Aryl Diazonium Chemistry for the Surface Functionalization of Glassy Biosensors
Abstract
:1. Introduction
2. Results and Discussion
2.1. SiCN Film Composition
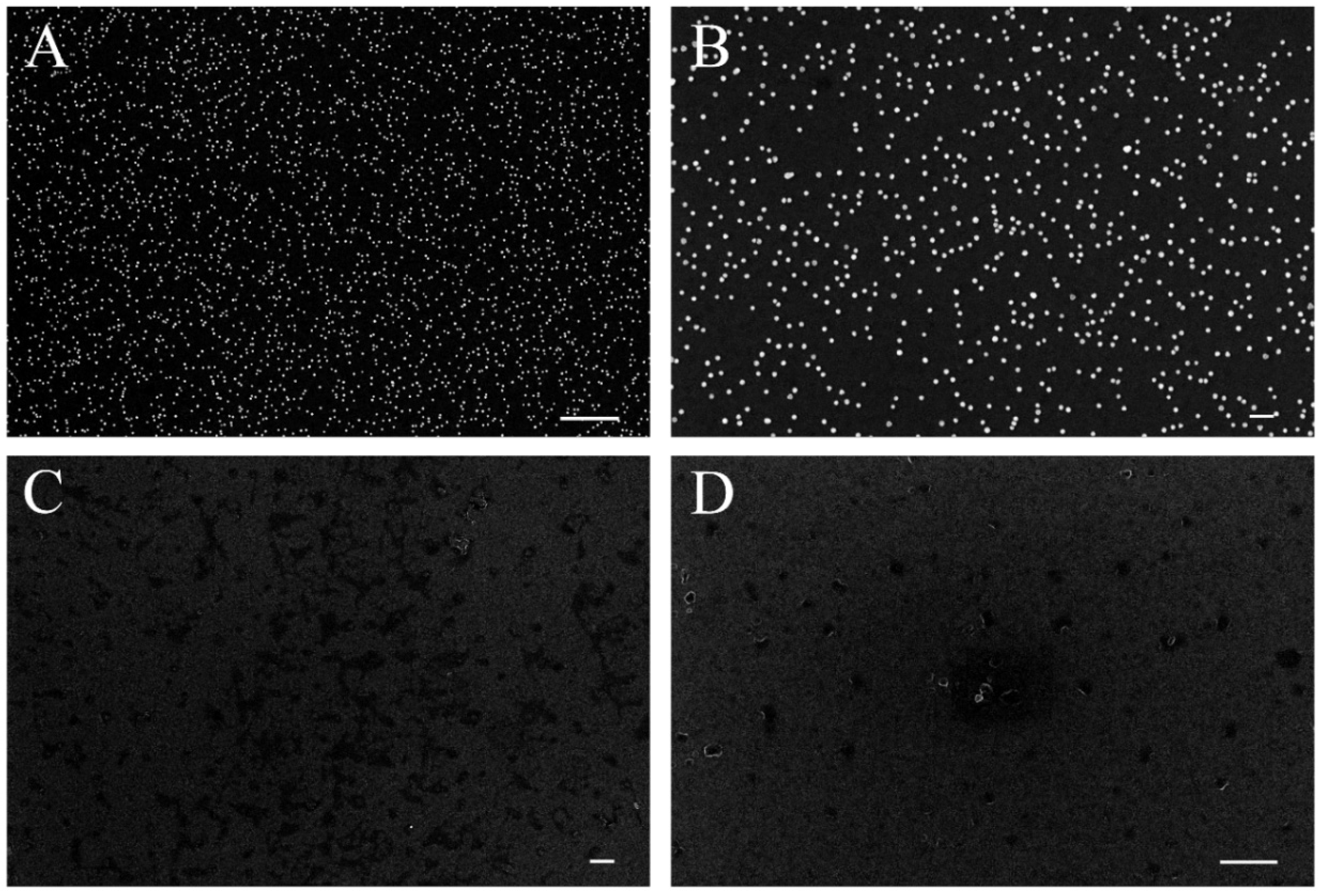
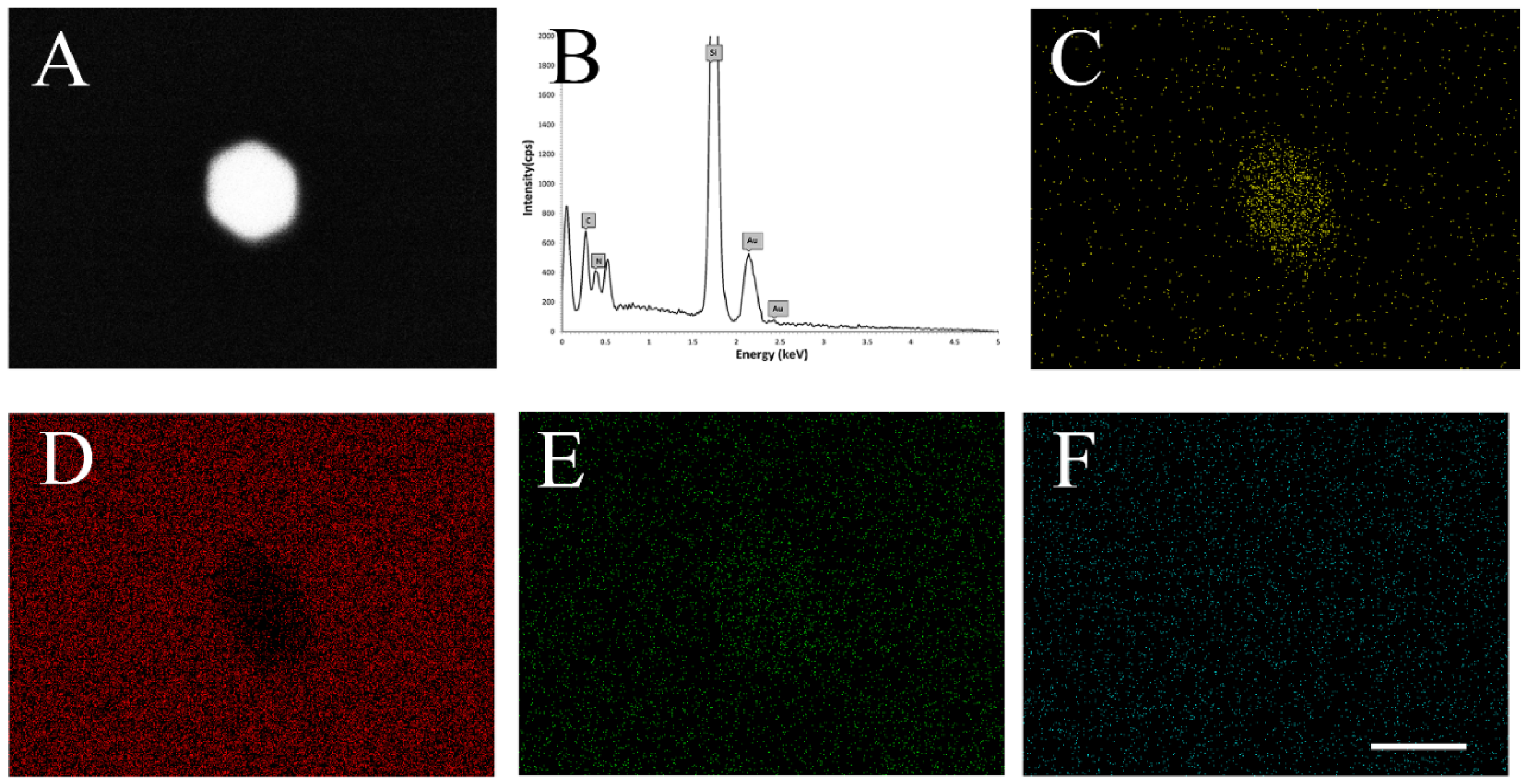
2.2. SEM and EDX of AuNPs
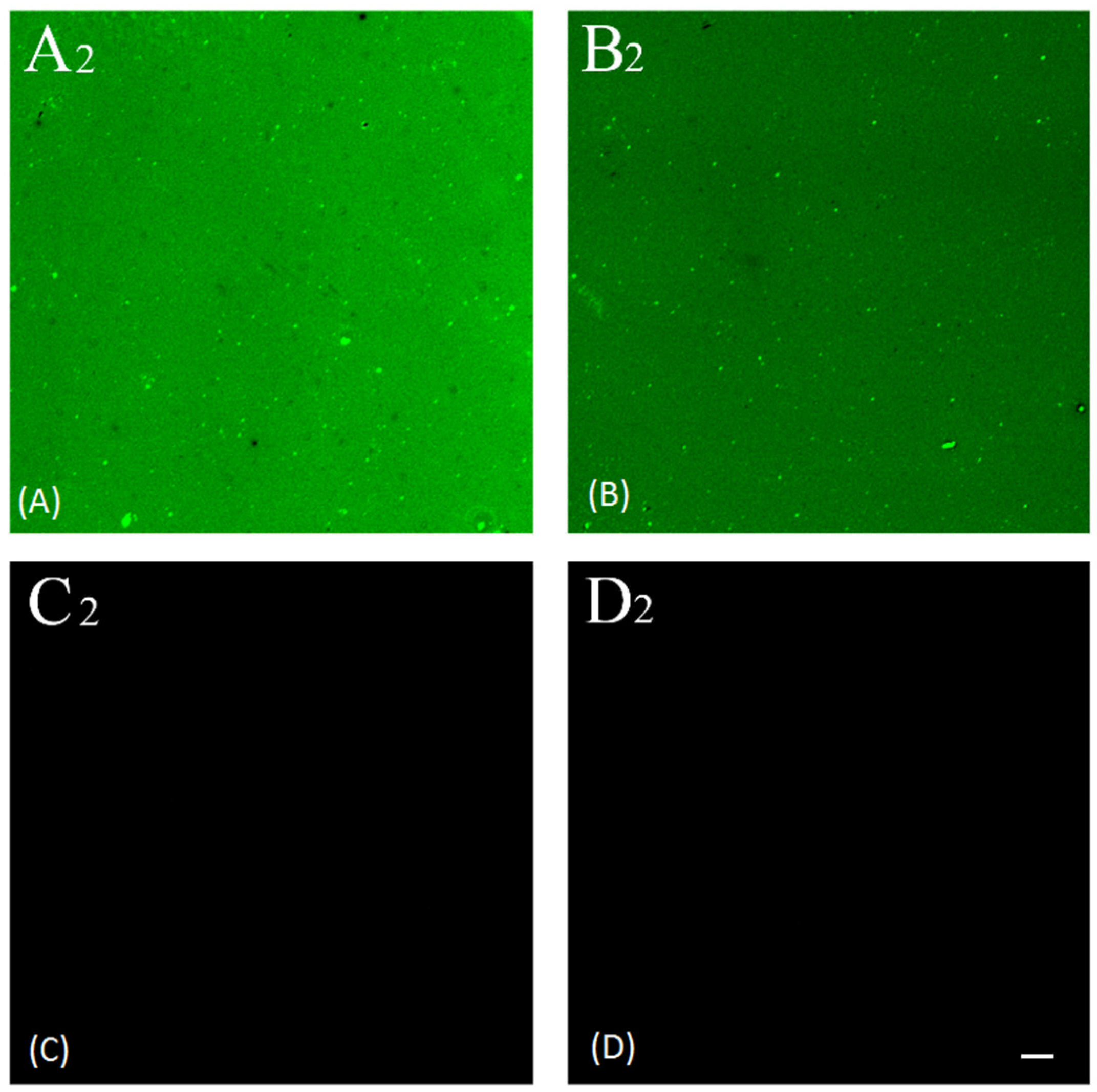
2.3. Fluorescence Imaging
3. Materials and Methods
3.1. SiCN Film Deposition
3.2. XPS Element Analysis
3.3. Diazonium Modification of SiCN
3.4. Gold Nanoparticle Labelled Sandwich Rabbit IgG Assay
3.4.1. Immobilization of AuNP Labelled Sandwich Rabbit IgG
3.4.2. SEM and EDX
3.5. FITC Labelled Sandwich Rabbit IgG Assay
3.5.1. Immobilization of FITC Labelled Sandwich Rabbit IgG
3.5.2. Confocal Microscopy Imaging
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| NEMS | Nano-electromechanical Systems |
| AuNP | Gold nanoparticle |
| SEM | Scanning electron microscopy |
| XPS | X-ray photoelectron microscopy |
| PBST | Phosphate Buffered Saline Tween-20 |
| PECVD | Plasma enhanced chemical vapor deposition |
| FITC | Fluorescein-5-Isothiocyanate |
References
- Haes, A.J.; Chang, L.; Klein, W.L.; Van Duyne, R.P. Detection of a biomarker for alzheimer’s disease from synthetic and clinical samples using a nanoscale optical biosensor. J. Am. Chem. Soc. 2005, 127, 2264–2271. [Google Scholar] [CrossRef] [PubMed]
- Tothill, I.E. Biosensors for cancer markers diagnosis. Semin. Cell Dev. Biol. 2009, 20, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, K.A.; Taylor, D.L. Fluorescent-protein biosensors: New tools for drug discovery. Trends Biotechnol. 1998, 16, 135–140. [Google Scholar] [CrossRef]
- Keusgen, M. Biosensors: New approaches in drug discovery. Naturwissenschaften 2002, 89, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.A. Optical biosensors in drug discovery. Nat. Rev. Drug Discov. 2002, 1, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Gartia, M.R.; Braunschweig, B.; Chang, T.-W.; Moinzadeh, P.; Minsker, B.S.; Agha, G.; Wieckowski, A.; Keefer, L.L.; Liu, G.L. The microelectronic wireless nitrate sensor network for environmental water monitoring. J. Environ. Monit. 2012, 14, 3068–3075. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.C.; Mastali, M.; Gau, V.; Suchard, M.A.; Møller, A.K.; Bruckner, D.A.; Babbitt, J.T.; Li, Y.; Gornbein, J.; Landaw, E.M. Use of electrochemical DNA biosensors for rapid molecular identification of uropathogens in clinical urine specimens. J. Clin. Microbiol. 2006, 44, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Edgeworth, J.A.; Farmer, M.; Sicilia, A.; Tavares, P.; Beck, J.; Campbell, T.; Lowe, J.; Mead, S.; Rudge, P.; Collinge, J.; et al. Detection of prion infection in variant creutzfeldt-jakob disease: A blood-based assay. Lancet 2011, 377, 487–493. [Google Scholar] [CrossRef]
