Automatic and Accurate Sleep Stage Classification via a Convolutional Deep Neural Network and Nanomembrane Electrodes
Abstract
:1. Introduction
2. Materials and Methods
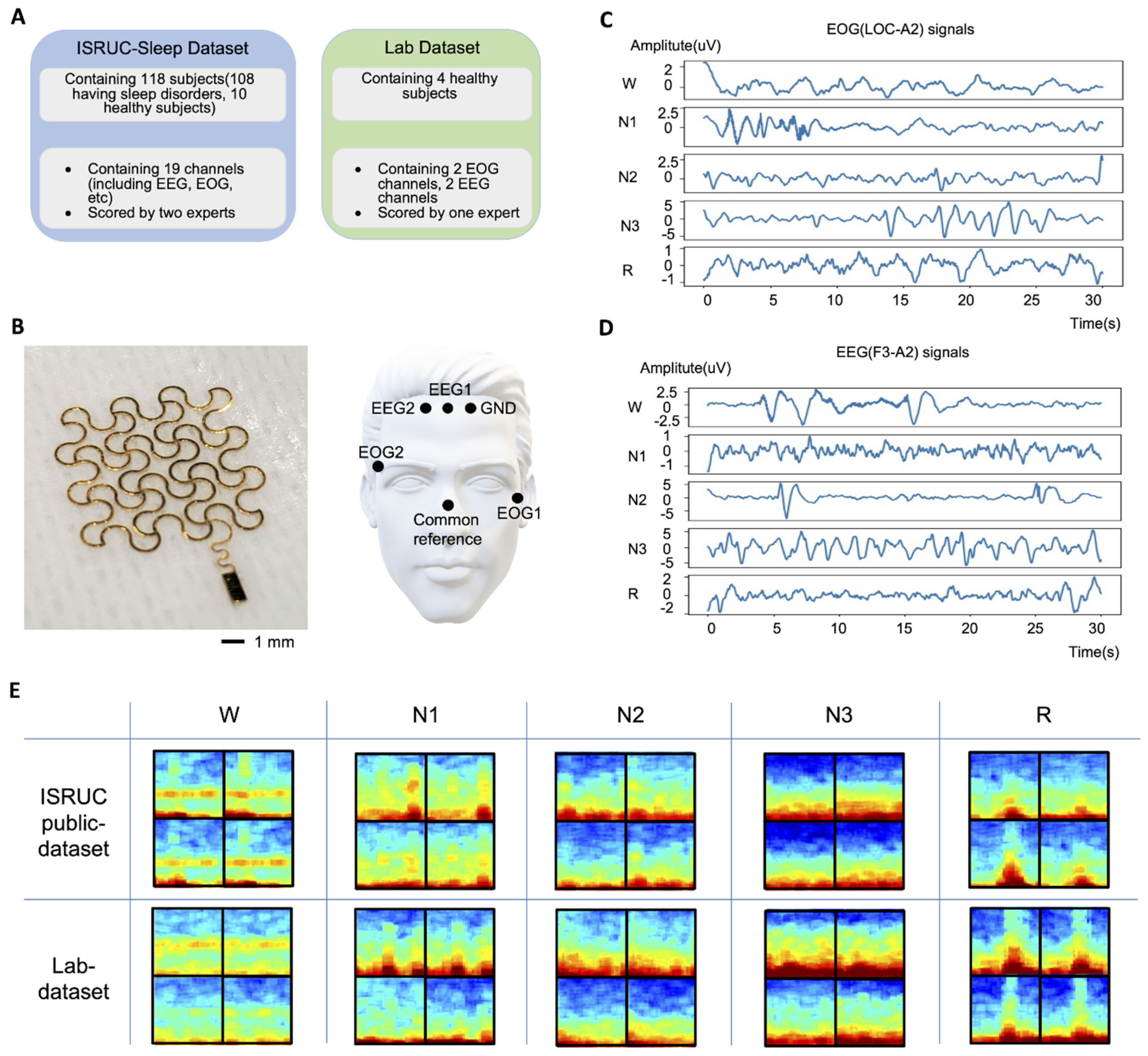
2.1. ISRUC Public Dataset
2.2. Measured Lab Dataset
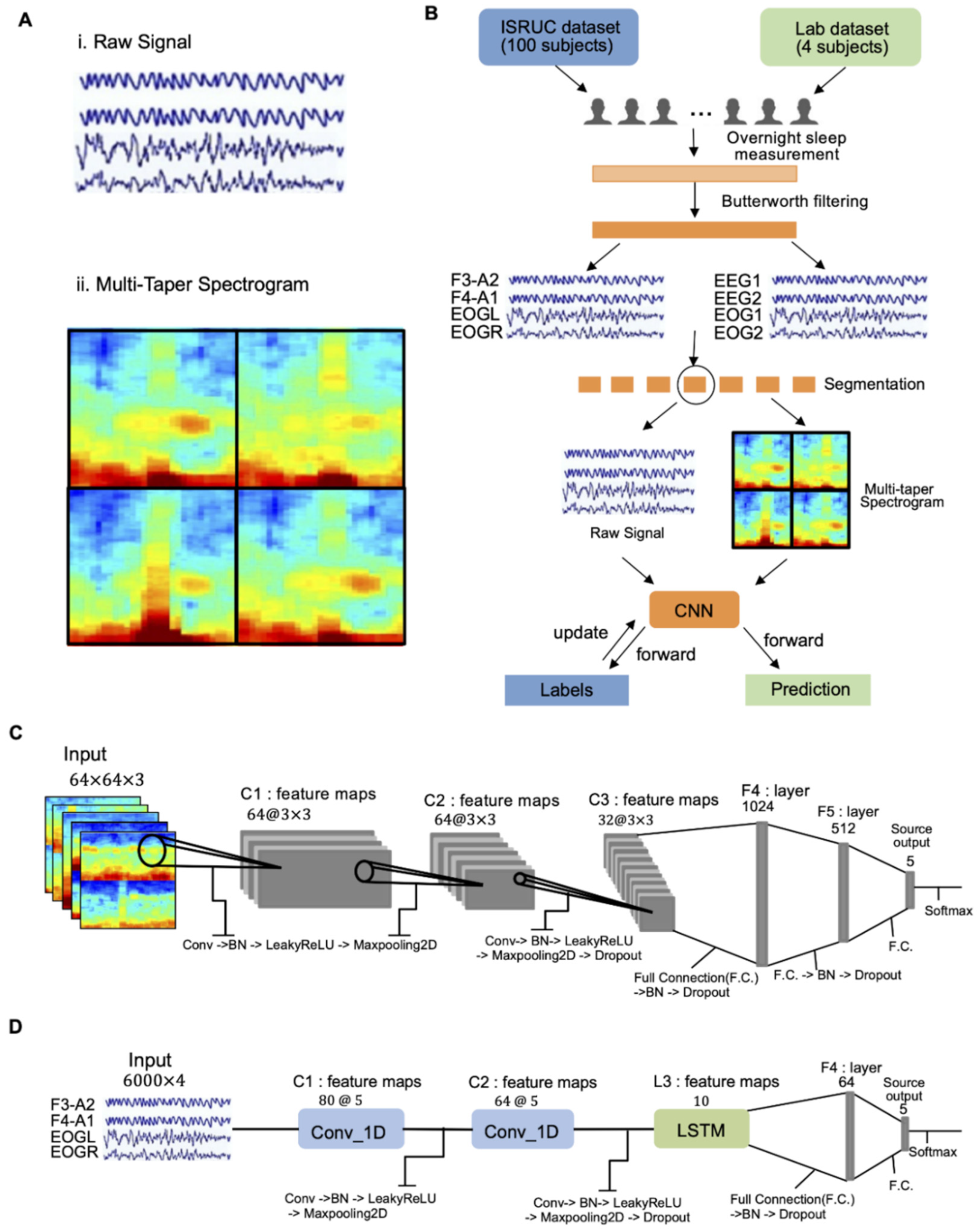
2.3. Data Pre-Processing
2.4. Input Dataset for Deep Learning
2.5. CNN-Based Classifier
3. Results and Discussion
3.1. Experimental Setup
3.2. Performance Comparison with Other Works
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rahman, M.M.; Bhuiyan, M.I.H.; Hassan, A.R. Sleep stage classification using single-channel EOG. Comput. Biol. Med. 2018, 102, 211–220. [Google Scholar] [CrossRef]
- Kim, H.; Kwon, S.; Kwon, Y.-T.; Yeo, W.-H. Soft Wireless Bioelectronics and Differential Electrodermal Activity for Home Sleep Monitoring. Sensors 2021, 21, 354. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.; Kim, H.; Yeo, W.-H. Recent advances in wearable sensors and portable electronics for sleep monitoring. Iscience 2021, 24, 102461. [Google Scholar] [CrossRef] [PubMed]
- Armon, C. Polysomnography. Medscape 2020, 31, 281–297. [Google Scholar]
- Zhu, G.; Li, Y.; Wen, P.P. Analysis and classification of sleep stages based on difference visibility graphs from a single-channel EEG signal. IEEE J. Biomed. Health Inform. 2014, 18, 1813–1821. [Google Scholar] [CrossRef]
- Ghimatgar, H.; Kazemi, K.; Helfroush, M.S.; Aarabi, A. An automatic single-channel EEG-based sleep stage scoring method based on hidden Markov Model. J. Neurosci. Methods 2019, 324, 108320. [Google Scholar] [CrossRef]
- Tzimourta, K.D.; Tsilimbaris, A.; Tzioukalia, K.; Tzallas, A.T.; Tsipouras, M.G.; Astrakas, L.G.; Giannakeas, N. EEG-based automatic sleep stage classification. Biomed. J. 2018, 1, 6. [Google Scholar]
- Khalighi, S.; Sousa, T.; Santos, J.M.; Nunes, U. ISRUC-Sleep: A comprehensive public dataset for sleep researchers. Comput. Methods Programs Biomed. 2016, 124, 180–192. [Google Scholar] [CrossRef]
- Lim, H.R.; Kim, H.S.; Qazi, R.; Kwon, Y.T.; Jeong, J.W.; Yeo, W.H. Advanced soft materials, sensor integrations, and applications of wearable flexible hybrid electronics in healthcare, energy, and environment. Adv. Mater. 2020, 32, 1901924. [Google Scholar] [CrossRef]
