The Distribution of Mobile Colistin-Resistant Genes, Carbapenemase-Encoding Genes, and Fluoroquinolone-Resistant Genes in Escherichia coli Isolated from Natural Water Sources in Upper Northeast Thailand
Abstract
:1. Introduction
2. Results
2.1. Microbiological Quality
2.2. Distribution of the Antibiotic Resistance Gene Profiles of the E. coli Isolates
2.3. Antibiotic Resistance Phenotypes of the E. coli Isolates
2.4. Phylogenetic Group of the E. coli Isolates
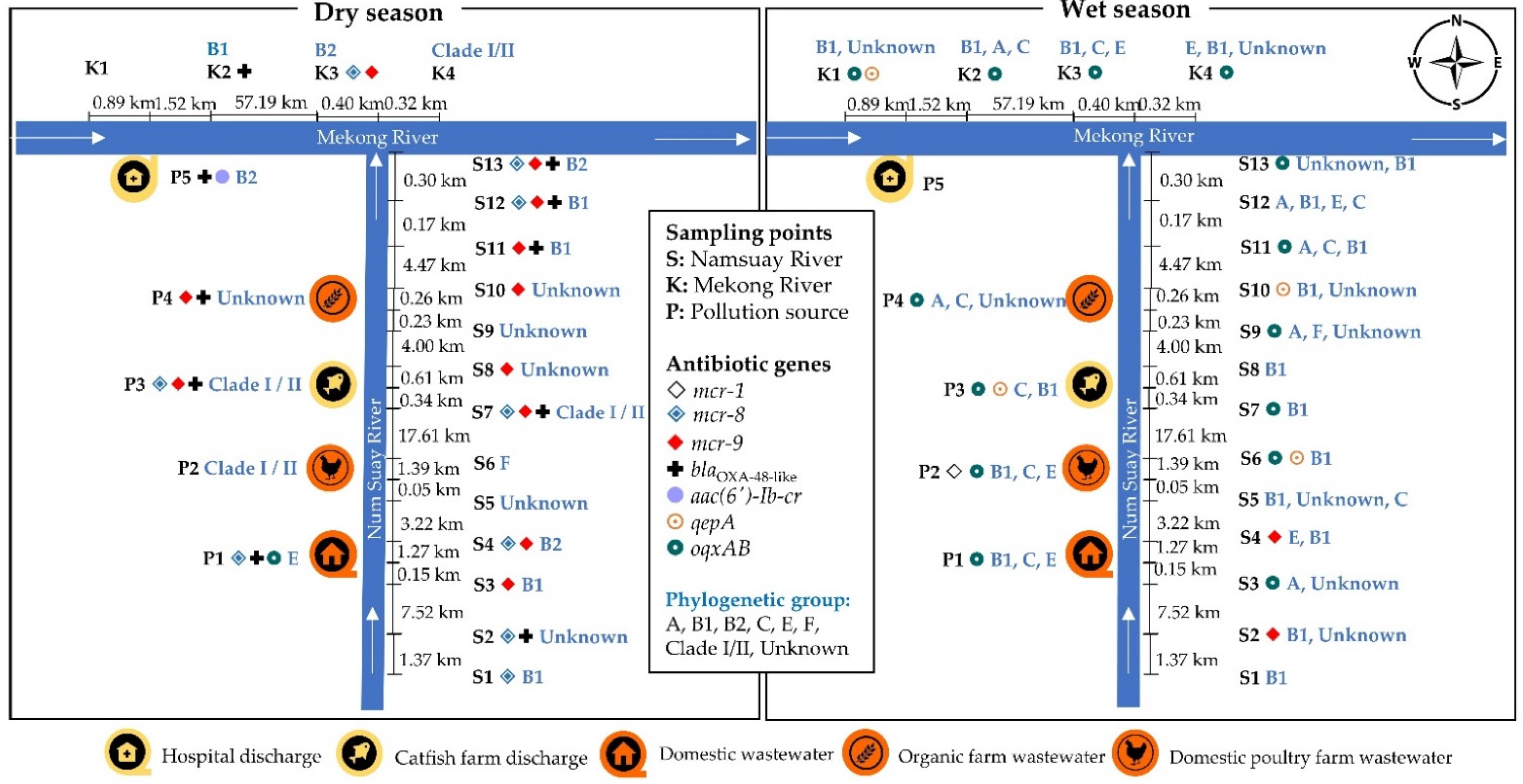
2.5. Location of the Antibiotic-Resistant E. coli Isolates
2.6. Associations of Phenotypic and Genotypic Antibiotic Resistance in E. coli with the Phylogenetic Groups and Seasons
3. Discussion
4. Materials and Methods
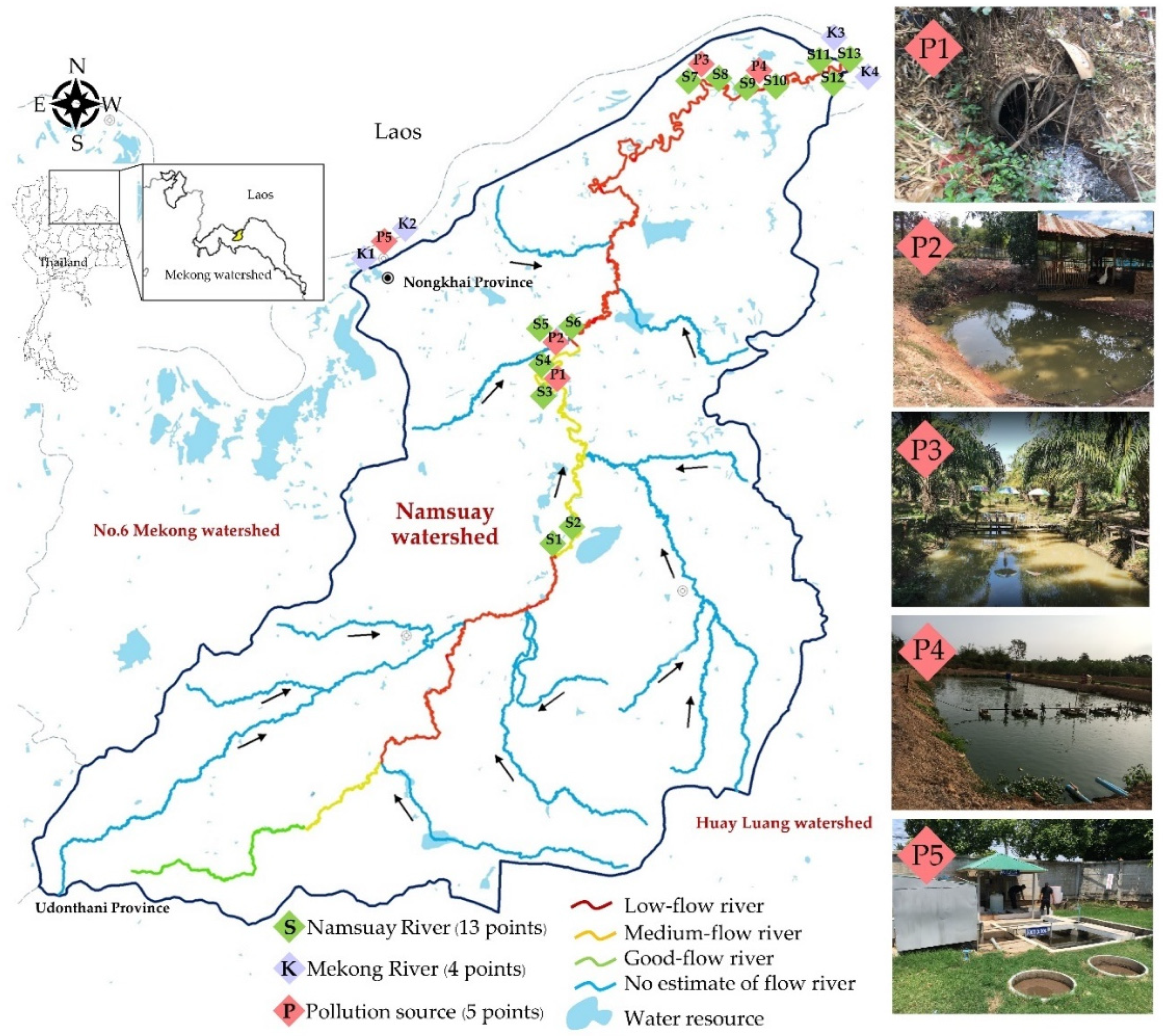
4.1. Study Area and Sampling Sites
4.2. Microbiological Quality Assessment
4.3. Detection of the Antibioic Resistance Genes in the E. coli Isolates
4.4. Antibiotic Susceptibility Profiles of the E. coli Isolates
4.5. E. coli Phylogenetic Group
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2020; World Health Organization: Geneva, Switzerland, 2020; pp. 109–115. [Google Scholar]
- Sapkota, A.R. Other Water Pollutants: Antibiotic-Resistant Bacteria. In Water and Sanitation-Related Diseases and the Environment; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2011; pp. 329–335. [Google Scholar]
- Anthony, A.A.; Adekunle, C.F.; Thor, A.S. Residual antibiotics, antibiotic resistant superbugs and antibiotic resistance genes in surface water catchments: Public health impact. Phys. Chem. Earth 2018, 105, 177–183. [Google Scholar] [CrossRef]
- Association, A.P.H. Microbiological examination. In Standard Methods for the Examination of Water and Wastewater, 23rd ed.; Baird, R.B., Eaton, A.D., Rice, E.W., Eds.; American Public Health Association: Washington, DC, USA, 2017; pp. 1–99. [Google Scholar]
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bungau, S.; Tit, D.M.; Behl, T.; Aleya, L.; Zaha, D.C. Aspects of excessive antibiotic consumption and environmental influences correlated with the occurrence of resistance to antimicrobial agents. Curr. Opin. Environ. Sci. 2021, 19, 100224. [Google Scholar] [CrossRef]
