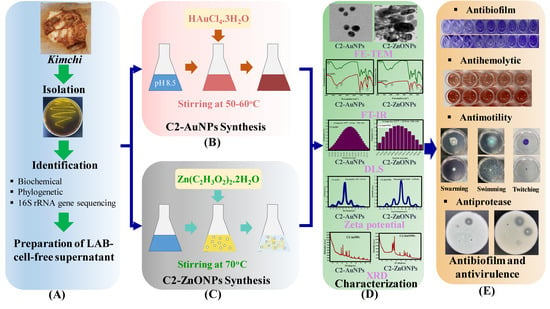
Antibiofilm and Antivirulence Activities of Gold and Zinc Oxide Nanoparticles Synthesized from Kimchi-Isolated Leuconostoc sp. Strain C2
Abstract
:1. Introduction
2. Results
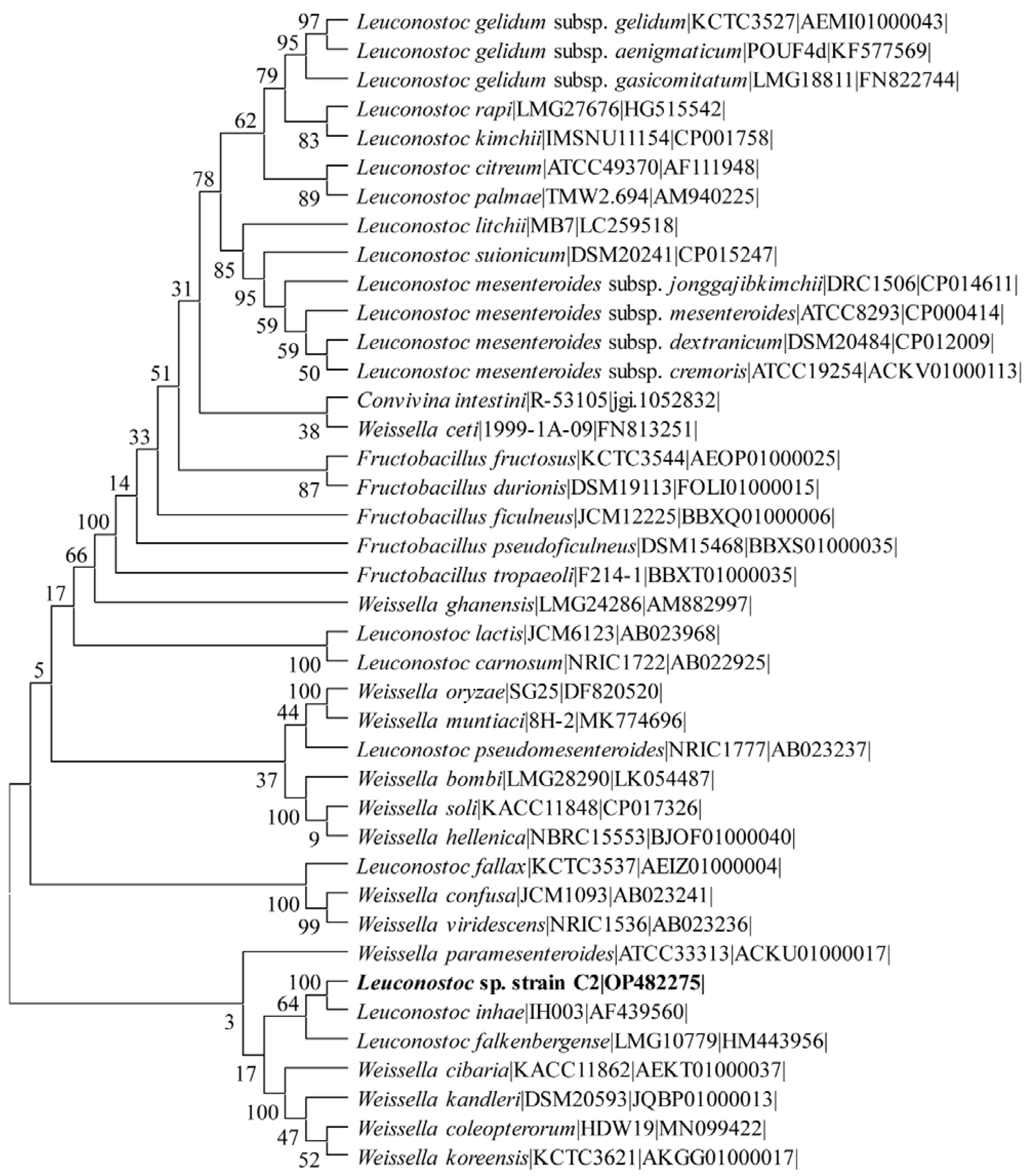
2.1. Isolation and Identification of the Lactic Acid Bacteria
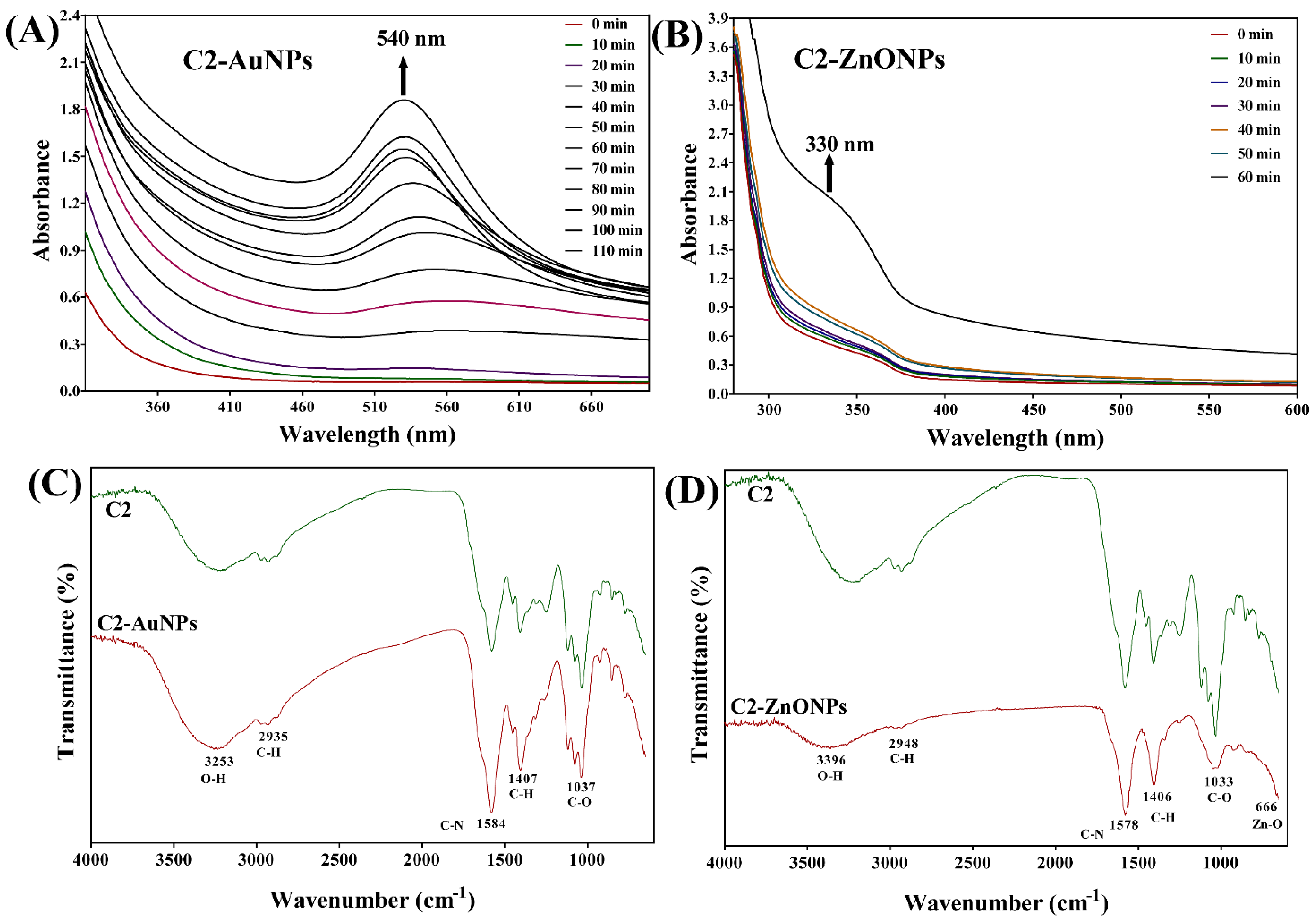
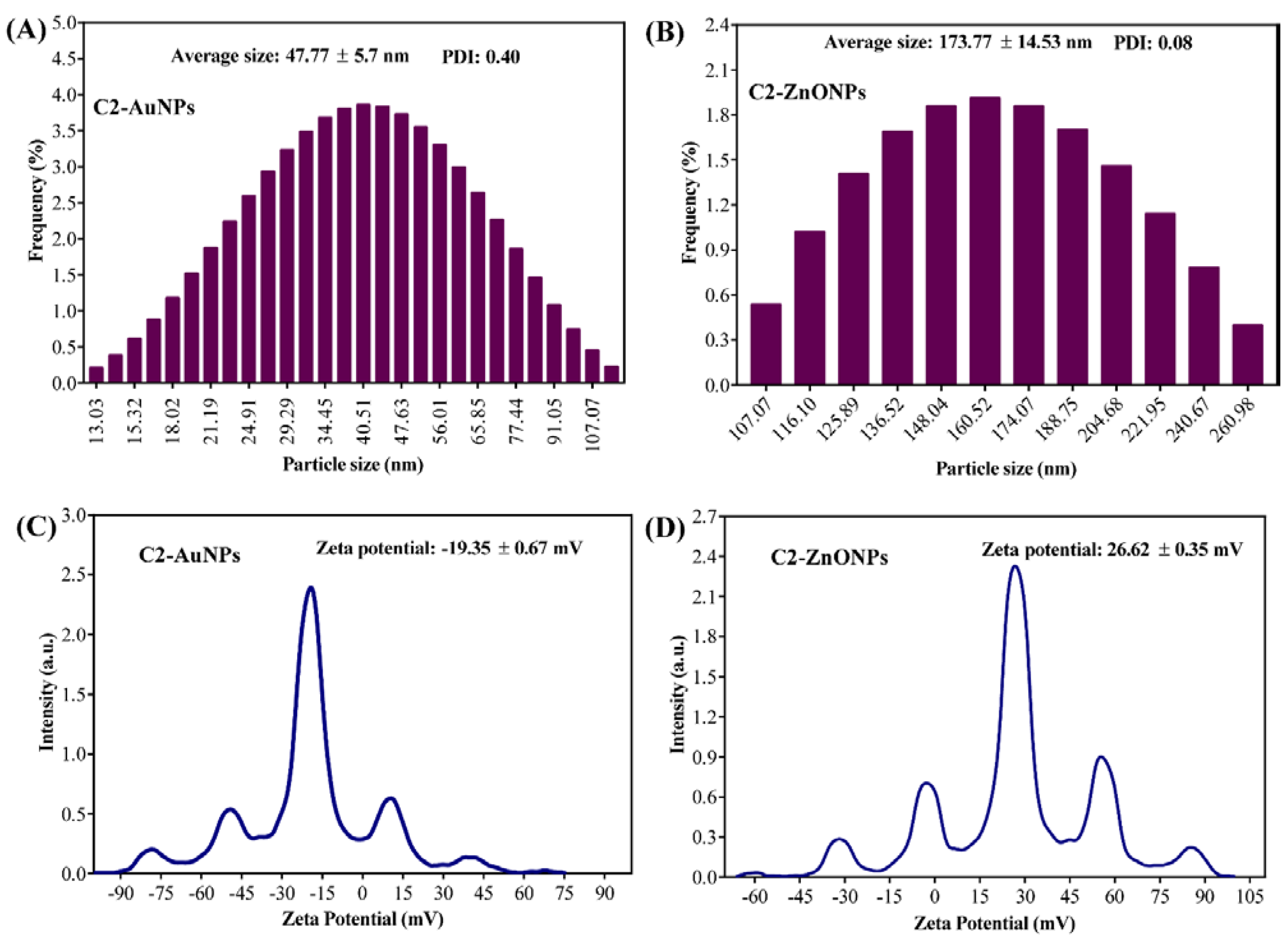
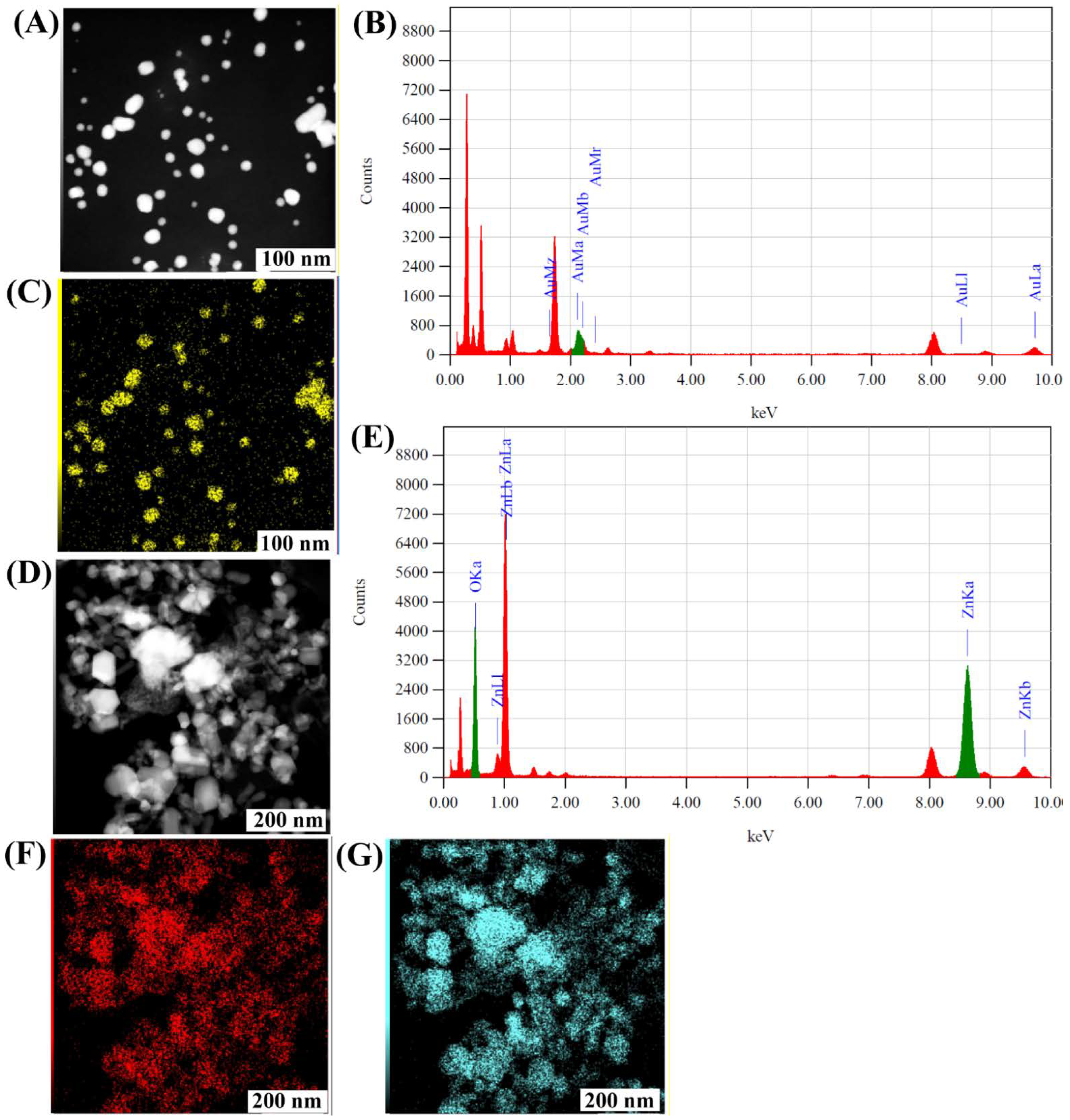
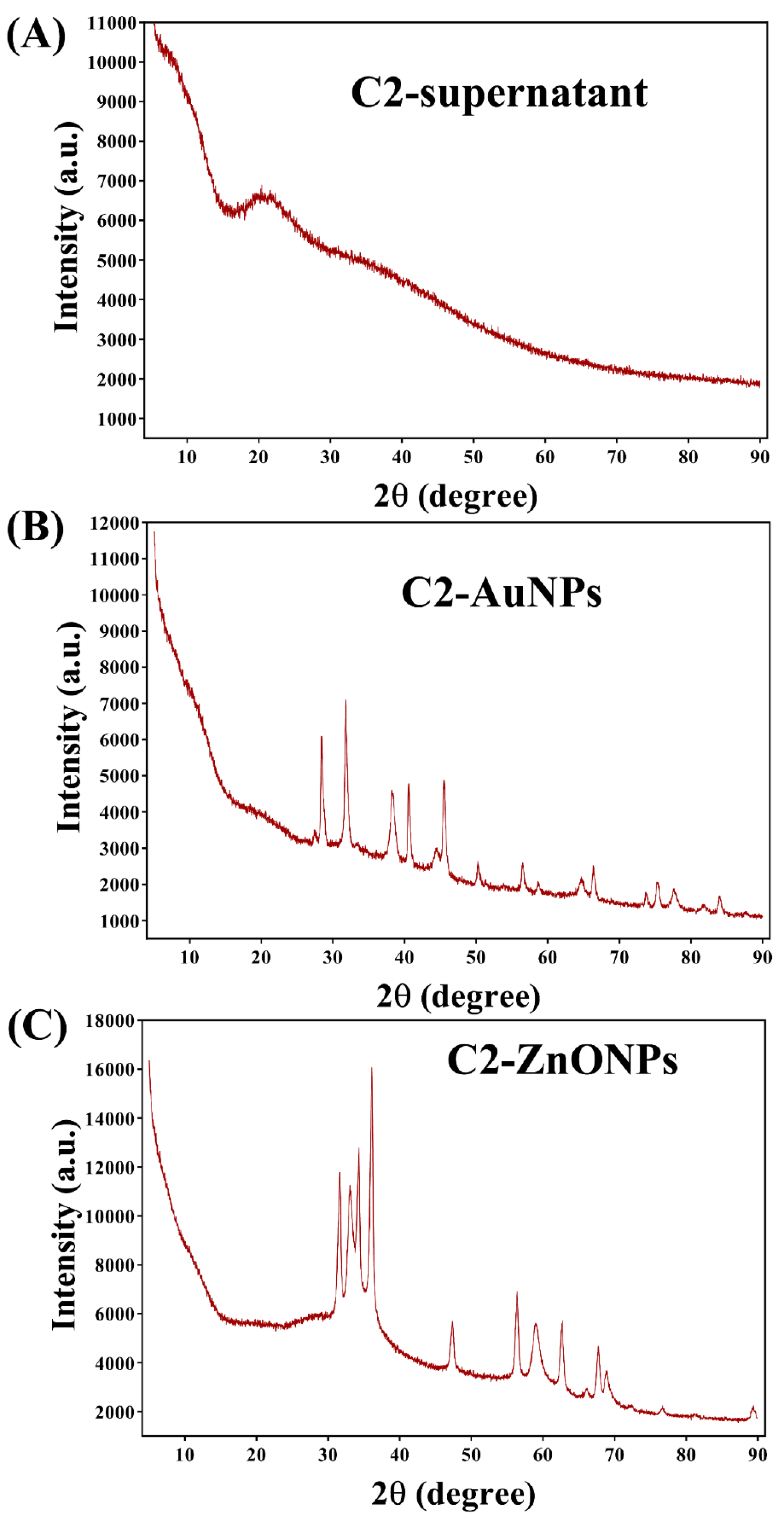
2.2. Biosynthesis and Characterization of the Gold and Zinc Oxide Nanoparticles
2.3. Antimicrobial Effect of LAB Supernatant and Synthesized Nanoparticles
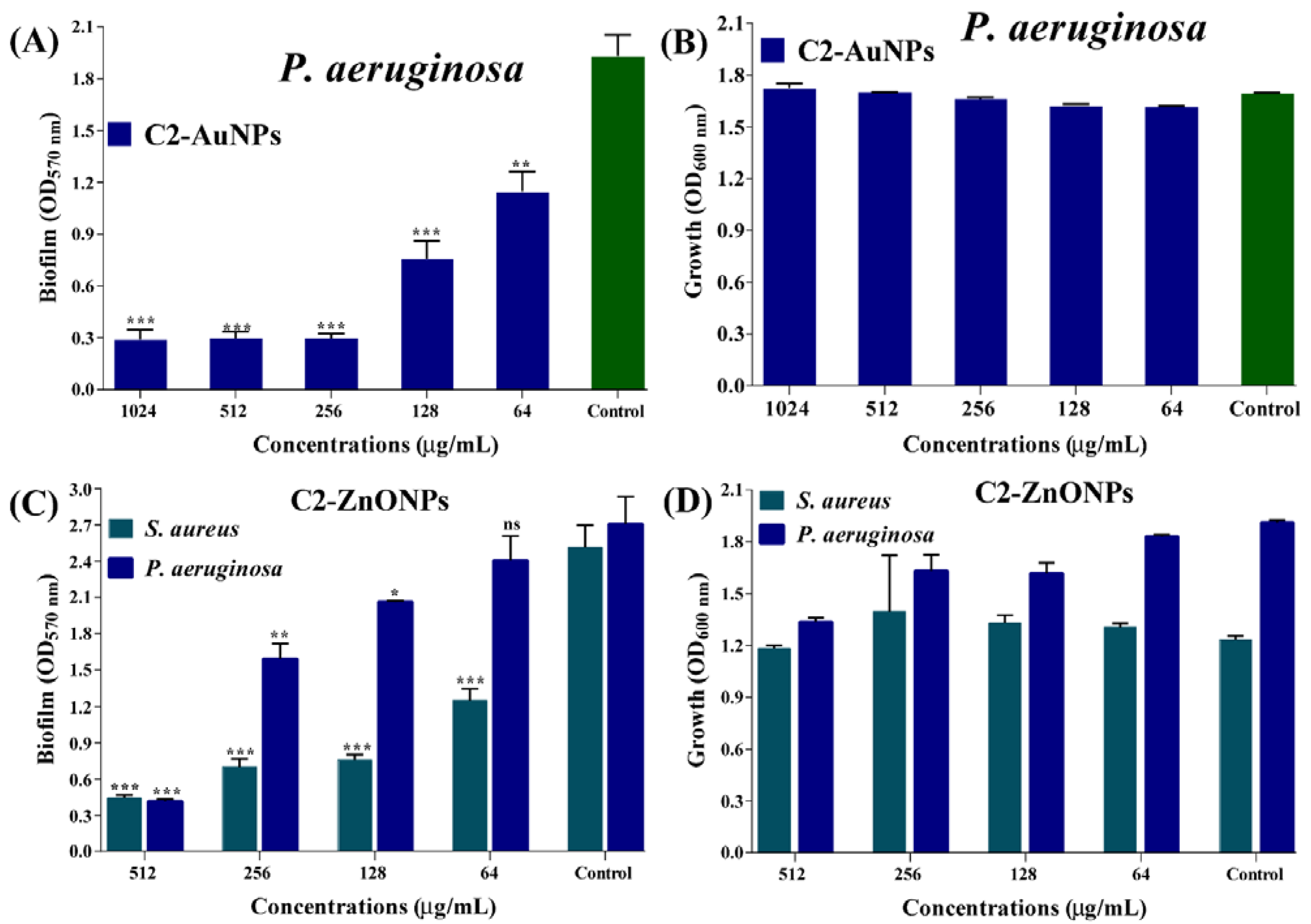
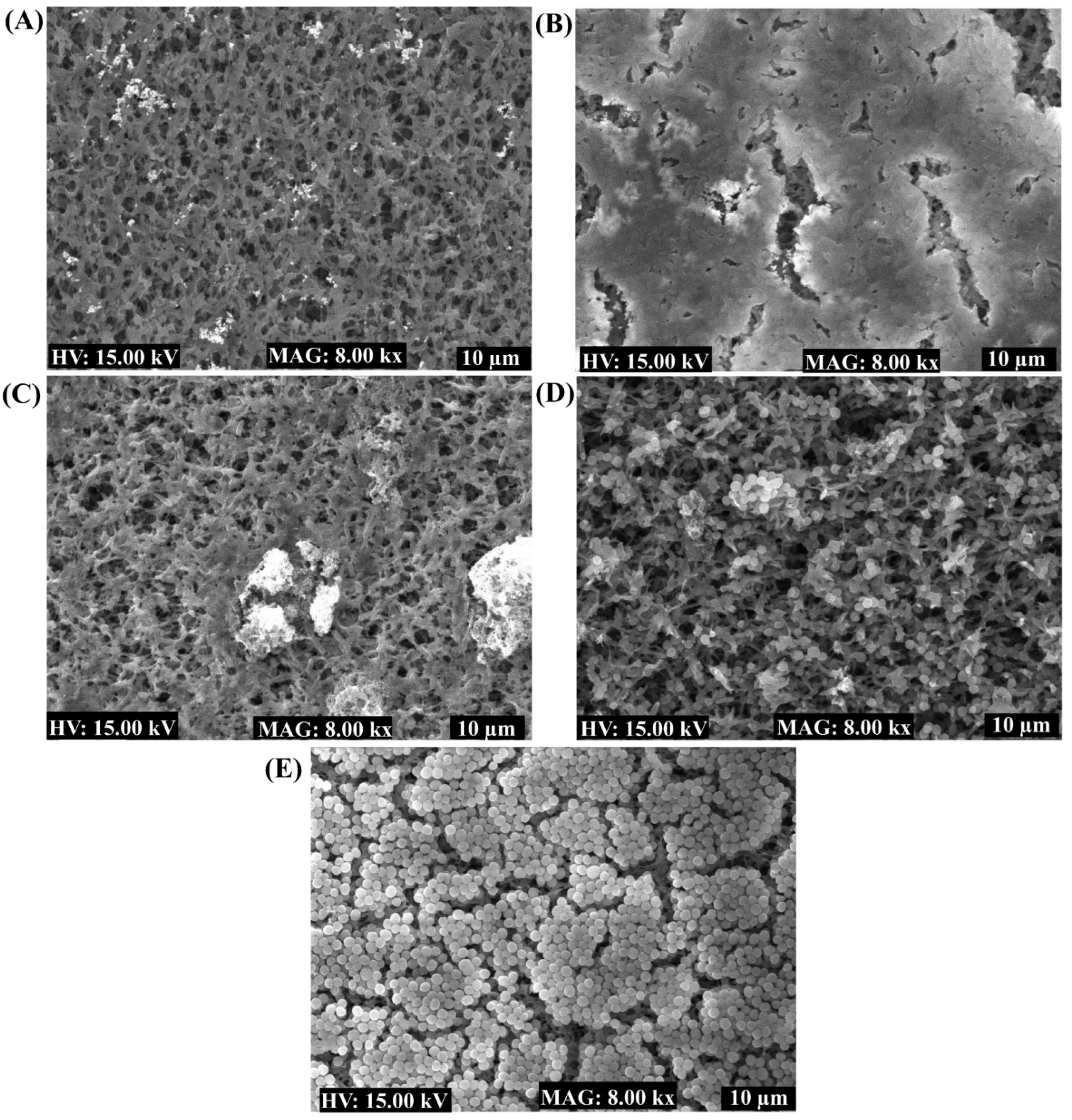
2.4. Antibiofilm Properties of the Nanoparticles toward Bacterial Pathogens
2.5. Eradication of the Established Mature Biofilm
2.6. Inhibitory Effect of NPs on P. aeruginosa Motilities
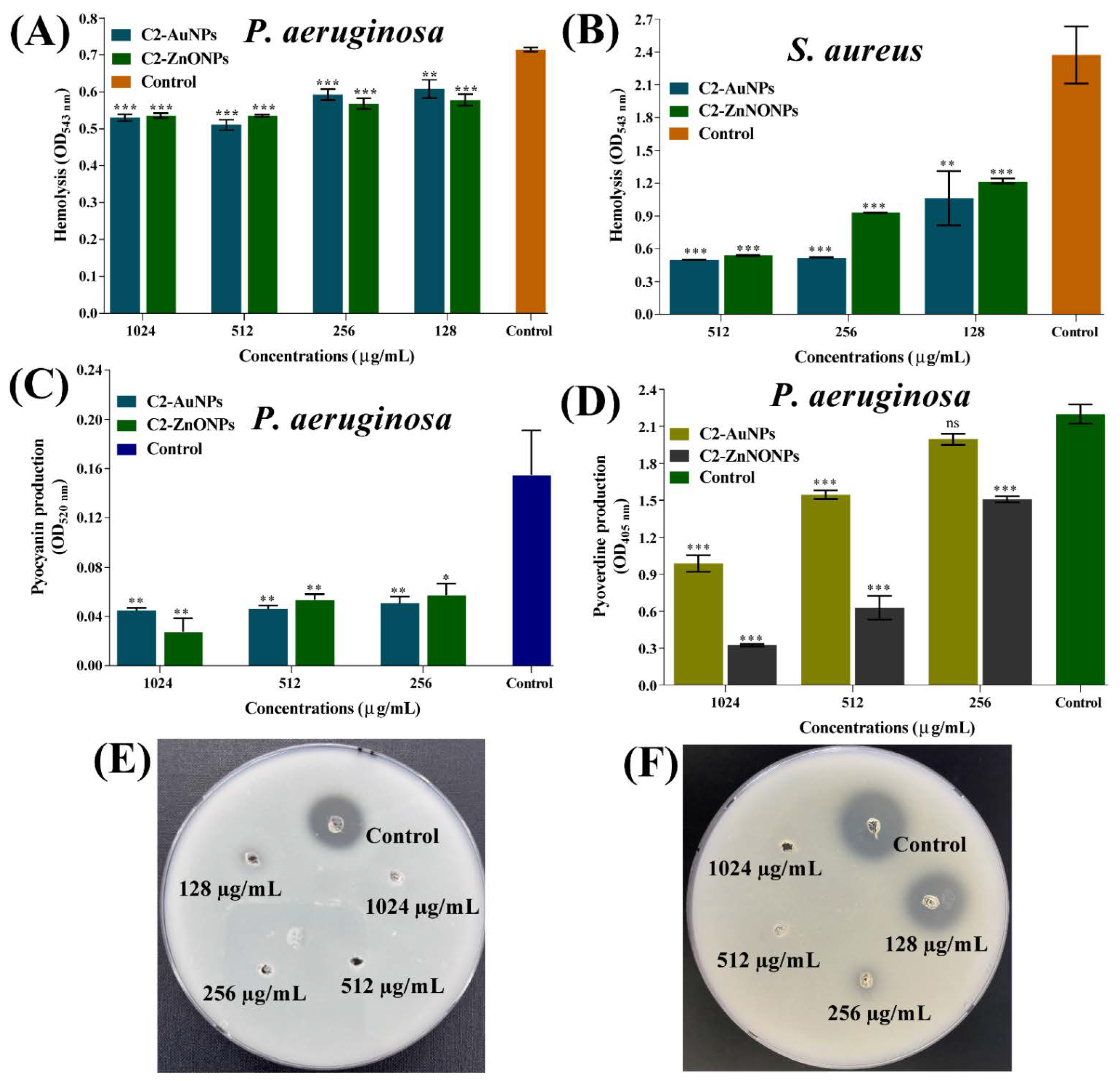
2.7. Virulence-Inhibitory Properties of the Nanoparticles
3. Discussion
4. Materials and Methods
4.1. Microbial Pathogens, Culture Media, and Growth Conditions
4.2. Isolation and Identification of LAB Strains
4.3. Preparation of LAB Cell-Free Culture Supernatant (CFCS)
4.4. Synthesis of LAB CFCS-AuNPs and ZnONPs
4.5. Metabolomic Analysis of LAB-Supernatant by GC–MS
4.6. Physiochemical Characterization of C2-AuNPs and C2-ZnONPs
4.7. Antibacterial Effects of LAB-Supernatant and Nanoparticles
4.8. Biofilm Inhibition and Eradication of Established Mature Biofilm
4.9. Examination of Biofilms Cells Using SEM
4.10. Assays for the Inhibition of Multiple Virulence Properties
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.; Wertheim, H.F.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef] [Green Version]
- Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [CrossRef]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Beatson, S.A.; Walker, M.J. Microbiology. Tracking antibiotic resistance. Science 2014, 345, 1454–1455. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. Biomed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef] [Green Version]
- de la Fuente-Núñez, C.; Reffuveille, F.; Fernández, L.; Hancock, R.E. Bacterial biofilm development as a multicellular adaptation: Antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 2013, 16, 580–589. [Google Scholar] [CrossRef]
- Mah, T.-F.C.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Khan, F.; Pham, D.T.N.; Tabassum, N.; Oloketuyi, S.F.; Kim, Y.M. Treatment strategies targeting persister cell formation in bacterial pathogens. Crit. Rev. Microbiol. 2020, 46, 665–688. [Google Scholar] [CrossRef]
- Lin, Q.; Deslouches, B.; Montelaro, R.C.; Di, Y.P. Prevention of ESKAPE pathogen biofilm formation by antimicrobial peptides WLBU2 and LL37. Int. J. Antimicrob. Agents 2018, 52, 667–672. [Google Scholar] [CrossRef]
- Jeong, G.J.; Khan, S.; Tabassum, N.; Khan, F.; Kim, Y.M. Marine-Bioinspired Nanoparticles as Potential Drugs for Multiple Biological Roles. Mar. Drugs 2022, 20, 527. [Google Scholar] [CrossRef]
- Cheng, Z.; Li, M.; Dey, R.; Chen, Y. Nanomaterials for cancer therapy: Current progress and perspectives. J. Hematol. Oncol. 2021, 14, 85. [Google Scholar] [CrossRef]
