Biosensor Applications of MAPLE Deposited Lipase
Abstract
:1. Introduction
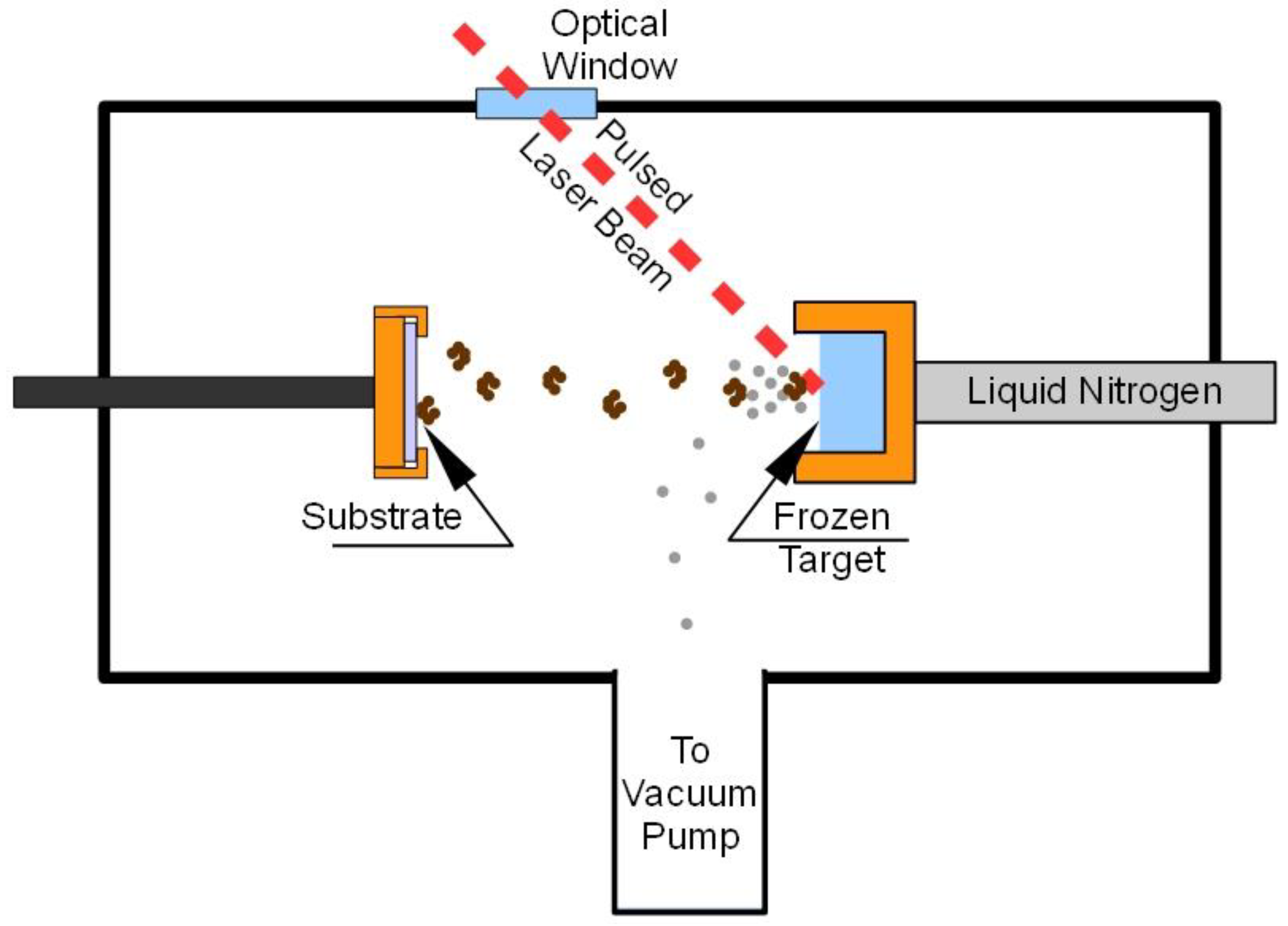
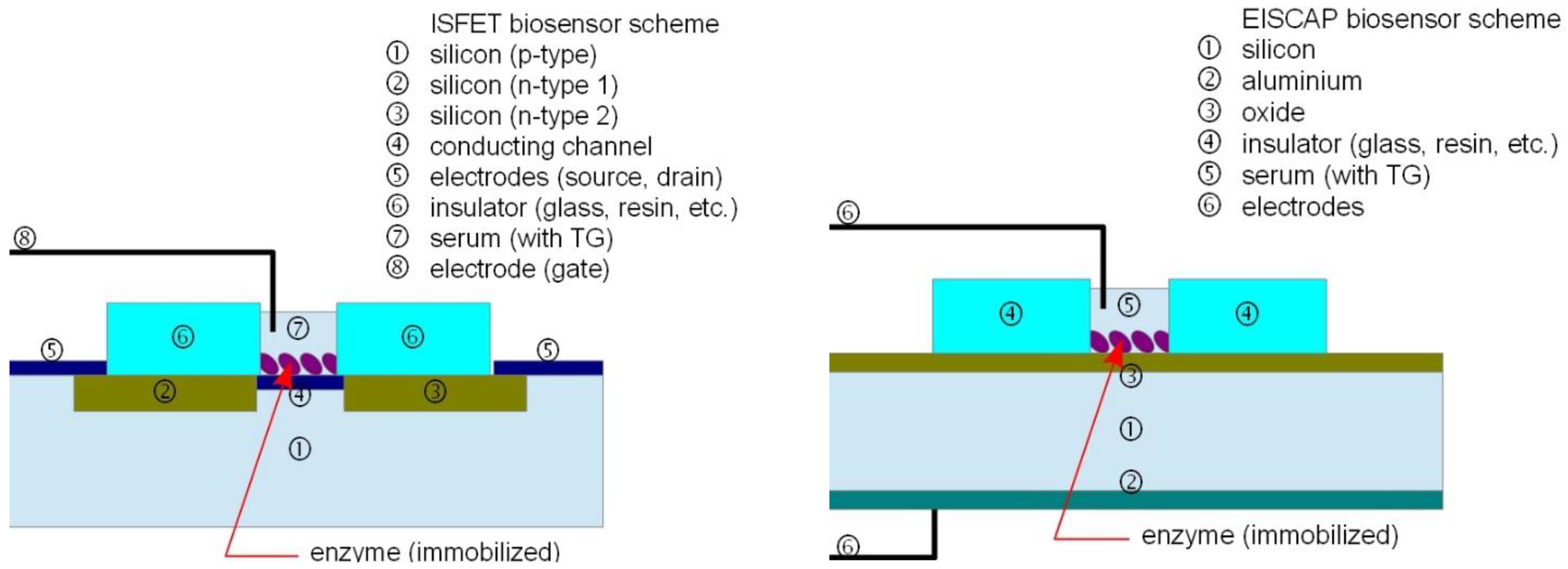
2. Experimental Section
3. Results and Discussion
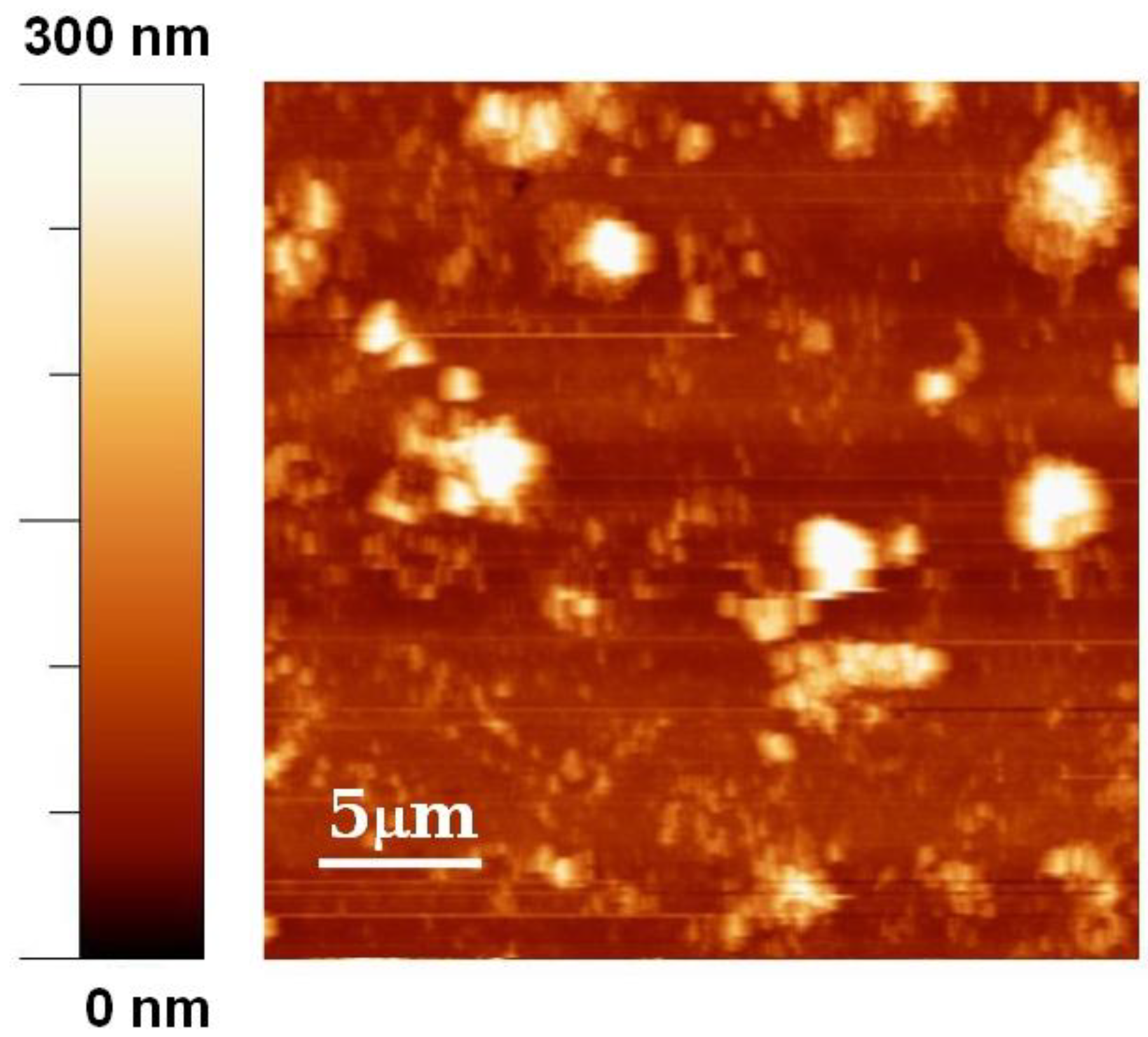

4. Conclusions
Author Contributions
Conflicts of Interest
References
- Pique, A.; McGill, R.A.; Chrisey, D.B.; Leonhardt, D.; Msln, T.E.; Spargo, B.J.; Callahan, J.H.; Vachet, R.W.; Chung, R.; Bucaro, M.A. Growth of organic thin films by the matrix assisted pulsed laser evaporation (MAPLE) technique. Thin Solid Films 1999, 355–356, 536–541. [Google Scholar] [CrossRef]
- Eason, R. Pulsed Laser Deposition of Thin Films; John Wiley & Sons: New York, NY, USA, 2007. [Google Scholar]
- Chrisey, D.B.; Pique, A.; McGill, R.A.; Horwitz, J.S.; Ringeisen, B.R.; Bubb, D.M.; Wu, P.K. Laser deposition of polymer and biomaterial films. Chem. Rev. 2003, 103, 553–576. [Google Scholar] [CrossRef] [PubMed]
- Bloisi, F.; Barra, M.; Cassinese, A.; Vicari, L.R.M. Matrix-assisted pulsed laser thin film deposition by using Nd:YAG laser. J. Nanomater. 2012, 2012. [Google Scholar] [CrossRef]
- Fryčeka, R.; Jelínek, M.; Kocourek, T.; Fitl, P.; Vrňata, M.; Myslík, V.; Vrbová, M. Thin organic layers prepared by MAPLE for gas sensor application. Thin Solid Films 2006, 495, 308–311. [Google Scholar] [CrossRef]
- Houser, E.J.; Chrisey, D.B.; Bercu, M.; Scarisoreanu, N.D.; Purice, A.; Colceag, D.; Constantinescu, C.; Moldovan, A.; Dinescu, M. Functionalized polysiloxane thin films deposited by matrix-assisted pulsed laser evaporation for advanced chemical sensor applications. Appl. Surf. Sci. 2006, 252, 4871–4876. [Google Scholar] [CrossRef]