- Zarei-Baygi, A.; Smith, A.L. Intracellular versus extracellular antibiotic resistance genes in the environment: Prevalence, horizontal transfer, and mitigation strategies. Bioresour. Technol. 2021, 319, 124181. [Google Scholar] [CrossRef]
- Elbediwi, M.; Li, Y.; Paudyal, N.; Pan, H.; Li, X.; Xie, S.; Rajkovic, A.; Feng, Y.; Fang, W.; Rankin, S.C.; et al. Global burden of colistin-resistant bacteria: Mobilized colistin resistance genes study (1980–2018). Microorganisms 2019, 7, 461. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Dai, X.; Zeng, J.; Gao, Y.; Zhang, Z.; Zhang, L. Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Sci. Rep. 2020, 10, 8113. [Google Scholar] [CrossRef] [PubMed]
- Hooban, B.; Joyce, A.; Fitzhenry, K.; Chique, C.; Morris, D. The role of the natural aquatic environment in the dissemination of extended spectrum beta-lactamase and carbapenemase encoding genes: A scoping review. Water Res. 2020, 180, 115880. [Google Scholar] [CrossRef]
- Yassine, I.; Rafei, R.; Osman, M.; Mallat, H.; Dabboussi, F.; Hamze, M. Plasmid-mediated quinolone resistance: Mechanisms, detection, and epidemiology in the Arab countries. Infect. Genet. Evol. 2019, 76, 104020. [Google Scholar] [CrossRef]
- Varela, A.R.; Macedo, G.N.; Nunes, O.C.; Manaia, C.M. Genetic characterization of fluoroquinolone resistant Escherichia coli from urban streams and municipal and hospital effluents. FEMS Microbiol. Ecol. 2015, 91, fiv015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, D.W.; Bergeron, G.; Bourassa, M.W.; Dickson, J.; Gomes, F.; Howe, A.; Kahn, L.H.; Morley, P.S.; Scott, H.M.; Simjee, S.; et al. Complexities in understanding antimicrobial resistance across domesticated animal, human, and environmental systems. Ann. N. Y. Acad. Sci. 2019, 1441, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.C.; Feng, W.Q.; Han, Y.; Zheng, J.; Chen, T.; Wei, Y.Y.; Gillings, M.; Zhu, Y.G.; Chen, H. Prevalence and transmission of antibiotic resistance and microbiota between humans and water environments. Environ. Int. 2018, 121, 1155–1161. [Google Scholar] [CrossRef]