- Pandit, C.; Roy, A.; Ghotekar, S.; Khusro, A.; Islam, M.N.; Emran, T.B.; Lam, S.E.; Khandaker, M.U.; Bradley, D.A. Biological agents for synthesis of nanoparticles and their applications. J. King Saud Univ.-Sci. 2022, 34, 101869. [Google Scholar] [CrossRef]
- Singh, J.; Dutta, T.; Kim, K.-H.; Rawat, M.; Samddar, P.; Kumar, P. ‘Green’ synthesis of metals and their oxide nanoparticles: Applications for environmental remediation. J. Nanobiotechnol. 2018, 16, 84. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Jeong, G.-J.; Singh, P.; Tabassum, N.; Mijakovic, I.; Kim, Y.-M. Retrospective analysis of the key molecules involved in the green synthesis of nanoparticles. Nanoscale 2022, 14, 14824–14857. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Bai, Y.; Zha, L.; Ullah, N.; Ullah, H.; Shah, S.R.H.; Sun, H.; Zhang, C. Mechanism of the Gut Microbiota Colonization Resistance and Enteric Pathogen Infection. Front. Cell. Infect. Microbiol. 2021, 11, 716299. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kim, S.; Kim, Y.J.; Perumalsamy, H.; Lee, S.; Hwang, E.; Yi, T.H. Green synthesis of gold nanoparticles using Euphrasia officinalis leaf extract to inhibit lipopolysaccharide-induced inflammation through NF-κB and JAK/STAT pathways in RAW 264.7 macrophages. Int. J. Nanomed. 2019, 14, 2945–2959. [Google Scholar] [CrossRef] [Green Version]
- Barani, M.; Masoudi, M.; Mashreghi, M.; Makhdoumi, A.; Eshghi, H. Cell-free extract assisted synthesis of ZnO nanoparticles using aquatic bacterial strains: Biological activities and toxicological evaluation. Int. J. Pharm. 2021, 606, 120878. [Google Scholar] [CrossRef]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 2002, 292, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Driffield, K.; Miller, K.; Bostock, J.M.; O’Neill, A.J.; Chopra, I. Increased mutability of Pseudomonas aeruginosa in biofilms. J. Antimicrob. Chemother. 2008, 61, 1053–1056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirtane, A.R.; Verma, M.; Karandikar, P.; Furin, J.; Langer, R.; Traverso, G. Nanotechnology approaches for global infectious diseases. Nat. Nanotechnol. 2021, 16, 369–384. [Google Scholar] [CrossRef]
- Kechagia, M.; Basoulis, D.; Konstantopoulou, S.; Dimitriadi, D.; Gyftopoulou, K.; Skarmoutsou, N.; Fakiri, E.M. Health benefits of probiotics: A review. ISRN Nutr. 2013, 2013, 481651. [Google Scholar] [CrossRef] [Green Version]
- Khan, F.; Kang, M.G.; Jo, D.M.; Chandika, P.; Jung, W.K.; Kang, H.W.; Kim, Y.M. Phloroglucinol-Gold and -Zinc Oxide Nanoparticles: Antibiofilm and Antivirulence Activities towards Pseudomonas aeruginosa PAO1. Mar. Drugs 2021, 19, 601. [Google Scholar] [CrossRef]
- Singh, M.; Kalaivani, R.; Manikandan, S.; Sangeetha, N.; Kumaraguru, A.K. Facile green synthesis of variable metallic gold nanoparticle using Padina gymnospora, a brown marine macroalga. Appl. Nanosci. 2013, 3, 145–151. [Google Scholar] [CrossRef] [Green Version]
- He, S.; Guo, Z.; Zhang, Y.; Zhang, S.; Wang, J.; Gu, N. Biosynthesis of gold nanoparticles using the bacteria Rhodopseudomonas capsulata. Mater. Lett. 2007, 61, 3984–3987. [Google Scholar] [CrossRef]