- Herbert, R.; Kim, J.-H.; Kim, Y.S.; Lee, H.M.; Yeo, W.-H. Soft material-enabled, flexible hybrid electronics for medicine, healthcare, and human-machine interfaces. Materials 2018, 11, 187. [Google Scholar] [CrossRef] [Green Version]
- Kwon, S.; Kwon, Y.-T.; Kim, Y.-S.; Lim, H.-R.; Mahmood, M.; Yeo, W.-H. Skin-conformal, soft material-enabled bioelectronic system with minimized motion artifacts for reliable health and performance monitoring of athletes. Biosens. Bioelectron. 2020, 151, 111981. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-S.; Mahmood, M.; Kwon, S.; Herbert, R.; Yeo, W.-H. Wireless Stretchable Hybrid Electronics for Smart, Connected, and Ambulatory Monitoring of Human Health. In Proceedings of the Meeting Abstracts; IOP Publishing: Bristol, UK, 2019; p. 2293. [Google Scholar]
- George, N.; Jemel, B.; Fiori, N.; Renault, B. Face and shape repetition effects in humans: A spatio-temporal ERP study. NeuroReport 1997, 8, 1417–1422. [Google Scholar] [CrossRef] [PubMed]
- Teplan, M. Fundamentals of EEG measurement. Meas. Sci. Rev. 2002, 2, 1–11. [Google Scholar]
- Yao, D.; Qin, Y.; Hu, S.; Dong, L.; Vega, M.L.B.; Sosa, P.A.V. Which reference should we use for EEG and ERP practice? Brain Topogr. 2019, 32, 530–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Regan, S.; Faul, S.; Marnane, W. Automatic detection of EEG artefacts arising from head movements. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, Buenos Aires, Argentina, 31 August–4 September 2010; pp. 6353–6356. [Google Scholar]
- Norton, J.J.; Lee, D.S.; Lee, J.W.; Lee, W.; Kwon, O.; Won, P.; Jung, S.-Y.; Cheng, H.; Jeong, J.-W.; Akce, A. Soft, curved electrode systems capable of integration on the auricle as a persistent brain–computer interface. Proc. Natl. Acad. Sci. USA 2015, 112, 3920–3925. [Google Scholar] [CrossRef] [Green Version]
- Mahmood, M.; Mzurikwao, D.; Kim, Y.-S.; Lee, Y.; Mishra, S.; Herbert, R.; Duarte, A.; Ang, C.S.; Yeo, W.-H. Fully portable and wireless universal brain–machine interfaces enabled by flexible scalp electronics and deep learning algorithm. Nat. Mach. Intell. 2019, 1, 412–422. [Google Scholar] [CrossRef] [Green Version]
- Tian, L.; Zimmerman, B.; Akhtar, A.; Yu, K.J.; Moore, M.; Wu, J.; Larsen, R.J.; Lee, J.W.; Li, J.; Liu, Y. Large-area MRI-compatible epidermal electronic interfaces for prosthetic control and cognitive monitoring. Nat. Biomed. Eng. 2019, 3, 194–205. [Google Scholar] [CrossRef]