- Dawangpa, A.; Lertwatcharasarakul, P.; Ramasoota, P.; Boonsoongnern, A.; Ratanavanichrojn, N.; Sanguankiat, A.; Phatthanakunanan, S.; Tulayakul, P. Genotypic and phenotypic situation of antimicrobial drug resistance of Escherichia coli in water and manure between biogas and non-biogas swine farms in central Thailand. J. Environ. Manag. 2021, 279, 111659. [Google Scholar] [CrossRef]
- Tansawai, U.; Walsh, T.R.; Niumsup, P.R. Extended spectrum ß-lactamase-producing Escherichia coli among backyard poultry farms, farmers, and environments in Thailand. Poult. Sci. 2019, 98, 2622–2631. [Google Scholar] [CrossRef]
- Thamlikitkul, V.; Tiengrim, S.; Thamthaweechok, N.; Buranapakdee, P.; Chiemchaisri, W. Contamination by Antibiotic-Resistant Bacteria in Selected Environments in Thailand. Int. J. Environ. Res. Public Health 2019, 16, 3753. [Google Scholar] [CrossRef] [Green Version]
- Amarasiri, M.; Sano, D.; Suzuki, S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: Current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 2020, 50, 2016–2059. [Google Scholar] [CrossRef]
- ResistanceMap: Antibiotic Resistance. Available online: https://resistancemap.cddep.org/AntibioticResistance.php (accessed on 12 September 2022).
- Kenyon, C. Positive Association between the Use of Quinolones in Food Animals and the Prevalence of Fluoroquinolone Resistance in E. coli and K. pneumoniae, A. baumannii and P. aeruginosa: A Global Ecological Analysis. Antibiotics 2021, 10, 1193. [Google Scholar] [CrossRef]
- Hooban, B.; Fitzhenry, K.; Cahill, N.; Joyce, A.; O’ Connor, L.; Bray, J.E.; Brisse, S.; Passet, V.; Abbas Syed, R.; Cormican, M.; et al. A Point Prevalence Survey of Antibiotic Resistance in the Irish Environment, 2018–2019. Environ. Int. 2021, 152, 106466. [Google Scholar] [CrossRef]
- Huijbers, P.M.C.; Larsson, D.G.J.; Flach, C.-F. Surveillance of antibiotic resistant Escherichia coli in human populations through urban wastewater in ten European countries. Environ. Pollut. 2020, 261, 114200. [Google Scholar] [CrossRef]
- Kunhikannan, S.; Thomas, C.J.; Franks, A.E.; Mahadevaiah, S.; Kumar, S.; Petrovski, S. Environmental hotspots for antibiotic resistance genes. Microbiologyopen 2021, 10, e1197. [Google Scholar] [CrossRef]