- Składanowski, M.; Wypij, M.; Laskowski, D.; Golińska, P.; Dahm, H.; Rai, M. Silver and gold nanoparticles synthesized from Streptomyces sp. isolated from acid forest soil with special reference to its antibacterial activity against pathogens. J. Clust. Sci. 2017, 28, 59–79. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Li, Q.; Ma, X.; Tian, B.; Li, T.; Yu, J.; Dai, S.; Weng, Y.; Hua, Y. Biosynthesis of gold nanoparticles by the extreme bacterium Deinococcus radiodurans and an evaluation of their antibacterial properties. Int. J. Nanomed. 2016, 11, 5931–5944. [Google Scholar] [CrossRef]
- Al-Kordy, H.M.H.; Sabry, S.A.; Mabrouk, M.E.M. Statistical optimization of experimental parameters for extracellular synthesis of zinc oxide nanoparticles by a novel haloalaliphilic Alkalibacillus sp.W7. Sci. Rep. 2021, 11, 10924. [Google Scholar] [CrossRef] [PubMed]
- Punjabi, K.; Mehta, S.; Chavan, R.; Chitalia, V.; Deogharkar, D.; Deshpande, S. Efficiency of Biosynthesized Silver and Zinc Nanoparticles Against Multi-Drug Resistant Pathogens. Front. Microbiol. 2018, 9, 2207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohd Yusof, H.; Abdul Rahman, N.A.; Mohamad, R.; Zaidan, U.H.; Samsudin, A.A. Biosynthesis of zinc oxide nanoparticles by cell-biomass and supernatant of Lactobacillus plantarum TA4 and its antibacterial and biocompatibility properties. Sci. Rep. 2020, 10, 19996. [Google Scholar] [CrossRef] [PubMed]
- Jain, D.; Shivani; Bhojiya, A.A.; Singh, H.; Daima, H.K.; Singh, M.; Mohanty, S.R.; Stephen, B.J.; Singh, A. Microbial Fabrication of Zinc Oxide Nanoparticles and Evaluation of Their Antimicrobial and Photocatalytic Properties. Front. Chem. 2020, 8, 778. [Google Scholar] [CrossRef]
- Ziklo, N.; Bibi, M.; Salama, P. The Antimicrobial Mode of Action of Maltol and Its Synergistic Efficacy with Selected Cationic Surfactants. Cosmetics 2021, 8, 86. [Google Scholar] [CrossRef]
- Guo, L.; Wang, A.; Sun, Y.; Xu, C. Evaluation of antioxidant and immunity function of tetramethylpyrazine phosphate tablets in vivo. Molecules 2012, 17, 5412–5421. [Google Scholar] [CrossRef] [Green Version]
- Edeeva, S.E.; Kopylova, G.N.; Bakaeva, Z.V.; Samonina, G.E.; Umarova, B.A.; Guseva, A.A. Protective and therapeutic effects of glyprolines in psychoemotional stress induced by cholecystokinin-4 injection. Bull. Exp. Biol. Med. 2008, 145, 302–306. [Google Scholar] [CrossRef]
- Abinaya, M.; Vaseeharan, B.; Divya, M.; Sharmili, A.; Govindarajan, M.; Alharbi, N.S.; Kadaikunnan, S.; Khaled, J.M.; Benelli, G. Bacterial exopolysaccharide (EPS)-coated ZnO nanoparticles showed high antibiofilm activity and larvicidal toxicity against malaria and Zika virus vectors. J. Trace Elem. Med. Biol. 2018, 45, 93–103. [Google Scholar] [CrossRef]
- Khan, F.; Manivasagan, P.; Lee, J.W.; Pham, D.T.N.; Oh, J.; Kim, Y.M. Fucoidan-Stabilized Gold Nanoparticle-Mediated Biofilm Inhibition, Attenuation of Virulence and Motility Properties in Pseudomonas aeruginosa PAO1. Mar. Drugs 2019, 17, 208. [Google Scholar] [CrossRef] [Green Version]
- Khatoon, Z.; McTiernan, C.D.; Suuronen, E.J.; Mah, T.F.; Alarcon, E.I. Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 2018, 4, e01067. [Google Scholar] [CrossRef]