- Mahmood, M.; Kwon, S.; Berkmen, G.K.; Kim, Y.-S.; Scorr, L.; Jinnah, H.; Yeo, W.-H. Soft nanomembrane sensors and flexible hybrid bioelectronics for wireless quantification of blepharospasm. IEEE Trans. Biomed. Eng. 2020, 67, 3094–3100. [Google Scholar] [CrossRef]
- Zavanelli, N.; Kim, H.; Kim, J.; Herbert, R.; Mahmood, M.; Kim, Y.-S.; Kwon, S.; Bolus, N.B.; Torstrick, F.B.; Lee, C.S. At-home wireless monitoring of acute hemodynamic disturbances to detect sleep apnea and sleep stages via a soft sternal patch. Sci. Adv. 2021, 7, eabl4146. [Google Scholar] [CrossRef]
- Kandel, E.R.; Schwartz, J.H.; Jessell, T.M.; Siegelbaum, S.; Hudspeth, A.J.; Mack, S. Principles of Neural Science; McGraw-Hill: New York, NY, USA, 2000; Volume 4. [Google Scholar]
- Vrbancic, G.; Podgorelec, V. Automatic classification of motor impairment neural disorders from EEG signals using deep convolutional neural networks. Elektron. Ir Elektrotechnika 2018, 24, 3–7. [Google Scholar] [CrossRef]
- Kuanar, S.; Athitsos, V.; Pradhan, N.; Mishra, A.; Rao, K.R. Cognitive analysis of working memory load from EEG, by a deep recurrent neural network. In Proceedings of the 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Calgary, AB, Canada, 15–20 April 2018; pp. 2576–2580. [Google Scholar]
- Vilamala, A.; Madsen, K.H.; Hansen, L.K. Deep convolutional neural networks for interpretable analysis of EEG sleep stage scoring. In Proceedings of the 2017 IEEE 27th international workshop on machine learning for signal processing (MLSP), Tokyo, Japan, 25–28 September 2017; pp. 1–6. [Google Scholar]
- Jiao, Z.; Gao, X.; Wang, Y.; Li, J.; Xu, H. Deep convolutional neural networks for mental load classification based on EEG data. Pattern Recognit. 2018, 76, 582–595. [Google Scholar] [CrossRef]
- Prerau, M.J.; Brown, R.E.; Bianchi, M.T.; Ellenbogen, J.M.; Purdon, P.L. Sleep neurophysiological dynamics through the lens of multitaper spectral analysis. Physiology 2017, 32, 60–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, Y.-T.; Kim, Y.-S.; Kwon, S.; Mahmood, M.; Lim, H.-R.; Park, S.-W.; Kang, S.-O.; Choi, J.J.; Herbert, R.; Jang, Y.C. All-printed nanomembrane wireless bioelectronics using a biocompatible solderable graphene for multimodal human-machine interfaces. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.W.; Yeo, W.H.; Akhtar, A.; Norton, J.J.; Kwack, Y.J.; Li, S.; Jung, S.Y.; Su, Y.; Lee, W.; Xia, J. Materials and optimized designs for human-machine interfaces via epidermal electronics. Adv. Mater. 2013, 25, 6839–6846. [Google Scholar] [CrossRef] [PubMed]