- Jucker, M.; Walker, L.C. Pathogenic protein seeding in alzheimer disease and other neurodegenerative disorders. Ann. Neurol. 2011, 70, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Stohr, J.; Watts, J.C.; Mensinger, Z.L.; Oehler, A.; Grillo, S.K.; DeArmond, S.J.; Prusiner, S.B.; Giles, K. Purified and synthetic alzheimer’s amyloid beta (a beta) prions. PNAS 2012, 109, 11025–11030. [Google Scholar] [CrossRef] [PubMed]
- Rushworth, J.V.; Ahmed, A.; Griffiths, H.H.; Pollock, N.M.; Hooper, N.M.; Millner, P.A. A label-free electrical impedimetric biosensor for the specific detection of alzheimer's amyloid-beta oligomers. Biosens. Bioelectron. 2014, 56, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Sreekumar, A.; Nyati, M.K.; Varambally, S.; Barrette, T.R.; Ghosh, D.; Lawrence, T.S.; Chinnaiyan, A.M. Profiling of cancer cells using protein microarrays: Discovery of novel radiation-regulated proteins. Cancer Res. 2001, 61, 7585–7593. [Google Scholar] [PubMed]
- Woodbury, R.L.; Varnum, S.M.; Zangar, R.C. Elevated hgf levels in sera from breast cancer patients detected using a protein microarray elisa. J. Proteome Res. 2002, 1, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Saborio, G.P.; Permanne, B.; Soto, C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 2001, 411, 810–813. [Google Scholar] [CrossRef] [PubMed]
- Castilla, J.; Saa, P.; Soto, C. Detection of prions in blood. Nat. Med. 2005, 11, 982–985. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.A.; Morales, R.; Barria, M.A.; Soto, C. Estimating prion concentration in fluids and tissues by quantitative pmca. Nat. Methods 2010, 7, U519–U547. [Google Scholar] [CrossRef] [PubMed]
- Moudjou, M.; Sibille, P.; Fichet, G.; Reine, F.; Chapuis, J.; Herzog, L.; Jaumain, E.; Laferriere, F.; Richard, C.A.; Laude, H.; et al. Highly infectious prions generated by a single round of microplate-based protein misfolding cyclic amplification. mBio 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hamid, I.; Ivnitski, D.; Atanasov, P.; Wilkins, E. Flow-through immunofiltration assay system for rapid detection of e. Coli o157:H7. Biosens. Bioelectron. 1999, 14, 309–316. [Google Scholar] [CrossRef]
- Lazcka, O.; Del Campo, F.J.; Munoz, F.X. Pathogen detection: A perspective of traditional methods and biosensors. Biosens. Bioelectron. 2007, 22, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Fung, Y.S.; Wong, Y.Y. Self-assembled monolayers as the coating in a quartz piezoelectric crystal immunosensor to detect salmonella in aqueous solution. Anal. Chem. 2001, 73, 5302–5309. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.P.; Liu, W.C.; Tseng, M.C.; Chu, Y.H. Ionic liquids tailored for reaction-based gas sensing on quartz crystal microbalance. Rev. Anal. Chem. 2015, 34, 77–86. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, M.; He, J.; Yang, Q. Progress of different sensing materials modified qcm gas sensors. Prog. Chem. 2015, 27, 251–266. [Google Scholar]
- Guo, H. Progress of quartz crystal microbalance in chiral analysis. J. Nanosci. Nanotechnol. 2014, 14, 1884–1897. [Google Scholar] [CrossRef]