- Bloisi, F.; Vicari, L.; Papa, R.; Califano, V.; Pedrazzani, R.; Bontempi, E.; Depero, L.E. Biomaterial thin film deposition and characterization by means of MAPLE technique. Mater. Sci. Eng. C 2007, 27, 1185–1190. [Google Scholar] [CrossRef]
- Caricato, A.P.; Lomascolo, M.; Luches, A.; Mandoj, F.; Manera, M.G.; Mastroianni, M.; Martino, M.; Paolesse, R.; Rella, R.; Romano, F.; Tunno, T.; Valerini, D. MAPLE deposition of methoxy Ge triphenylcorrole thin films. Appl. Phys. A 2008, 93, 651–654. [Google Scholar] [CrossRef]
- Califano, V.; Bloisi, F.; Vicari, L.R.M.; Bretcanu, O.; Boccaccini, A.R. Matrix-assisted pulsed laser evaporation of poly(D,L-lactide) for biomedical applications: Effect of near infrared radiation. J. Biomed. Optic. 2008, 13. [Google Scholar] [CrossRef]
- Cristescu, R.; Mihaiescu, D.; Socol, G.; Stamatin, I.; Mihailescu, I.N.; Chrisey, D.B. Deposition of biopolymer thin films by matrix assisted pulsed laser evaporation. Appl. Phys. A 2004, 79, 1023–1026. [Google Scholar] [CrossRef]
- Stamatin, L.; Cristescu, R.; Socol, G.; Moldovan, A.; Mihaiescu, D.; Stamatin, I.; Mihailescu, I.N.; Chrisey, D.B. Laser deposition of fibrinogen blood proteins thin films by matrix assisted pulsed laser evaporation. Appl. Surf. Sci. 2005, 248, 422–427. [Google Scholar] [CrossRef]
- Ringeisen, B.R.; Callahan, J.; Wu, P.K.; Piqué, A.; Spargo, B.; McGill, R.A.; Bucaro, M.; Kim, H.; Bubb, D.M.; Chrisey, D.B. Novel laser-based deposition of active protein thin films. Langmuir 2001, 17, 3472–3479. [Google Scholar] [CrossRef]