- Cheng, P.; Yang, Y.; Li, F.; Li, X.; Liu, H.; Fazilani, S.A.; Guo, W.; Xu, G.; Zhang, X. The prevalence and mechanism of fluoroquinolone resistance in Escherichia coli isolated from swine farms in China. BMC Vet. Res. 2020, 16, 258. [Google Scholar] [CrossRef]
- Lekagul, A.; Tangcharoensathien, V.; Liverani, M.; Mills, A.; Rushton, J.; Yeung, S. Understanding antibiotic use for pig farming in Thailand: A qualitative study. Antimicrob. Resist. Infect. Control 2021, 10, 3. [Google Scholar] [CrossRef]
- Zhang, X.; Zhi, X.; Chen, L.; Shen, Z. Spatiotemporal variability and key influencing factors of river fecal coliform within a typical complex watershed. Water Res. 2020, 178, 115835. [Google Scholar] [CrossRef]
- Assawatheptawee, K.; Treebupachatsakul, P.; Luangtongkum, T.; Niumsup, P.R. Risk Factors for Community-Acquired Urinary Tract Infections Caused by Multidrug-Resistant Enterobacterales in Thailand. Antibiotics 2022, 11, 1039. [Google Scholar] [CrossRef]
- Khanawapee, A.; Kerdsin, A.; Chopjitt, P.; Boueroy, P.; Hatrongjit, R.; Akeda, Y.; Tomono, K.; Nuanualsuwan, S.; Hamada, S. Distribution and Molecular Characterization of Escherichia coli Harboring mcr Genes Isolated from Slaughtered Pigs in Thailand. Microb. Drug Resist. 2021, 27, 971–979. [Google Scholar] [CrossRef]
- Yasmin, S.; Karim, A.M.; Lee, S.H.; Zahra, R. Temporal Variation of Meropenem Resistance in E. coli Isolated from Sewage Water in Islamabad, Pakistan. Antibiotics 2022, 11, 635. [Google Scholar] [CrossRef]
- Asaduzzaman, M.; Rousham, E.; Unicomb, L.; Islam, M.R.; Amin, M.B.; Rahman, M.; Hossain, M.I.; Mahmud, Z.H.; Szegner, M.; Wood, P.; et al. Spatiotemporal distribution of antimicrobial resistant organisms in different water environments in urban and rural settings of Bangladesh. Sci. Total Environ. 2022, 831, 154890. [Google Scholar] [CrossRef]
- Duriez, P.; Clermont, O.; Bonacorsi, S.; Bingen, E.; Chaventré, A.; Elion, J.; Picard, B.; Denamur, E. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 2001, 147, 1671–1676. [Google Scholar] [CrossRef] [Green Version]