- Bresch, E.; Großekathöfer, U.; Garcia-Molina, G. Recurrent deep neural networks for real-time sleep stage classification from single channel EEG. Front. Comput. Neurosci. 2018, 12, 85. [Google Scholar] [CrossRef] [Green Version]
- Schaltenbrand, N.; Lengelle, R.; Macher, J.-P. Neural network model: Application to automatic analysis of human sleep. Comput. Biomed. Res. 1993, 26, 157–171. [Google Scholar] [CrossRef]
- Anderer, P.; Gruber, G.; Parapatics, S.; Woertz, M.; Miazhynskaia, T.; Klösch, G.; Saletu, B.; Zeitlhofer, J.; Barbanoj, M.J.; Danker-Hopfe, H. An E-health solution for automatic sleep classification according to Rechtschaffen and Kales: Validation study of the Somnolyzer 24 × 7 utilizing the Siesta database. Neuropsychobiology 2005, 51, 115–133. [Google Scholar] [CrossRef]
- Tsinalis, O.; Matthews, P.M.; Guo, Y.; Zafeiriou, S. Automatic sleep stage scoring with single-channel EEG using convolutional neural networks. arXiv 2016, arXiv:1610.01683. [Google Scholar]
- Tsinalis, O.; Matthews, P.M.; Guo, Y. Automatic sleep stage scoring using time-frequency analysis and stacked sparse autoencoders. Ann. Biomed. Eng. 2016, 44, 1587–1597. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Supratak, A.; Pan, W.; Wu, C.; Matthews, P.M.; Guo, Y. Mixed neural network approach for temporal sleep stage classification. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 26, 324–333. [Google Scholar] [CrossRef] [Green Version]
- Supratak, A.; Dong, H.; Wu, C.; Guo, Y. DeepSleepNet: A model for automatic sleep stage scoring based on raw single-channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 1998–2008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambon, S.; Galtier, M.N.; Arnal, P.J.; Wainrib, G.; Gramfort, A. A deep learning architecture for temporal sleep stage classification using multivariate and multimodal time series. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 758–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malafeev, A.; Laptev, D.; Bauer, S.; Omlin, X.; Wierzbicka, A.; Wichniak, A.; Jernajczyk, W.; Riener, R.; Buhmann, J.; Achermann, P. Automatic human sleep stage scoring using deep neural networks. Front. Neurosci. 