- Ilic, B.; Czaplewski, D.; Zalalutdinov, M.; Craighead, H.G.; Neuzi, P.; Campagnolo, C.; Batt, C. Single cell detection with micromechanical oscillators. J. Vac. Sci. Technol. B 2001, 19, 2825–2828. [Google Scholar] [CrossRef]
- Ilic, B.; Yang, Y.; Craighead, H.G. Virus detection using nanoelectromechanical devices. Appl. Phys. Lett. 2004, 85, 2604–2606. [Google Scholar] [CrossRef]
- Poshtiban, S.; Singh, A.; Fitzpatrick, G.; Evoy, S. Bacteriophage tail-spike protein derivitized microresonator arrays for specific detection of pathogenic bacteria. Sens. Actuators B Chem. 2013, 181, 410–416. [Google Scholar] [CrossRef]
- Campbell, G.A.; Medina, M.B.; Mutharasan, R. Detection of staphylococcus enterotoxin b at picogram levels using piezoelectric-excited millimeter-sized cantilever sensors. Sens. Actuators B Chem. 2007, 126, 354–360. [Google Scholar] [CrossRef]
- Fischer, L.M.; Wright, V.A.; Guthy, C.; Yang, N.; McDermott, M.T.; Buriak, J.M.; Evoy, S. Specific detection of proteins using nanomechanical resonators. Sens. Actuators B Chem. 2008, 134, 613–617. [Google Scholar] [CrossRef]
- Quitoriano, N.J.; Belov, M.; Evoy, S.; Kamins, T.I. Single-crystal, si nanotubes, and their mechanical resonant properties. Nano. Lett. 2009, 9, 1511–1516. [Google Scholar] [CrossRef] [PubMed]
- Bausells, J. Piezoresistive cantilevers for nanomechanical sensing. Microelectron. Eng. 2015, 145, 9–20. [Google Scholar] [CrossRef]
- Abdel-Hamid, I.; Ghindidlis, A.L.; Atanasov, P.; Wilkins, E. Development of a flow-through immunoassay system. Sens. Actuators B 1998, 49, 202–210. [Google Scholar] [CrossRef]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow cytometry: Basic principles and applications. Crit. Rev. Biotechnol. 2016, 36. [Google Scholar] [CrossRef] [PubMed]
- Mross, S.; Pierrat, S.; Zimmermann, T.; Kraft, M. Microfluidic enzymatic biosensing systems: A review. Biosens. Bioelectron. 2015, 70, 376–391. [Google Scholar] [CrossRef] [PubMed]
- Gau, J.J.; Lan, E.H.; Dunn, B.; Ho, C.M.; Woo, J.C. A MEMS based amperometric detector for E. coli bacteria using self-assembled monolayers. Biosens. Bioelectron. 2001, 16, 745–755. [Google Scholar] [CrossRef]
- Gervals, L.; Gel, M.; Allain, B.; Tolba, M.; Brovko, L.; Zourob, M.; Mandeville, R.; Griffiths, M.; Evoy, S. Immobilization of biotinylated bacteriophages on biosensor surfaces. Sens. Actuators B Chem. 2007, 125, 615–621. [Google Scholar] [CrossRef]
- Xu, Y.; Xie, X.; Duan, Y.; Wang, L.; Cheng, Z.; Cheng, J. A review of impedance measurements of whole cells. Biosens. Bioelectron. 2016, 77, 824–836. [Google Scholar] [CrossRef] [PubMed]
- Chillawar, R.R.; Tadi, K.K.; Motghare, R.V. Voltammetric techniques at chemically modified electrodes. J. Anal. Chem. 2015, 70, 399–418. [Google Scholar] [CrossRef]
- Amatore, C.; Delacotte, J.; Guille-Collignon, M.; Lemaître, F. Vesicular exocytosis and microdevices-microelectrode arrays. Analyst 2015, 140, 3687–3695. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.D.; Yu, Q.; Chen, S.; Homola, J.; Jiang, S. Comparison of E. coli O157:H7 preparation methods used for detection with surface plasmon resonance sensor. Sens. Actuators B 2005, 107, 202–208. [Google Scholar] [CrossRef]