- Dongari-Bagtzoglou, A. Pathogenesis of mucosal biofilm infections: Challenges and progress. Expert Rev. Anti Infect. 2008, 6, 201–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Husain, F.M.; Qais, F.A.; Ahmad, I.; Hakeem, M.J.; Baig, M.H.; Masood Khan, J.; Al-Shabib, N.A. Biosynthesized Zinc Oxide Nanoparticles Disrupt Established Biofilms of Pathogenic Bacteria. Appl. Sci. 2022, 12, 710. [Google Scholar] [CrossRef]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P.; et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003, 22, 3803–3815. [Google Scholar] [CrossRef]
- Lee, J.-H.; Cho, H.S.; Kim, Y.; Kim, J.-A.; Banskota, S.; Cho, M.H.; Lee, J. Indole and 7-benzyloxyindole attenuate the virulence of Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2013, 97, 4543–4552. [Google Scholar] [CrossRef]
- Salam, A.M.; Quave, C.L. Targeting Virulence in Staphylococcus aureus by Chemical Inhibition of the Accessory Gene Regulator System In Vivo. mSphere 2018, 3, e00500-17. [Google Scholar] [CrossRef] [Green Version]
- Khan, F.; Pham, D.T.N.; Oloketuyi, S.F.; Kim, Y.M. Regulation and controlling the motility properties of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 2020, 104, 33–49. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, Y.-G.; Cho, M.H.; Lee, J. ZnO nanoparticles inhibit Pseudomonas aeruginosa biofilm formation and virulence factor production. Microbiol. Res. 2014, 169, 888–896. [Google Scholar] [CrossRef]
- Dwivedi, S.; Wahab, R.; Khan, F.; Mishra, Y.K.; Musarrat, J.; Al-Khedhairy, A.A. Reactive oxygen species mediated bacterial biofilm inhibition via zinc oxide nanoparticles and their statistical determination. PLoS ONE 2014, 9, e111289. [Google Scholar] [CrossRef]
- Kaur, T.; Putatunda, C.; Vyas, A.; Kumar, G. Zinc oxide nanoparticles inhibit bacterial biofilm formation via altering cell membrane permeability. Prep. Biochem. Biotechnol. 2021, 51, 309–319. [Google Scholar] [CrossRef]
- Perveen, K.; Husain, F.M.; Qais, F.A.; Khan, A.; Razak, S.; Afsar, T.; Alam, P.; Almajwal, A.M.; Abulmeaty, M.M.A. Microwave-Assisted Rapid Green Synthesis of Gold Nanoparticles Using Seed Extract of Trachyspermum ammi: ROS Mediated Biofilm Inhibition and Anticancer Activity. Biomolecules 2021, 11, 197. [Google Scholar] [CrossRef]
- Lin, Y.K.; Yang, S.C.; Hsu, C.Y.; Sung, J.T.; Fang, J.Y. The Antibiofilm Nanosystems for Improved Infection Inhibition of Microbes in Skin. Molecules 2021, 26, 6392. [Google Scholar] [CrossRef] [PubMed]
- Won, S.-M.; Chen, S.; Park, K.W.; Yoon, J.-H. Isolation of lactic acid bacteria from kimchi and screening of Lactobacillus sakei ADM14 with anti-adipogenic effect and potential probiotic properties. LWT 2020, 126, 109296. [Google Scholar] [CrossRef]
- Fazlurrahman; Batra, M.; Pandey, J.; Suri, C.R.; Jain, R.K. Isolation and characterization of an atrazine-degrading Rhodococcus sp. strain MB-P1 from contaminated soil. Lett. Appl. Microbiol. 2009, 49, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Zhang, X.; Xu, Z. Identification of antibacterial substances of Lactobacillus plantarum DY-6 for bacteriostatic action. Food Sci. Nutr. 2020, 8, 2854–2863. [Google Scholar] [CrossRef] [PubMed]
- Jo, D.-M.; Park, S.-K.; Khan, F.; Kang, M.-G.; Lee, J.-H.; Kim, Y.-M. An approach to extend the shelf life of ribbonfish fillet using lactic acid bacteria cell-free culture supernatant. Food Control 2021, 123, 107731. [Google Scholar] [CrossRef]