- Purice, A.; Schou, J.; Kingshott, P.; Pryds, N.; Dinescu, M. Characterization of lysozyme films produced by matrix assisted pulsed laser evaporation (MAPLE). Appl. Surf. Sci. 2007, 253, 6451–6455. [Google Scholar] [CrossRef]
- Trojanowicz, M. Enantioselective electrochemical sensors and biosensors: A mini-review. Electrochem. Comm. 2014, 38, 47–52. [Google Scholar] [CrossRef]
- Cardosi, M.; Haggett, B. Biosensor Devices. In Sensor Systems for Environmental Monitoring; Campbell, M., Ed.; Chapman & Hall: London, UK, 1997. [Google Scholar]
- Eckel, R.H. Lipoprotein lipase. A multifunctional enzyme relevant to common metabolic desease. New Engl. J. Med. 1989, 320, 1060–1068. [Google Scholar] [CrossRef] [PubMed]
- Nasratun, M.; Said, H.A.; Noraziah, A.; Alla, A.N.A. Immobilization of lipase from Candida Rugosa on chitosan beads for transesterification reaction. Am. J. Appl. Sci. 2009, 6, 1653–1657. [Google Scholar] [CrossRef]
- Veeramani, M.S.; Shyam, K.P.; Ratchagar, N.P.; Chadha, A.; Bhattacharya, E. Miniaturised silicon biosensors for the detection of triglyceride in blood serum. Anal. Meth. 2014, 6, 1728–1735. [Google Scholar] [CrossRef]
- Kullick, T.; Ulber, R.; Meyer, H.H.; Scheper, T.; Schugerl, K. Biosensors for enantioselective analysis. Anal. Chim. Acta 1994, 293, 271–276. [Google Scholar] [CrossRef]
- Pirozzi, D.; Fanelli, E.; Aronne, A.; Pernice, P.; Mingione, A. Lipase entrapment in a zirconia matrix: Sol–gel synthesis and catalytic properties. J. Mol. Catal. B Enzym. 2009, 59, 116–120. [Google Scholar] [CrossRef]
- Lee, D.H.; Park, C.H.; Yeo, J.M.; Kim, S.W. Lipase immobilization on silica gel using a cross-linking method. J. Ind. Eng. Chem. 2006, 12, 777–782. [Google Scholar]