- Mojaz-Dalfardi, N.; Kalantar-Neyestanaki, D.; Hashemizadeh, Z.; Mansouri, S. Comparison of virulence genes and phylogenetic groups of Escherichia coli isolates from urinary tract infections and normal fecal flora. Gene Rep. 2020, 20, 100709. [Google Scholar] [CrossRef]
- Redha, M.A.; Al Sweih, N.; Albert, M.J. Virulence and phylogenetic groups of Escherichia coli cultured from raw sewage in Kuwait. Gut Pathog. 2022, 14, 18. [Google Scholar] [CrossRef]
- Molina, F.; López-Acedo, E.; Tabla, R.; Roa, I.; Gómez, A.; Rebollo, J.E. Improved detection of Escherichia coli and coliform bacteria by multiplex PCR. BMC Biotechnol. 2015, 15, 48. [Google Scholar] [CrossRef]
- Hatrongjit, R.; Kerdsin, A.; Akeda, Y.; Hamada, S. Detection of plasmid-mediated colistin-resistant and carbapenem-resistant genes by multiplex PCR. MethodsX 2018, 5, 532–536. [Google Scholar] [CrossRef]
- Ciesielczuk, H.; Hornsey, M.; Choi, V.; Woodford, N.; Wareham, D.W. Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone-resistance determinants. J. Med. Microbiol. 2013, 62, 1823–1827. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; Clinical Laboratory Standard Institute: Wayne, PA, USA, 2020; pp. 1–41. [Google Scholar]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Yates, F. Tests of Significance for 2 × 2 Contingency Tables. J. R. Stat. Soc. Ser. A Gen. 1984, 147, 426–449. [Google Scholar] [CrossRef]
| Season | Sample Type | Number of Samples | Total Coliform Bacteria (MPN/100 mL, Number of Samples) | Faecal Coliform Bacteria (MPN/100 mL, Number of Samples) | ||
|---|---|---|---|---|---|---|
| Maximum (N) | Minimum (N) | Maximum (N) | Minimum (N) | |||
| Dry | Surface water | 17 | >1600.00, (1) | 22.00, (1) | 350.00, (1) | 2.00, (2) |
| Wastewater | 3 | >1600.00, (3) | - | >1600.00, (2) | 350.00, (1) | |
| Discharge water | 2 | >1600.00, (2) | - | >1600.00, (1) | 79.00, (1) | |
| Wet | Surface water | 17 | >1600.00, (7) | 41.00, (1) | >1600.00, (4) | 79.00, (4) |
| Wastewater | 3 | >1600.00, (2) | 350.00, (1) | >1600.00, (3) | - | |
| Discharge water | 2 | >1600.00, (1) | 79.00, (1) | >1600.00, (1) | 0.00, (1) | |