- Khan, F.; Lee, J.W.; Pham, D.T.N.; Lee, J.H.; Kim, H.W.; Kim, Y.K.; Kim, Y.M. Streptomycin mediated biofilm inhibition and suppression of virulence properties in Pseudomonas aeruginosa PAO1. Appl. Microbiol. Biotechnol. 2020, 104, 799–816. [Google Scholar] [CrossRef]
- Essar, D.W.; Eberly, L.; Hadero, A.; Crawford, I.P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: Interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 1990, 172, 884–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilardoni, G.; Clericuzio, M.; Tosi, S.; Zanoni, G.; Vidari, G. Antifungal acylcyclopentenediones from fruiting bodies of Hygrophorus chrysodon. J. Nat. Prod. 2007, 70, 137–139. [Google Scholar] [CrossRef] [Green Version]
- Mokhtari, M.; Jackson, M.D.; Brown, A.S.; Ackerley, D.F.; Ritson, N.J.; Keyzers, R.A.; Munkacsi, A.B. Bioactivity-Guided Metabolite Profiling of Feijoa (Acca sellowiana) Cultivars Identifies 4-Cyclopentene-1,3-dione as a Potent Antifungal Inhibitor of Chitin Synthesis. J. Agric. Food Chem. 2018, 66, 5531–5539. [Google Scholar] [CrossRef]
- Maier, N.K.; Leppla, S.H.; Moayeri, M. The cyclopentenone prostaglandin 15d-PGJ2 inhibits the NLRP1 and NLRP3 inflammasomes. J. Immunol. 2015, 194, 2776–2785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, A.; Alonso, M.; Castro, A.; Dorronsoro, I.; Gelpí, J.L.; Luque, F.J.; Pérez, C.; Moreno, F.J. SAR and 3D-QSAR studies on thiadiazolidinone derivatives: Exploration of structural requirements for glycogen synthase kinase 3 inhibitors. J. Med. Chem. 2005, 48, 7103–7112. [Google Scholar] [CrossRef] [PubMed]
- Lizardi, L.; Garcia, M.C.; Sanchez, J.A.; Zuazaga, C. Sulfhydryl alkylating agents induce calcium current in skeletal muscle fibers of a crustacean (Atya lanipes). J. Membr. Biol. 1992, 129, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.; Straus, D.S. Effects of cyclopentenone prostaglandins and related compounds on insulin-like growth factor-I and Waf1 gene expression. Biochim. Biophys. Acta 1998, 1397, 31–42. [Google Scholar] [CrossRef]
- Uto, Y.; Nagasawa, H.; Jin, C.Z.; Nakayama, S.; Tanaka, A.; Kiyoi, S.; Nakashima, H.; Shimamura, M.; Inayama, S.; Fujiwara, T.; et al. Design of antiangiogenic hypoxic cell radiosensitizers: 2-nitroimidazoles containing a 2-aminomethylene-4-cyclopentene-1,3-dione moiety. Bioorg. Med. Chem. 2008, 16, 6042–6053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, L.; King, J.B.; Morrow, B.H.; Shen, J.K.; Miller, A.N.; Cichewicz, R.H. Diarylcyclopentendione metabolite obtained from a Preussia typharum isolate procured using an unconventional cultivation approach. J. Nat. Prod. 2012, 75, 1819–1823. [Google Scholar] [CrossRef] [Green Version]
- Neumann, P.; Brodhun, F.; Sauer, K.; Herrfurth, C.; Hamberg, M.; Brinkmann, J.; Scholz, J.; Dickmanns, A.; Feussner, I.; Ficner, R. Crystal structures of Physcomitrella patens AOC1 and AOC2: Insights into the enzyme mechanism and differences in substrate specificity. Plant Physiol. 2012, 160, 1251–1266. [Google Scholar] [CrossRef] [Green Version]
- Braga, S.F.; Alves, É.V.; Ferreira, R.S.; Fradico, J.R.; Lage, P.S.; Duarte, M.C.; Ribeiro, T.G.; Júnior, P.A.; Romanha, A.J.; Tonini, M.L.; et al. Synthesis and evaluation of the antiparasitic activity of bis-(arylmethylidene) cycloalkanones. Eur. J. Med. Chem. 2014, 71, 282–289. [Google Scholar] [CrossRef]