2018, 12, 781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sors, A.; Bonnet, S.; Mirek, S.; Vercueil, L.; Payen, J.-F. A convolutional neural network for sleep stage scoring from raw single-channel EEG. Biomed. Signal Process. Control 2018, 42, 107–114. [Google Scholar] [CrossRef]
- Stephansen, J.B.; Olesen, A.N.; Olsen, M.; Ambati, A.; Leary, E.B.; Moore, H.E.; Carrillo, O.; Lin, L.; Han, F.; Yan, H. Neural network analysis of sleep stages enables efficient diagnosis of narcolepsy. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Cui, Z.; Zheng, X.; Shao, X.; Cui, L. Automatic sleep stage classification based on convolutional neural network and fine-grained segments. Complexity 2018, 2018. [Google Scholar] [CrossRef] [Green Version]
- Yildirim, O.; Baloglu, U.B.; Acharya, U.R. A deep learning model for automated sleep stages classification using PSG signals. Int. J. Environ. Res. Public Health 2019, 16, 599. [Google Scholar] [CrossRef] [Green Version]
- Phan, H.; Andreotti, F.; Cooray, N.; Chén, O.Y.; De Vos, M. Joint classification and prediction CNN framework for automatic sleep stage classification. IEEE Trans. Biomed. Eng. 2018, 66, 1285–1296. [Google Scholar] [CrossRef]
- Shen, H.; Ran, F.; Xu, M.; Guez, A.; Li, A.; Guo, A. An Automatic Sleep Stage Classification Algorithm Using Improved Model Based Essence Features. Sensors 2020, 20, 4677. [Google Scholar] [CrossRef]
- Lee, T.; Hwang, J.; Lee, H. Trier: Template-guided neural networks for robust and interpretable sleep stage identification from eeg recordings. arXiv 2020, arXiv:2009.05407. [Google Scholar]
- Zhang, X.; Xu, M.; Li, Y.; Su, M.; Xu, Z.; Wang, C.; Kang, D.; Li, H.; Mu, X.; Ding, X. Automated multi-model deep neural network for sleep stage scoring with unfiltered clinical data. Sleep Breath. 2020, 24, 581–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Essl, M.; Rappelsberger, P. EEG cohererence and reference signals: Experimental results and mathematical explanations. Med. Biol. Eng. Comput. 1998, 36, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, L.T.; Peterson, M.A.; Kaszniak, A.W.; Allen, J.J. EEG phase synchrony differences across visual perception conditions may depend on recording and analysis methods. Clin. Neurophysiol. 2005, 116, 172–189. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.; Afghah, F.; Acharya, U.R. SleepEEGNet: Automated sleep stage scoring with sequence to sequence deep learning approach. PLoS ONE 2019, 14, e0216456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melek, M.; Manshouri, N.; Kayikcioglu, T. An automatic EEG-based sleep staging system with introducing NAoSP and NAoGP as new metrics for sleep staging systems. Cogn. Neurodynamics 2021, 15, 405–423. [Google Scholar] [CrossRef]

| Input Type | Number of Epochs (ISRUC Public Dataset) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Training Set | Validation Set | Test Set | |||||||||||||
| Aw | N1 | N2 | N3 | R | Aw | N1 | N2 | N3 | R | Aw | N1 | N2 | N3 | R | |
| Raw signal | 10,968 | 3299 | 13,366 | 8264 | 6197 | 3591 | 1148 | 4570 | 2692 | 2031 | 3617 | 1065 | 4492 | 2739 | 2119 |
| Spectrogram | 11,013 | 3275 | 13,210 | 8189 | 6207 | 3568 | 1073 | 4729 | 2675 | 1987 | 3575 | 1144 | 4469 | 2791 | 2053 |
| Input Type | ISRUC Public Dataset | |
|---|---|---|
| Test Accuracy | Cohen’s Kappa | |
| Raw signal | 87.05% | 0.829 |
| Multi-taper spectrogram | 88.85% | 0.854 |
| Input Type | Lab Dataset | Number of Epochs (Lab Dataset) | |||||
|---|---|---|---|---|---|---|---|
| Prediction Accuracy | Cohen’s Kappa | Prediction Set | |||||
| Aw | N1 | N2 | N3 | R | |||
| Raw signal | 72.94% | 0.608 | 230 | 111 | 1091 | 721 | 554 |
| Multi-taper spectrogram | 81.52% | 0.734 | 230 | 111 | 1091 | 721 | 554 |
| Ref. | Year | Data Type | Input Data | Number of Subjects | Public Dataset | Private Dataset | Number of Channels | Classification Method |
|---|---|---|---|---|---|---|---|---|
| Accuracy (%) /Kappa | Accuracy (%) /Kappa | |||||||
| This work | 2022 | ISRUC and Lab dataset | Multi-taper spectrogram and Raw data | 100 | 88.85/0.854 87.05/0.829 | 81.52/0.734 72.94/0.608 | 2 EEG, 2 EOG | CNN |
| [31] | 1993 | Private data | Extracted features | 12 | - | 80.60/- | 2 EEG, 1 EOG, 1 EMG | Multilayer Neural Network |
| [32] | 2005 | SIESTA | Extracted features | 590 | 79.6/0.72 | - | 1 EEG, 2 EOG, 1 EMG | LDA, Decision tree |
| [5] | 2014 | Sleep-EDF | Extracted features | 1 | 88.9/- | - | 1 EEG | SVM |
| [33] | 2016 | Sleep-EDF | Raw data | 20 | 74/0.65 | - | 1 EEG | CNN |
| [34] | 2016 | Sleep-EDF | Extracted features | 20 | 78/- | - | 1 EEG | Stacked Sparse Autoencoders |