- Zaidan, U.H.; Rahman, M.B.A.; Othman, S.S.; Basri, M.; Abdulmalek, E.; Rahman, R.N.Z.R.A.; Salleh, A.B. Biocatalytic production of lactose ester catalysed by mica-based immobilised lipase. Food Chem. 2012, 131, 199–205. [Google Scholar] [CrossRef]
- Petersen, M.T.N.; Fojan, P.; Petersen, S.B. How do lipases and esterases work: The electrostatic contribution. J. Biotechnol. 2001, 85, 115–147. [Google Scholar] [CrossRef] [PubMed]
- Malcata, F.X.; Reyes, H.R.; Garcia, H.S.; Hill, C.G., Jr.; Amundson, C.H. Immobilized lipase reactors for modification of fats and oils—A review. J. Am. Oil Chem. Soc. (JAOCS) 1990, 67, 890–910. [Google Scholar] [CrossRef]
- Bosley, J. Turning lipases into industrial biocatalyst. Biochem. Soc. Trans. 1997, 2, 174–178. [Google Scholar]
- Bosley, J.A.; Peilow, A.D. Immobilization of lipases on porous polypropylene: Reduction in esterification efficiency at low loading. J. Am. Oil Chem. Soc. (JAOCS) 1977, 74, 107–111. [Google Scholar] [CrossRef]
- Balcao, V.M.; Paiva, A.L.; Malcata, F.X. Bioreactors with immobilized lipases: State-of-the-art. Enzym. Microb. Technol. 1996, 18, 392–416. [Google Scholar] [CrossRef]
- Knežević, Z.D.; Šiler-Marinković, S.S.; Mojović, L.V. Immobilized lipases as practical catalysts. Acta Periodica Technologica 2004, 35, 151–164. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzym. Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Greer, J.A. Design challenges for matrix assisted pulsed laser evaporation and infrared resonant laser evaporation equipment. Appl. Phys. A 2011, 105, 661–671. [Google Scholar] [CrossRef]