- Beranič, N.; Stefane, B.; Brus, B.; Gobec, S.; Rižner, T.L. New enzymatic assay for the AKR1C enzymes. Chem. Biol. Interact. 2013, 202, 204–209. [Google Scholar] [CrossRef]
- Li, J.; Zhang, D.; Wu, X. Synthesis and biological evaluation of novel exo-methylene cyclopentanone tetracyclic diterpenoids as antitumor agents. Bioorg. Med. Chem. Lett. 2011, 21, 130–132. [Google Scholar] [CrossRef]
- Yamada, K.; Shimizu, A.; Komatsu, H.; Sakata, R.; Ohta, A. Effects of 2,5-dimethylpyrazine on plasma testosterone and polyamines- and acid phosphatase-levels in the rat prostate. Biol. Pharm. Bull. 1994, 17, 730–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, K.; Watanabe, Y.; Aoyagi, Y.; Ohta, A. Effect of alkylpyrazine derivatives on the duration of pentobarbital-induced sleep, picrotoxicin-induced convulsion and gamma-aminobutyric acid (GABA) levels in the mouse brain. Biol. Pharm. Bull. 2001, 24, 1068–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, K.; Kobayashi, Y.; Fujihara, H.; Ohta, A. Inhibitory effect of 2,5-dimethylpyrazine on oxytocic agent-induced uterine hypercontraction of normal or pregnant female rats. Biol. Pharm. Bull. 1998, 21, 538–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Liu, Q.; Zhao, Z.; Bai, B.; Sun, Z.; Cai, L.; Fu, Y.; Ma, Y.; Wang, Q.; Xi, G. Effect of hydroxyl on antioxidant properties of 2,3-dihydro-3,5-dihydroxy-6-methyl-4H-pyran-4-one to scavenge free radicals. RSC Adv. 2021, 11, 34456–34461. [Google Scholar] [CrossRef]
- Yu, X.; Zhao, M.; Liu, F.; Zeng, S.; Hu, J. Identification of 2,3-dihydro-3,5-dihydroxy-6-methyl-4H-pyran-4-one as a strong antioxidant in glucose–histidine Maillard reaction products. Food Res. Int. 2013, 51, 397–403. [Google Scholar] [CrossRef]
- Čechovská, L.; Cejpek, K.; Konečný, M.; Velíšek, J. On the role of 2,3-dihydro-3,5-dihydroxy-6-methyl-(4H)-pyran-4-one in antioxidant capacity of prunes. Eur. Food Res. Technol. 2011, 233, 367–376. [Google Scholar] [CrossRef]
- Suganuma, H.; Inakuma, T.; Kikuchi, Y. Amelioratory Effect of Barley Tea Drinking on Blood Fluidity. J. Nutr. Sci. Vitaminol. 2002, 48, 165–168. [Google Scholar] [CrossRef]
- Yanagimoto, K.; Lee, K.G.; Ochi, H.; Shibamoto, T. Antioxidative activity of heterocyclic compounds found in coffee volatiles produced by Maillard reaction. J. Agric. Food Chem. 2002, 50, 5480–5484. [Google Scholar] [CrossRef]
- Hidalgo, F.J.; Nogales, F.; Zamora, R. Effect of the Pyrrole Polymerization Mechanism on the Antioxidative Activity of Nonenzymatic Browning Reactions. J. Agric. Food Chem. 2003, 51, 5703–5708. [Google Scholar] [CrossRef]
- Devadas, S.M.; Nayak, U.Y.; Narayan, R.; Hande, M.H.; Ballal, M. 2,5-Dimethyl-4-hydroxy-3(2H)-furanone as an Anti-biofilm Agent Against Non-Candida albicans Candida Species. Mycopathologia 2019, 184, 403–411. [Google Scholar] [CrossRef]
