | 2.070 | T0–T8: approx. −2log10 T8–T24: plateau & mild increase |
| C. albicans clinical | M+ | 0.648 | 5.680 | 10.534 | 0.001 | 2.157 | T0–T8: approx. −2log10 T8–T24: plateau & mild increase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daoud, F.C.; M’Zali, F.; Zabala, A.; Moore, N.; Rogues, A.-M. Do Different Sutures with Triclosan Have Different Antimicrobial Activities? A Pharmacodynamic Approach. Antibiotics 2022, 11, 1195. https://doi.org/10.3390/antibiotics11091195
Daoud FC, M’Zali F, Zabala A, Moore N, Rogues A-M. Do Different Sutures with Triclosan Have Different Antimicrobial Activities? A Pharmacodynamic Approach. Antibiotics. 2022; 11(9):1195. https://doi.org/10.3390/antibiotics11091195
Chicago/Turabian StyleDaoud, Frederic C., Fatima M’Zali, Arnaud Zabala, Nicholas Moore, and Anne-Marie Rogues. 2022. "Do Different Sutures with Triclosan Have Different Antimicrobial Activities? A Pharmacodynamic Approach" Antibiotics 11, no. 9: 1195. https://doi.org/10.3390/antibiotics11091195