| Genotypic and Phenotypic Patterns | Dry Season (n = 21) | Wet Season (n = 92) | Total (n = 113) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Surface Water (n = 16) | Wastewater (n = 3) | Discharge Water (n = 2) | Surface Water (n = 73) | Wastewater (n = 14) | Discharge Water (n = 5) | Surface Water (n = 89) | Wastewater (n = 17) | Discharge Water (n = 7) | |
| ARG | 13 (81.25) | 2 (66.67) | 2 (100.00) | 27 (36.99) | 6 (42.86) | 2 (40.00) | 40 (44.94) | 8 (47.06) | 4 (57.14) |
| mcr-8 | 1 (6.25) | 1 (1.12) | |||||||
| mcr-9 | 4 (25.00) | 4 (5.48) | 8 (8.99) | ||||||
| oqxAB | 20 (27.4) | 5 (35.71) | 1 (20.00) | 20 (22.47) | 5 (29.41) | 1 (14.29) | |||
| qepA | 1 (1.37) | 1 (20.00) | 1 (1.12) | 1 (14.29) | |||||
| blaoxa-48-like | 1 (6.25) | 1 (1.12) | |||||||
| mcr-8,mcr-9 | 2 (12.5) | 2 (2.25) | |||||||
| oqxAB, qepA | 2 (2.74) | 2 (2.25) | |||||||
| mcr-1, oqxAB | 1 (7.14) | 1 (5.88) | |||||||
| mcr-8,blaoxa-48-like | 1 (6.25) | 1 (1.12) | |||||||
| mcr-9, blaoxa-48-like | 1 (6.25) | 1 (33.33) | 1 (1.12) | 1 (5.88) | |||||
| aac(6′)-Ib-cr, blaoxa-48-like | 1 (50.00) | 1 (14.29) | |||||||
| mcr-8,mcr-9, blaoxa-48-like | 3 (18.75) | 1 (50.00) | 3 (3.37) | 1 (14.29) | |||||
| mcr-9, oqxAB, blaoxa-48-like | 1 (33.33) | 1 (5.88) | |||||||
| Not detect ARGs | 3 (18.75) | 1 (33.33) | 46 (63.01) | 8 (57.14) | 3 (60) | 49 (55.06) | 9 (52.94) | 3 (42.86) | |
| AR | 10 (62.50) | 3 (100.00) | 2 (100.00) | 17 (23.29) | 6 (42.86) | 3 (60.00) | 27 (30.34) | 9 (52.94) | 5 (71.43) |
| CIP | 3 (18.75) | 1 (33.33) | 1 (50.00) | 12 (16.44) | 4 (28.57) | 3 (60.00) | 15 (16.85) | 5 (29.41) | 4 (57.14) |
| COL | 5 (31.25) | 1 (33.33) | 5 (5.62) | 1 (5.88) | |||||
| IMP | 2 (2.74) | 2 (2.25) | |||||||
| CTX + COL | 1 (6.25) | 1 (1.12) | |||||||
| IMP + COL | 1 (6.25) | 1 (1.12) | |||||||
| CIP + COL | 1 (33.33) | 1 (5.88) | |||||||
| CIP + CTX | 1 (1.37) | 1 (1.12) | |||||||
| IMP + CTX | 1 (1.37) | 1 (1.12) | |||||||
| CIP + CTZ + CTX | 1 (50.00) | 1 (1.37) | 2 (14.29) | 1 (1.12) | 2 (11.76) | 1 (14.29) | |||
| Susceptibility | 6 (37.5) | 56 (76.71) | 8 (57.14) | 2 (40) | 62 (69.66) | 8 (47.06) | 2 (28.57) | ||
| Antibiotic Resistance Gene Patterns | Fluoroquinolone | Carbapenem | 3rd Generation of Cephalosporins | Polymyxin | |
|---|---|---|---|---|---|
| Ciprofloxacin | Imipenem | Ceftazidime | Cefotaxime | Colistin | |