- Sung, W.S.; Jung, H.J.; Park, K.; Kim, H.S.; Lee, I.-S.; Lee, D.G. 2,5-dimethyl-4-hydroxy-3(2H)-furanone (DMHF); antimicrobial compound with cell cycle arrest in nosocomial pathogens. Life Sci. 2007, 80, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Chuang, P.-H.; Lee, C.-W.; Chou, J.-Y.; Murugan, M.; Shieh, B.-J.; Chen, H.-M. Anti-fungal activity of crude extracts and essential oil of Moringa oleifera Lam. Bioresour. Technol. 2007, 98, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-q.; Zhang, L.; Tao, X.-g.; Wei, L.; Liu, B.; Huang, L.-l.; Chen, Y.-g. Tetramethylpyrazine upregulates the aquaporin 8 expression of hepatocellular mitochondria in septic rats. J. Surg. Res. 2013, 185, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jiang, J.; Zhang, Z.; Yu, P.; Wang, L.; Xu, C.; Liu, W.; Wang, Y. Antioxidative and thrombolytic TMP nitrone for treatment of ischemic stroke. Bioorganic Med. Chem. 2008, 16, 8868–8874. [Google Scholar] [CrossRef]
- Sun, Y.; Song, M.; Niu, L.; Bai, X.; Sun, N.; Zhao, X.; Jiang, J.; He, J.; Li, H. Antiviral effects of the constituents derived from Chinese herb medicines on infectious bursal disease virus. Pharm. Biol. 2013, 51, 1137–1143. [Google Scholar] [CrossRef] [Green Version]
- Ren, Z.; Ma, J.; Zhang, P.; Luo, A.; Zhang, S.; Kong, L.; Qian, C. The effect of ligustrazine on L-type calcium current, calcium transient and contractility in rabbit ventricular myocytes. J. Ethnopharmacol. 2012, 144, 555–561. [Google Scholar] [CrossRef]
- Wang, H.; Jenner, A.M.; Lee, C.-Y.J.; Shui, G.; Tang, S.Y.; Whiteman, M.; Wenk, M.R.; Halliwell, B. The identification of antioxidants in dark soy sauce. Free Radic. Res. 2007, 41, 479–488. [Google Scholar] [CrossRef]
- Chaves, S.; Canário, S.; Carrasco, M.P.; Mira, L.; Santos, M.A. Hydroxy(thio)pyrone and hydroxy(thio)pyridinone iron chelators: Physico-chemical properties and anti-oxidant activity. J. Inorg. Biochem. 2012, 114, 38–46. [Google Scholar] [CrossRef]
- Yang, M.-L.; Kuo, P.-C.; Hwang, T.-L.; Wu, T.-S. Anti-inflammatory Principles from Cordyceps sinensis. J. Nat. Prod. 2011, 74, 1996–2000. [Google Scholar] [CrossRef]
- Kandioller, W.; Hartinger, C.G.; Nazarov, A.A.; Bartel, C.; Skocic, M.; Jakupec, M.A.; Arion, V.B.; Keppler, B.K. Maltol-Derived Ruthenium–Cymene Complexes with Tumor Inhibiting Properties: The Impact of Ligand–Metal Bond Stability on Anticancer Activity In Vitro. Chem.-Eur. J. 2009, 15, 12283–12291. [Google Scholar] [CrossRef]
- Amatori, S.; Ambrosi, G.; Fanelli, M.; Formica, M.; Fusi, V.; Giorgi, L.; Macedi, E.; Micheloni, M.; Paoli, P.; Pontellini, R.; et al. Synthesis, Basicity, Structural Characterization, and Biochemical Properties of Two [(3-Hydroxy-4-pyron-2-yl)methyl]amine Derivatives Showing Antineoplastic Features. J. Org. Chem. 2012, 77, 2207–2218. [Google Scholar] [CrossRef] [PubMed]
- Watanabe-Akanuma, M.; Inaba, Y.; Ohta, T. Mutagenicity of UV-irradiated maltol in Salmonella typhimurium. Mutagenesis 2007, 22, 43–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, K.S.; Tanaka, T.; Cho, E.J.; Yokozawa, T. Evaluation of the Peroxynitrite Scavenging Activity of Heat-Processed Ginseng. J. Med. Food 2009, 12, 124–130. [Google Scholar] [CrossRef]
- Lalitha, N.; Sadashivaiah, B.; Ramaprasad, T.R.; Singh, S.A. Anti-hyperglycemic activity of myricetin, through inhibition of DPP-4 and enhanced GLP-1 levels, is attenuated by co-ingestion with lectin-rich protein. PLoS ONE 2020, 15, e0231543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichimura, T.; Yamanaka, A.; Otsuka, T.; Yamashita, E.; Maruyama, S. Antihypertensive Effect of Enzymatic Hydrolysate of Collagen and Gly-Pro in Spontaneously Hypertensive Rats. Biosci. Biotechnol. Biochem. 2009, 73, 2317–2319. [Google Scholar] [CrossRef]