- Liu, X.; Hu, Y.; Zheng, S.; Liu, Y.; He, Z.; Luo, F. Surface plasmon resonance immunosensor for fast, highly sensitive, and in situ detection of the magnetic nanoparticles-enriched Salmonella enteritidis. Sens. Actuators B Chem. 2016, 230, 191–198. [Google Scholar] [CrossRef]
- Arya, S.K.; Singh, A.; Naidoo, R.; Wu, P.; McDermott, M.T.; Evoy, S. Chemically immobilized t4-bacteriophage for specific escherichia coli detection using surface plasmon resonance. Analyst 2011, 136, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Arya, S.K.; Glass, N.; Hanifi-Moghaddam, P.; Naidoo, R.; Szymanski, C.M.; Tanha, J.; Evoy, S. Bacteriophage tailspike proteins as molecular probes for sensitive and selective bacterial detection. Biosens. Bioelectron. 2010, 26, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Butet, J.; Brevet, P.F.; Martin, O.J.F. Optical second harmonic generation in plasmonic nanostructures: From fundamental principles to advanced applications. Acs. Nano. 2015, 9, 10545–10562. [Google Scholar] [CrossRef] [PubMed]
- Langer, J.; Novikov, S.M.; Liz-Marzán, L.M. Sensing using plasmonic nanostructures and nanoparticles. Nanotechnology 2015, 26. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface plasmon resonance: A versatile technique for biosensor applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Bo, L.; Semenova, Y.; Farrell, G.; Brambilla, G. Optical microfibre based photonic components and their applications in label-free biosensing. Biosensors 2015, 5, 471–499. [Google Scholar] [CrossRef] [PubMed]
- Buswell, S.C.; Wright, V.A.; Buriak, J.M.; Van, V.; Evoy, S. Specific detection of proteins using photonic crystal waveguides. Opt. Express 2008, 16, 15949–15957. [Google Scholar] [CrossRef] [PubMed]
- Mukundan, H.; Kubicek, J.Z.; Holt, A.; Shively, J.E.; Martinez, J.S.; Grace, K.; Grace, W.K.; Swanson, B.I. Planar optical waveguide-based biosensor for the quantitative detection of tumor markers. Sens. Actuators B Chem. 2009, 138, 453–460. [Google Scholar] [CrossRef]
- Schmitt, K.; Schirmer, B.; Hoffmann, C.; Brandenburg, A.; Meyrueis, P. Interferometric biosensor based on planar optical waveguide sensor chips for label-free detection of surface bound bioreactions. Biosens. Bioelectron. 2007, 22, 2591–2597. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.C.; Ho, J.A.A. Attomole DNA electrochemical sensor for the detection of escherichia coli o157. Anal. Chem. 2009, 81, 2470–2476. [Google Scholar] [CrossRef] [PubMed]
- Joung, H.A.; Lee, N.R.; Lee, S.K.; Ahn, J.; Shin, Y.B.; Choi, H.S.; Lee, C.S.; Kim, S.; Kim, M.G. High sensitivity detection of 16s rrna using peptide nucleic acid probes and a surface plasmon resonance biosensor. Anal. Chim. Acta 2008, 630, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.D.; Yu, Q.M.; Chen, S.F.; Homola, J.; Jiang, S.Y. Comparison of E coli o157:H7 preparation methods used for detection with surface plasmon resonance sensor. Sens. Actuators B Chem. 2005, 107, 202–208. [Google Scholar] [CrossRef]