- Foresti, M.L.; Ferreira, M.L. Frequent analytical/experimental problems in lipase-mediated synthesis in solvent-free systems and how to avoid them. Anal. Bioanal. Chem. 2005, 381, 1408–1425. [Google Scholar] [CrossRef] [PubMed]
- Aronne, A.; Bloisi, F.; Calabria, R.; Califano, V.; Fanelli, E.; Massoli, P.; Vicari, L.R.M. Structural characterization of MAPLE deposited lipase biofilm. Appl. Surf. Sci. 2014, in press. [Google Scholar]
- Gutiérrez-Llorente, A.; Horowitz, G.; Péres-Casero, R.; Perriére, J.; Fave, J.L.; Yassar, A.; Sant, C. Growth of polyalkylthiophene films by matrix assisted pulsed laser evaporation. Org. Electron. 2004, 5, 29–34. [Google Scholar] [CrossRef]
- Gupta, R.K.; Ghosh, K.; Kahol, P.K.; Yoon, J.; Guha, S. Pulsed laser thin film growth of di-octyl substituted polyfluorene and its co-polymers. Appl. Surf. Sci. 2008, 254, 7069–7073. [Google Scholar] [CrossRef]
- Bubb, D.M.; Corga, J.; Yi, S.Y.; Khan, M.; Hughes, L.; Gurudas, U.; Papantonakis, M.; McGill, R.A. An experimental investigation of inhomogeneities in resonant infrared matrix-assisted pulsed-laser deposited thin polymer films. Appl. Phys. A 2010, 100, 523–531. [Google Scholar] [CrossRef]
- Califano, V.; Bloisi, F.; Vicari, L.; Colombi, P.; Bontempi, E.; Depero, L.E. MAPLE deposition of biomaterial multilayers. Appl. Surf. Sci. 2008, 254, 7143–7148. [Google Scholar] [CrossRef]
