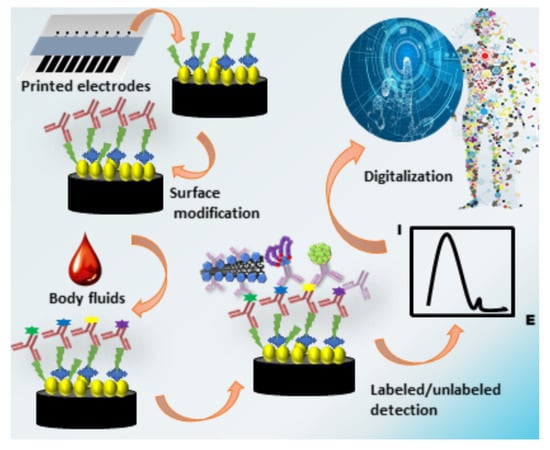
Printed Electrodes in Microfluidic Arrays for Cancer Biomarker Protein Detection
Abstract
:1. Introduction
2. Immunoassay Techniques for Printed Electrodes
2.1. Immunoassay Protocol
2.2. Ultrasensitive Detection
2.2.1. Dissolvable Nanoparticles
2.2.2. Multi-Enzyme Conjugates
2.3. Label-Free Detection
3. Screen Printing
3.1. Nanoparticle Surface Coatings of SPEs
3.2. SPEs and Molecular Imprinting
3.3. SPE in μPADs
4. Inkjet Printing
5. Challenges for Printed Electrodes in Microfluidic Assays
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Cuntries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Dixit, C.K.; Kadimisetty, K.; Otieno, B.A.; Tang, C.; Malla, S.; Krause, C.E.; Rusling, J.F. Electrochemistry-Based Approaches to Low Cost, High Sensitivity, Automated, Multiplexed Protein Immunoassays for Cancer Diagnostics. Analyst 2016, 141, 536–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalia, M. Biomarkers for Personalized Oncology: Recent Advances and Future Challenges. Metabolism 2015, 64, S16–S21. [Google Scholar] [CrossRef] [PubMed]
- Borrebaeck, C.A.K. Precision Diagnostics: Moving towards Protein Biomarker Signatures of Clinical Utility in Cancer. Nat. Rev. Cancer 2017, 17, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Rusling, J.F.; Kumar, C.V.; Gutkind, J.S.; Patel, V. Measurement of Biomarker Proteins for Point-of-Care Early Detection and Monitoring of Cancer. Analyst 2010, 135, 2496–2511. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Hassan, E.M.; DeRosa, M.C. Recent Advances in Cancer Early Detection and Diagnosis: Role of Nucleic Acid Based Aptasensors. TrAC—Trends Anal. Chem. 2020, 124, 115806. [Google Scholar] [CrossRef]
- Rusling, J.F. Multiplexed Electrochemical Protein Detection and Translation to Personalized Cancer Diagnostics. Anal. Chem. 2013, 85, 5304–5310. [Google Scholar] [CrossRef] [Green Version]
- Rusling, J.F.; Bishop, G.W.; Doan, N.M.; Papadimitrakopoulos, F. Nanomaterials and Biomaterials in Electrochemical Arrays for Protein Detection. J. Mater. Chem. B 2014, 2, 12–30. [Google Scholar] [CrossRef]
- Munge, B.S.; Stracensky, T.; Gamez, K.; DiBiase, D.; Rusling, J.F. Multiplex Immunosensor Arrays for Electrochemical Detection of Cancer Biomarker Proteins. Electroanalysis 2016, 28, 2644–2658. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, J.A.; Weinstein, J.N. Biomarkers in Cancer Staging, Prognosis and Treatment Selection. Nat. Rev. Cancer 2005, 5, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Jayanthi, V.S.P.K.S.A.; Das, A.B.; Saxena, U. Recent Advances in Biosensor Development for the Detection of Cancer Biomarkers. Biosens. Bioelectron. 2017, 91, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Füzéry, A.K.; Levin, J.; Chan, M.M.; Chan, D.W. Translation of Proteomic Biomarkers into FDA Approved Cancer Diagnostics: Issues and Challenges. Clin. Proteom. 2013, 10, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertok, T.; Lorencova, L.; Chocholova, E.; Jane, E.; Vikartovska, A.; Kasak, P.; Tkac, J. Electrochemical Impedance Spectroscopy Based Biosensors: Mechanistic Principles, Analytical Examples and Challenges towards Commercialization for Assays of Protein Cancer Biomarkers. ChemElectroChem 2019, 6, 989–1003. [Google Scholar] [CrossRef] [Green Version]
- Duffy, D. Short Keynote Paper: Single Molecule Detection of Protein Biomarkers to Define the Continuum from Health to Disease. IEEE J. Biomed. Health Inform. 2020, 24, 1864–1868. [Google Scholar] [CrossRef]
- Dhanapala, L.; Jones, A.L.; Czarnecki, P.; Rusling, J.F. Sub-Zeptomole Detection of Biomarker Proteins Using a Microfluidic Immunoarray with Nanostructured Sensors. Anal. Chem 2020, 92, 8021–8025. [Google Scholar] [CrossRef]
- Duffy, D.C.; Walt, D.R. Protein Detection by Counting Molecules. Clin. Chem. 2019, 65, 809–810. [Google Scholar] [CrossRef]
- Wu, C.; Maley, A.M.; Walt, D.R. Single-Molecule Measurements in Microwells for Clinical Applications. Crit. Rev. Clin. Lab. Sci. 2020, 57, 270–290. [Google Scholar] [CrossRef]
- MSD Assays on Meso Scale Discovery Platform (MSD-E). Available online: https://www.bioagilytix.com/meso-scale-discovery-electrochemiluminescence (accessed on 22 June 2020).