- Oh, B.K.; Lee, W.; Chun, B.S.; Bae, Y.M.; Lee, W.H.; Choi, J.W. The fabrication of protein chip based on surface plasmon resonance for detection of pathogens. Biosens. Bioelectron. 2005, 20, 1847–1850. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.D.; Ladd, J.; Yu, Q.M.; Chen, S.F.; Homola, J.; Jiang, S.Y. Quantitative and simultaneous detection of four foodborne bacterial pathogens with a multi-channel spr sensor. Biosens. Bioelectron. 2006, 22, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Sorokulova, I.B.; Vodyanoy, V.J.; Simonian, A.L. Lytic phage as a specific and selective probe for detection of staphylococcus aureus—A surface plasmon resonance spectroscopic study. Biosens. Bioelectron. 2007, 22, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.H.; Johnson, M.L.; Guntupalli, R.; Petrenko, V.A.; Chin, B.A. Detection of bacillus anthracis spores in liquid using phage-based magnetoelastic micro-resonators. Sens. Actuators B Chem. 2007, 127, 559–566. [Google Scholar] [CrossRef]
- Singh, A.; Arutyunov, D.; Szymanski, C.M.; Evoy, S. Bacteriophage based probes for pathogen detection. Analyst 2012, 137, 3405–3421. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Arutyunov, D.; McDermott, M.T.; Szymanski, C.M.; Evoy, S. Specific detection of campylobacter jejuni using the bacteriophage nctc 12673 receptor binding protein as a probe. Analyst 2011, 136, 4780–4786. [Google Scholar] [CrossRef] [PubMed]
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Surface plasmon resonance detection of E. coli and methicillin-resistant s. Aureus using bacteriophages. Biosens. Bioelectron. 2012, 37, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Chibli, H.; Ghali, H.; Park, S.; Peter, Y.A.; Nadeau, J.L. Immobilized phage proteins for specific detection of staphylococci. Analyst 2014, 139, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Mehta, G.; Srivastava, S. Label-free detection techniques for protein microarrays: Prospects, merits and challenges. Proteomics 2010, 10, 731–748. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Lee, B.; Liu, G.L.; Agarwal, A.; Lee, L.P. Label-free and highly sensitive biomolecular detection using sers and electrokinetic preconcentration. Lab Chip 2009, 9, 3360–3363. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.X.; Densmore, A.; Delâge, A.; Waldron, P.; McKinnon, R.; Janz, S.; Lapointe, J.; Lopinski, G.; Mischki, T.; Post, E.; et al. Folded cavity soi microring sensors for highsensitivity and real time measurement ofbiomolecular binding. Opt. Express 2008, 16, 15137–15148. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.X.; Vachon, M.; Densmore, A.; Ma, R.; Delâge, A.; Janz, S.; Lapointe, J.; Li, Y.; Lopinski, G.; Zhang, D.; et al. Label-free biosensor array based on silicon-on-insulator ring resonators addressed using a WDM approach. Opt. Lett. 2010, 35, 2771–2773. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.L.; Lee, L.P. Nanowell surface enhanced raman scattering arrays fabricated by soft-lithography for label-free biomolecular detections in integrated microfluidics. Appl. Phys. Lett. 2005, 87, 074101. [Google Scholar] [CrossRef]