| ARGs not detected | 15 (13.27) | 2 (1.77) | 2 (1.77) | 4 (3.54) | |
| mcr-8 | 1 (0.88) | ||||
| mcr-9 | 3 (2.65) | ||||
| oqxAB | 8 (7.08) | 1 (0.88) | 1 (0.88) | ||
| qepA | 1 (0.88) | 1 (0.88) | |||
| blaoxa-48-like | 1 (0.88) | ||||
| mcr-8 + mcr-9 | 1 (0.88) | ||||
| oqxAB + qepA | 1 (0.88) | ||||
| mcr-1+ oqxAB | 1 (0.88) | ||||
| mcr-8+ blaoxa-48-like | |||||
| mcr-9+ blaoxa-48-like | 1 (0.88) | ||||
| aac(6′)-Ib-cr + blaoxa-48-like | 1 (0.88) | 1 (0.88) | 1 (0.88) | ||
| mcr-8 + mcr-9 + blaoxa-48-like | 1 (0.88) | 1 (0.88) | 1 (0.88) | 2 (1.77) | |
| mcr-9 + oqxAB + blaoxa-48-like | 1 (0.88) | 1 (0.88) | |||
| Total | 30 (26.55) | 4 (3.54) | 4 (3.54) | 7 (6.19) | 9 (7.96) |
| Profile | A | B1 | B2 | C | E | F | Clade I or II | Unknown | Total |
|---|---|---|---|---|---|---|---|---|---|
| Number of isolates | 11 (9.73) | 48 (42.48) | 4 (3.54) | 12 (10.62) | 4 (3.54) | 11 (9.73) | 2 (1.77) | 21 (18.58) | 113 (100.00) |
| Antibiotic resistance genes | 5 (4.42) | 18 (15.93) | 4 (3.54) | 5 (4.42) | 2 (1.77) | 8 (7.08) | 1 (0.88) | 9 (7.96) | 52 (46.02) |
| mcr-8 | 1 (0.88) | 1 (0.88) | |||||||
| mcr-9 | 3 (2.65) | 2 (1.77) | 3 (2.65) | 8 (7.08) | |||||
| oqxAB | 4 (3.54) | 9 (7.96) | 4 (3.54) | 4 (3.54) | 1 (0.88) | 4 (3.54) | 26 (23.01) | ||
| qepA | 1 (0.88) | 1 (0.88) | 2 (1.77) | ||||||
| blaoxa-48-like | 1 (0.88) | 1 (0.88) | |||||||
| mcr-8 + mcr-9 | 2 (1.77) | 2 (1.77) | |||||||
| oqxAB + qepA | 2 (1.77) | 2 (1.77) | |||||||
| mcr-1 + oqxAB | 1 (0.88) | 1 (0.88) | |||||||
| mcr-8 + blaoxa-48-like | 1 (0.88) | 1 (0.88) | |||||||
| mcr-9 + blaoxa-48-like | 1 (0.88) | 1 (0.88) | 2 (1.77) | ||||||
| aac(6′)-Ib-cr + blaoxa-48-like | 1 (0.88) | 1 (0.88) | |||||||
| mcr-8 + mcr-9 + blaoxa-48-like | 1 (0.88) | 1 (0.88) | 2 (1.77) | 4 (3.54) | |||||
| mcr-9 + oqxAB + blaoxa-48-like | 1 (0.88) | 1 (0.88) | |||||||
| Undetectable ARGs | 6 (5.31) | 30 (26.55) | 7 (6.19) | 3 (2.65) | 1 (0.88) | 2 (1.77) | 12 (10.62) | 61 (53.98) | |
| Antibiotic resistance | 3 (2.65) | 8 (7.08) | 3 (2.65) | 6 (5.31) | 4 (3.54) | 6 (5.31) | 1 (0.88) | 10 (8.85) | 41 (36.28) |
| CIP | 1 (0.88) | 4 (3.54) | 6 (5.31) | 3 (2.65) | 5 (4.42) | 1 (0.88) | 4 (3.54) | 24 (21.24) | |