| [35] | 2017 | Montreal archive | Extracted features | 62 | 83.35/- | - | 1 EEG | Mixed Neural Network |
| [36] | 2017 | Sleep-EDF & Montreal | Raw data | 32 | 86.2/0.80 | - | 1 EEG | DeepSleepNET (CNN + LSTM) |
| [37] | 2018 | Montreal archive | Raw data | 61 | 78/0.80 | - | 6 EEG, 2 EOG, 3 EMG | Multivariate Network |
| [38] | 2018 | Private dataset | Extracted features | 76 | - | -/0.8 | 1 EEG, 2 EOG | Random Forest, CNN, LSTM |
| [39] | 2018 | SHHS | Raw data | 5728 | 87/0.81 | - | 1 EEG | CNN |
| [40] | 2018 | 12 sleep centers | Raw data | 1086 | 87/0.766 | - | 4 EEG, 2 EOG, 1 EMG | CNN |
| [7] | 2018 | ISRUC | Extracted features | 100 | 75.29/- | - | 6 EEG | Random Forest |
| [41] | 2018 | ISRUC | Raw data | 116 | 92.2/- | - | 6 EEG, 2 EOG, 3 EMG | CNN |
| [30] | 2018 | SIESTA/private data | Raw data | 147 | -/0.760 | -/0.703 | 1 EEG, 2 EOG | RNN |
| [6] | 2019 | ISRUC | Extracted features | 10 | 79.64/0.74 | - | 6 EEG | HMM |
| [42] | 2019 | Sleep-EDF | Raw data | 61 | 91.22/- | - | 1 EEG, 1 EOG | CNN |
| [43] | 2019 | Montreal archive | Extracted features | 200 | 83.6/- | - | 1 EEG, 1EOG, 1EMG | CNN |
| [44] | 2020 | ISRUC | Extracted features | 10 | 81.65/0.76 | - | 1 EEG | IMBEFs |
| [45] | 2020 | Sleep-EDF | Raw data | 100 | 85.52/- | - | 2 EEG | CNN |
| [46] | 2020 | ISRUC | Raw data | 294 | 81.8/0.72 | - | 2 EEG, 2 EOG, 1 EMG, 1 ECG | CNN + RNN |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, K.; Kwon, S.; Yeo, W.-H. Automatic and Accurate Sleep Stage Classification via a Convolutional Deep Neural Network and Nanomembrane Electrodes. Biosensors 2022, 12, 155. https://doi.org/10.3390/bios12030155
Kwon K, Kwon S, Yeo W-H. Automatic and Accurate Sleep Stage Classification via a Convolutional Deep Neural Network and Nanomembrane Electrodes. Biosensors. 2022; 12(3):155. https://doi.org/10.3390/bios12030155
Chicago/Turabian StyleKwon, Kangkyu, Shinjae Kwon, and Woon-Hong Yeo. 2022. "Automatic and Accurate Sleep Stage Classification via a Convolutional Deep Neural Network and Nanomembrane Electrodes" Biosensors 12, no. 3: 155. https://doi.org/10.3390/bios12030155
APA StyleKwon, K., Kwon, S., & Yeo, W.-H. (2022). Automatic and Accurate Sleep Stage Classification via a Convolutional Deep Neural Network and Nanomembrane Electrodes. Biosensors, 12(3), 155. https://doi.org/10.3390/bios12030155