- Furuki, M.; Kameoka, J.; Craighead, H.G.; Isaacson, M.S. Surface plasmon resonance sensors utilizing rnicrofabricated channels. Sens. Actuators B Chem. 2001, 79, 63–69. [Google Scholar] [CrossRef]
- Cui, Y.; Wei, Q.Q.; Park, H.K.; Lieber, C.M. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science 2001, 293, 1289–1292. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.D.; Wolfbeis, O.S. Fiber-optic chemical sensors and biosensors (2008–2012). Anal. Chem. 2013, 85, 487–508. [Google Scholar] [CrossRef] [PubMed]
- Mehrvar, M.; Bis, C.; Scharer, J.M.; Moo-Young, M.; Luong, J.H. Fiber-optic biosensors—Trends and advances. Anal. Sci. 2000, 16, 677–692. [Google Scholar] [CrossRef]
- Wang, Y.-M.; Pang, X.-F.; Zhang, Y.-Y. Recent advances in fiber-optic DNA biosensors. J. Biomed. Sci. Eng. 2009, 2, 312–317. [Google Scholar] [CrossRef]
- Sekaric, L.; Carr, D.W.; Evoy, S.; Parpia, J.M.; Craighead, H.G. Nanomechanical resonant structures in silicon nitride: Fabrication, operation and dissipation issues. Sens. Actuators A Phys. 2002, 101, 215–219. [Google Scholar] [CrossRef]
- Fischer, L.M.; Wilding, N.; Gel, M.; Evoy, S. Low-stress silicon carbonitride for the machining of high-frequency nanomechanical resonators. J. Vac. Sci. Technol. B 2007, 25, 33–37. [Google Scholar] [CrossRef]
- Verbridge, S.S.; Craighead, H.G.; Parpia, J.M. A megahertz nanomechanical resonator with room temperature quality factor over a million. Appl. Phys. Lett. 2008, 92. [Google Scholar] [CrossRef]
- Southworth, D.R.; Barton, R.A.; Verbridge, S.S.; Ilic, B.; Fefferman, A.D.; Craighead, H.G.; Parpia, J.M. Stress and silicon nitride: A crack in the universal dissipation of glasses. Phys. Rev. Lett. 2009, 102. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, M.A.; Dew, S.K.; Evoy, S.; Stepanova, M. Fabrication of sub-10 nm silicon carbon nitride resonators using a hydrogen silsesquioxane mask patterned by electron beam lithography. Microelectron. Eng. 2011, 88, 2338–2341. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Voros, J.; Reimhult, E. Electrochemical biosensors-sensor principles and architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef] [Green Version]
- Gopel, W.; Heiduschka, P. Interface analysis in biosensor design. Biosens. Bioelectron. 1995, 10, 853–883. [Google Scholar] [CrossRef]
- Credou, J.; Volland, H.; Dano, J.; Berthelot, T. A one-step and biocompatible cellulose functionalization for covalent antibody immobilization on immunoassay membranes. J. Mater. Chem. B 2013, 1, 3277–3286. [Google Scholar] [CrossRef]