| COL | 2 (1.77) | 1 (0.88) | 3 (2.65) | 6 (5.31) | |||||
| IMP | 2 (1.77) | 2 (1.77) | |||||||
| CTX + COL | 1 (0.88) | 1 (0.88) | |||||||
| IMP + COL | 1 (0.88) | 1 (0.88) | |||||||
| CIP + COL | 1 (0.88) | 1 (0.88) | |||||||
| CIP + CTX | 1 (0.88) | 1 (0.88) | |||||||
| IMP + CTX | 1 (0.88) | 1 (0.88) | |||||||
| CIP + CTZ + CTX | 1 (0.88) | 1 (0.88) | 2 (1.77) | 4 (3.54) | |||||
| Susceptibility | 8 (7.08) | 40 (35.4) | 1 (0.88) | 6 (5.31) | 5 (4.42) | 1 (0.88) | 11 (9.73) | 72 (63.72) |
| Phenotypic and Genotypic Antibiotic Resistance in E. coli | B1 (n = 48) | Non-B1 (n = 65) | p-Values |
|---|---|---|---|
| Antibiotic resistance | 8 | 33 | <0.001 |
| Antibiotic susceptibility | 40 | 32 | |
| Antibiotic resistance genes | 18 | 34 | 0.131 |
| Non-antibiotic resistance genes | 30 | 31 |
| Phenotypic and Genotypic Antibiotic Resistance in E. coli | Dry (n = 21) | Wet (n = 92) | p-Values |
|---|---|---|---|
| Antibiotic resistance | 15 | 26 | <0.001 |
| Antibiotic susceptibility | 6 | 66 | |
| Antibiotic resistance genes | 17 | 35 | <0.001 |
| Non-antibiotic resistance genes | 4 | 57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabut, P.; Yongyod, R.; Ungcharoen, R.; Kerdsin, A. The Distribution of Mobile Colistin-Resistant Genes, Carbapenemase-Encoding Genes, and Fluoroquinolone-Resistant Genes in Escherichia coli Isolated from Natural Water Sources in Upper Northeast Thailand. Antibiotics 2022, 11, 1760. https://doi.org/10.3390/antibiotics11121760
Tabut P, Yongyod R, Ungcharoen R, Kerdsin A. The Distribution of Mobile Colistin-Resistant Genes, Carbapenemase-Encoding Genes, and Fluoroquinolone-Resistant Genes in Escherichia coli Isolated from Natural Water Sources in Upper Northeast Thailand. Antibiotics. 2022; 11(12):1760. https://doi.org/10.3390/antibiotics11121760
Chicago/Turabian StyleTabut, Pongthep, Rapeepan Yongyod, Ratchadaporn Ungcharoen, and Anusak Kerdsin. 2022. "The Distribution of Mobile Colistin-Resistant Genes, Carbapenemase-Encoding Genes, and Fluoroquinolone-Resistant Genes in Escherichia coli Isolated from Natural Water Sources in Upper Northeast Thailand" Antibiotics 11, no. 12: 1760. https://doi.org/10.3390/antibiotics11121760