- Natalello, A.; Ami, D.; Brocca, S.; Lotti, M.; Doglia, S.M. Secondary structure, conformational stability and glycosylation of a recombinant Candida rugosa lipase studied by Fourier-transform infrared spectroscopy. Biochem. J. 2005, 385, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Ranaldi, S.; Belle, V.; Rodriguez, J.; Guigliarelli, B.; Sturgis, J.; Carriere, F.; Fournel, A. Lid opening and unfolding in Human pancreatic lipase at low pH revealed by site-directed spin labeling EPR and FTIR spectroscopy. Biochemistry 2009, 48, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Inayat, A.; Schwieger, W.; Hartmann, M. Improved activity and stability of lipase immobilized in cage-like large pore mesoporous organosilicas. Microporous Mesoporous Mater. 2012, 154, 133–141. [Google Scholar] [CrossRef]
- Noinville, S.; Revault, M.; Baron, M.-H.; Tiss, A.; Yapoudjian, S.; Ivanova, M.; Verger, R. Conformational changes and orientation of Humicola lanuginosa lipase on a solid hydrophobic surface: An in situ interface Fourier transform infrared-attenuated total reflection study. Biophys. J. 2002, 82, 2709–2719. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Califano, V.; Bloisi, F.; Aronne, A.; Federici, S.; Nasti, L.; Depero, L.E.; Vicari, L.R.M. Biosensor Applications of MAPLE Deposited Lipase. Biosensors 2014, 4, 329-339. https://doi.org/10.3390/bios4040329
Califano V, Bloisi F, Aronne A, Federici S, Nasti L, Depero LE, Vicari LRM. Biosensor Applications of MAPLE Deposited Lipase. Biosensors. 2014; 4(4):329-339. https://doi.org/10.3390/bios4040329
Chicago/Turabian StyleCalifano, Valeria, Francesco Bloisi, Antonio Aronne, Stefania Federici, Libera Nasti, Laura E. Depero, and Luciano R. M. Vicari. 2014. "Biosensor Applications of MAPLE Deposited Lipase" Biosensors 4, no. 4: 329-339. https://doi.org/10.3390/bios4040329