- Mevellec, V.; Roussel, S.; Tessier, L.; Chancolon, J.; Mayne-L'Hermite, M.; Deniau, G.; Viel, P.; Palacin, S. Grafting polymers on surfaces: A new powerful and versatile diazonium salt-based one-step process in aqueous media. Chem. Mater. 2007, 19, 6323–6330. [Google Scholar] [CrossRef]
- Mesnage, A.; Esnouf, S.; Jegou, P.; Deniau, G.; Palacin, S. Understanding the redox-induced polymer grafting process: A dual surface-solution analysis. Chem. Mater. 2010, 22, 6229–6239. [Google Scholar] [CrossRef]
- Guthy, C.; Das, R.M.; Drobot, B.; Evoy, S. Resonant characteristics of ultranarrow SiCN nanomechanical resonators. J. Appl. Phys. 2010, 108, 014306. [Google Scholar] [CrossRef]
- Guthy, C.; Belov, M.; Janzen, A.; Quitoriano, N.J.; Singh, A.; Wright, V.A.; Finley, E.; Kamins, T.I.; Evoy, S. Large-scale arrays of nanomechanical sensors for biomolecular fingerprinting. Sens. Actuators B Chem. 2013, 187, 111–117. [Google Scholar] [CrossRef]
- Shewchuk, D.M.; McDermott, M.T. Comparison of diazonium salt derived and thiol derived nitrobenzene layers on gold. Langmuir 2009, 25, 4556–4563. [Google Scholar] [CrossRef] [PubMed]
- El Hadj, F.A.; Amiar, A.; Cherkaoui, M.; Chazalviel, J.N.; Ozanam, F. Study of organic grafting of the silicon surface from 4-nitrobenzene diazonium tetrafluoroborate. Electrochim. Acta 2012, 70, 318–324. [Google Scholar] [CrossRef]
- Young, J.; Su, Y.; Lee, S.; Pak, C.; Kim, H. Enhanced durability of linker-containing carbon nanotube functionalized via diazonium reaction. Electrochim. Acta 2015, 154, 63–69. [Google Scholar]
- Solak, A.O.; Eichorst, L.R.; Clark, W.J.; McCreery, R.L. Modified carbon surfaces as "organic electrodes" that exhibit conductance switching. Anal. Chem. 2003, 75, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Combellas, C.; Kanoufi, F.; Pinson, J.; Podvorica, F.I. Time-of-flight secondary ion mass spectroscopy characterization of the covalent bonding between a carbon surface and aryl groups. Langmuir 2005, 21, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Assresahegn, B.D.; Brousse, T.; Bélanger, D. Advances on the use of diazonium chemistry for functionalization of materials used in energy storage systems. Carbon 2015, 92, 362–381. [Google Scholar] [CrossRef]
- Menanteau, T.; Benoît, C.; Breton, T.; Cougnon, C. Enhancing the performance of a diazonium-modified carbon supercapacitor by controlling the grafting process. Electrochem. Commun. 2016, 63, 70–73. [Google Scholar] [CrossRef]
- Bouša, D.; Pumera, M.; Sedmidubský, D.; Šturala, J.; Luxa, J.; Mazánek, V.; Sofer, Z. Fine tuning of graphene properties by modification with aryl halogens. Nanoscale 2016, 8, 1493–1502. [Google Scholar] [CrossRef] [PubMed]
- González, M.C.R.; Orive, A.G.; Salvarezza, R.C.; Creus, A.H. Electrodeposition of gold nanoparticles on aryl diazonium monolayer functionalized hopg surfaces. Phys. Chem. Chem. Phys. 2016, 18, 1953–1960. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.P.; Maya, F.; Kosynkin, D.V.; Dirk, S.M.; Stapleton, J.J.; McGuiness, C.L.; Allara, D.L.; Tour, J.M. Direct covalent grafting of conjugated molecules onto si, gaas, and pd surfaces from aryidiazonium salts. J. Am. Chem. Soc. 2004, 126, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Bengamra, M.; Khlifi, A.; Ktari, N.; Mahouche-Chergui, S.; Carbonnier, B.; Fourati, N.; Kalfat, R.; Chehimi, M.M. Silanized aryl layers through thiol-yne photo-click reaction. Langmuir 2015, 31, 10717–10724. [Google Scholar] [CrossRef] [PubMed]