- Abdurraafi, M.; Dermawan, A.; Julianti, E.; Putra, M.; Karim, F. Identification and Evaluation of Antibacterial Compounds from the Vibrio sp. associated with the Ascidian Pycnoclavella diminuta. Pharm. Sci. Res. 2019, 6, 2. [Google Scholar]
- Lee, S.; Eom, S.; Nguyen, K.V.A.; Lee, J.; Park, Y.; Yeom, H.D.; Lee, J.H. The Application of the Neuroprotective and Potential Antioxidant Effect of Ergotamine Mediated by Targeting N-Methyl-D-Aspartate Receptors. Antioxidants 2022, 11, 1471. [Google Scholar] [CrossRef]
- Vesely, D.L. Ergotamine and dihydroergotamine enhance guanylate cyclase activity. Res. Commun. Chem. Pathol. Pharm. 1983, 40, 245–254. [Google Scholar]
- Kruk-Slomka, M.; Michalak, A.; Biala, G. Antidepressant-like effects of the cannabinoid receptor ligands in the forced swimming test in mice: Mechanism of action and possible interactions with cholinergic system. Behav. Brain Res. 2015, 284, 24–36. [Google Scholar] [CrossRef]
- Nojima, H.; Ohba, Y.; Kita, Y. Oleamide derivatives are prototypical anti-metastasis drugs that act by inhibiting Connexin 26. Curr. Drug Saf. 2007, 2, 204–211. [Google Scholar] [CrossRef]
- Lechin, F.; Van Der Dijs, B.; Bentolila, A.; Peña, F. Antidiarrheal effects of dihydroergotamine. J. Clin. Pharm. 1977, 17, 339–349. [Google Scholar] [CrossRef] [PubMed]
- Silberstein, S.D. The pharmacology of ergotamine and dihydroergotamine. Headache 1997, 37 (Suppl. 1), S15–S25. [Google Scholar] [PubMed]



| Sample | PA | SA | SM | CA | EC | VV | KP | LM |
|---|---|---|---|---|---|---|---|---|
| C-2 supernatant (%) | ND | 25 | 25 | 50 | 25 | 12.5 | 25 | 25 |
| C2-AuNPs (µg/mL) | >2048 | 1024 | 512 | 2048 | 1024 | ND | 512 | 2048 |
| C2-ZnONPs (µg/mL) | >2048 | 512 | ND | 256 | >2048 | >2048 | >2048 | >2048 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, M.-G.; Khan, F.; Jo, D.-M.; Oh, D.; Tabassum, N.; Kim, Y.-M. Antibiofilm and Antivirulence Activities of Gold and Zinc Oxide Nanoparticles Synthesized from Kimchi-Isolated Leuconostoc sp. Strain C2. Antibiotics 2022, 11, 1524. https://doi.org/10.3390/antibiotics11111524
Kang M-G, Khan F, Jo D-M, Oh D, Tabassum N, Kim Y-M. Antibiofilm and Antivirulence Activities of Gold and Zinc Oxide Nanoparticles Synthesized from Kimchi-Isolated Leuconostoc sp. Strain C2. Antibiotics. 2022; 11(11):1524. https://doi.org/10.3390/antibiotics11111524
Chicago/Turabian StyleKang, Min-Gyun, Fazlurrahman Khan, Du-Min Jo, DoKyung Oh, Nazia Tabassum, and Young-Mog Kim. 2022. "Antibiofilm and Antivirulence Activities of Gold and Zinc Oxide Nanoparticles Synthesized from Kimchi-Isolated Leuconostoc sp. Strain C2" Antibiotics 11, no. 11: 1524. https://doi.org/10.3390/antibiotics11111524
APA StyleKang, M.-G., Khan, F., Jo, D.-M., Oh, D., Tabassum, N., & Kim, Y.-M. (2022). Antibiofilm and Antivirulence Activities of Gold and Zinc Oxide Nanoparticles Synthesized from Kimchi-Isolated Leuconostoc sp. Strain C2. Antibiotics, 11(11), 1524. https://doi.org/10.3390/antibiotics11111524