- Hurley, B.L.; McCreery, R.L. Covalent bonding of organic molecules to cu and al alloy 2024 t3 surfaces via diazonium ion reduction. J. Electrochem. Soc. 2004, 151, B252–B259. [Google Scholar] [CrossRef]
- Laurentius, L.; Stoyanov, S.R.; Gusarov, S.; Kovalenko, A.; Du, R.; Lopinski, G.P.; McDermott, M.T. Diazonium-derived aryl films on gold nanoparticles: Evidence for a carbon–gold covalent bond. Acs Nano 2011, 5, 4219–4227. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, C.L.; Landis, E.C. Electrochemical attachment of diazonium-generated films on nanoporous gold. Langmuir 2015, 31, 8633–8641. [Google Scholar] [CrossRef] [PubMed]
- Neal, S.N.; Workie, B.; McCandless, B.E.; Mohamed, A.A. Gold-organic thin films from the reductive grafting of diazonium gold(III) salts. J. Electroanal. Chem. 2015, 757, 73–79. [Google Scholar] [CrossRef]
- Zheng, W.; Du, R.B.; Cao, Y.; Mohammad, M.A.; Dew, S.K.; McDermott, M.T.; Evoy, S. Diazonium chemistry for the bio-functionalization of glassy nanostring resonator arrays. Sensors 2015, 15, 18724–18741. [Google Scholar] [CrossRef] [PubMed]


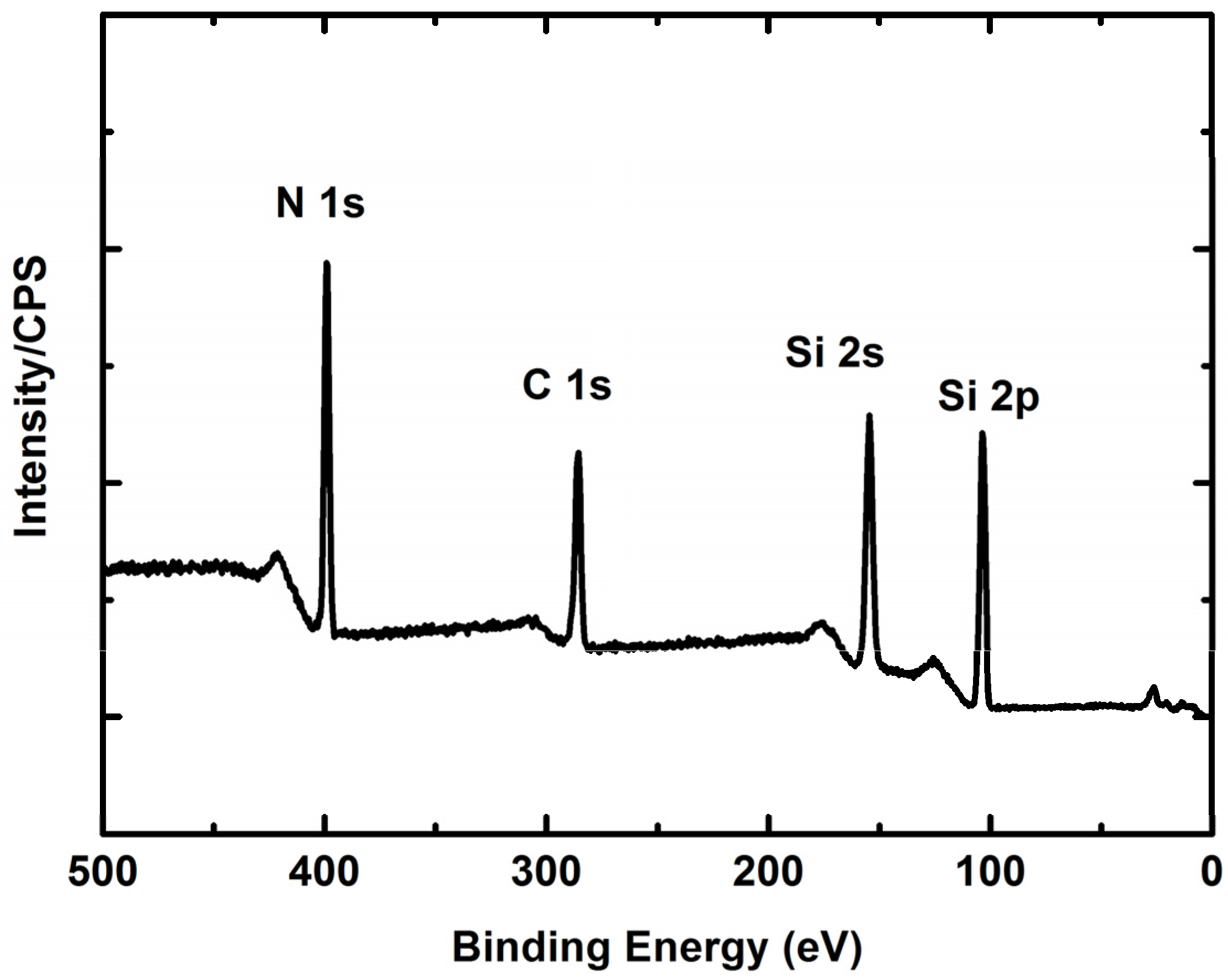


| Element | Atomic Concentration % | Mass Concentration % |
|---|---|---|
| N 1s | 29.69 | 21.94 |
| C 1s | 30.87 | 19.57 |
| Si 2p | 39.46 | 58.49 |
| Sample | Intensity (AU) | Signal to Noise Ratio | Dilution Ratio of FITC Conjugated Anti-Rabbit IgG |
|---|---|---|---|
| A2 | 37,967 | 185 | 1:200 |
| C2 | 205 | ||
| B2 | 25,791 | 105 | 1:400 |
| D2 | 245 |
| Sample | Control Type | Dilution Ratio | Label |
|---|---|---|---|
| A1 | positive | 1:1 | AuNP |
| B1 | positive | 1:3 | AuNP |
| C1 | negative | 1:1 | AuNP |
| D1 | negative | 1:3 | AuNP |
| A2 | positive | 1:200 | FITC |
| B2 | positive | 1:400 | FITC |
| C2 | negative | 1:200 | FITC |
| D2 | negative | 1:400 | FITC |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, W.; Van den Hurk, R.; Cao, Y.; Du, R.; Sun, X.; Wang, Y.; McDermott, M.T.; Evoy, S. Aryl Diazonium Chemistry for the Surface Functionalization of Glassy Biosensors. Biosensors 2016, 6, 8. https://doi.org/10.3390/bios6010008
Zheng W, Van den Hurk R, Cao Y, Du R, Sun X, Wang Y, McDermott MT, Evoy S. Aryl Diazonium Chemistry for the Surface Functionalization of Glassy Biosensors. Biosensors. 2016; 6(1):8. https://doi.org/10.3390/bios6010008
Chicago/Turabian StyleZheng, Wei, Remko Van den Hurk, Yong Cao, Rongbing Du, Xuejun Sun, Yiyu Wang, Mark T. McDermott, and Stephane Evoy. 2016. "Aryl Diazonium Chemistry for the Surface Functionalization of Glassy Biosensors" Biosensors 6, no. 1: 8. https://doi.org/10.3390/bios6010008