- Luminex Assays | Thermo Fisher Scientific US. Available online: https://www.thermofisher.com/us/en/home/life/science/antibodies/immunoassays/procartaplex-assays-luminex.html (accessed on 22 June 2020).
- Akkilic, N.; Geschwindner, S.; Höök, F. Single-Molecule Biosensors: Recent Advances and Applications. Biosens. Bioelectron. 2020, 151, 111944. [Google Scholar] [CrossRef]
- Jin, W.; Tang, Q.; Wan, M.; Cui, K.; Zhang, Y.; Ren, G.; Ni, B.; Sklar, J.; Przytycka, T.M.; Childs, R.; et al. Genome-Wide Detection of DNase i Hypersensitive Sites in Single Cells and FFPE Tissue Samples. Nature 2015, 528, 142–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, A.; Dhanapala, L.; Kankanamage, R.N.T.; Kumar, C.V.; Rusling, J.F. Multiplexed Immunosensors and Immunoarrays. Anal. Chem. 2020, 92, 345–362. [Google Scholar] [CrossRef] [PubMed]
- Arduini, F.; Micheli, L.; Moscone, D.; Palleschi, G.; Piermarini, S.; Ricci, F.; Volpe, G. Electrochemical Biosensors Based on Nanomodified Screen-Printed Electrodes: Recent Applications in Clinical Analysis. TrAC—Trends in Anal. Chem. 2016, 79, 114–126. [Google Scholar] [CrossRef] [Green Version]
- Dorothee Grieshaber, R.M.J.V.E.R. Electrochemical Biosensors—Sensor Principles and Architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Wang, X.; Gao, D.; Li, M.; Li, H.; Li, C.; Wu, X.; Yang, B. CVD graphene as an electrochemical sensing platform for simultaneous detection of biomolecules. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Lu, L.; Gunasekaran, S. Dual-channel ITO-microfluidic electrochemical immunosensor for simultaneous detection of two mycotoxins. Talanta 2019, 194, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Stromberg, L.R.; Hondred, J.A.; Sanborn, D.; Mendivelso-Perez, D.; Ramesh, S.; Rivero, I.V.; Kogot, J.; Smith, E.; Gomes, C.; Claussen, J.C. Stamped multilayer graphene laminates for disposable in-field electrodes: Application to electrochemical sensing of hydrogen peroxide and glucose. Microchim. Acta 2019, 186, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Rama, E.C.; Costa-García, A. Screen-printed Electrochemical Immunosensors for the Detection of Cancer and Cardiovascular Biomarkers. Electroanalysis 2016, 28, 1700–1715. [Google Scholar] [CrossRef]
- Jensen, G.C.; Krause, C.E.; Sotzing, G.A.; Rusling, J.F. Inkjet-printed gold nanoparticle electrochemical arrays on plastic. Application to immunodetection of a cancer biomarker protein. Phys. Chem. Chem. Phys. 2011, 13, 4888–4894. [Google Scholar] [CrossRef]
- Sui, Y.; Zorman, C.A. Review—Inkjet Printing of Metal Structures for Electrochemical Sensor Applications. J. Electrochem. Soc. 2020, 167, 037571. [Google Scholar] [CrossRef]
- Kim, D.; Lee, S.H.; Jeong, S.H.; Moon, J. All-Ink-Jet Printed Flexible Organic Thin-Film Transistors on Plastic Substrates. Electrochem. Solid State Lett. 2009, 12, H195–H197. [Google Scholar] [CrossRef]
- Taleat, Z.; Khoshroo, A.; Mazloum-Ardakani, M. Screen-printed electrodes for biosensing: A review (2008–2013). Microchim. Acta 2014, 181, 865–891. [Google Scholar] [CrossRef]
- Tao, R.; Ning, H.; Chen, J.; Zou, J.; Fang, Z.; Yang, C.; Zhou, Y.; Zhang, J.; Yao, R.; Peng, J. Inkjet Printed Electrodes in Thin Film Transistors. IEEE J. Electron. Devices Soc. 2018, 6, 774–790. [Google Scholar] [CrossRef]
- Beitollahi, H.; Mohammadi, S.Z.; Safaei, M.; Tajik, S. Applications of Electrochemical Sensors and Biosensors Based on Modified Screen-Printed Electrodes: A Review. Anal. Methods 2020, 12, 1547–1560. [Google Scholar] [CrossRef]
- Heineman, W.R.; Halsall, H.B. Strategies for electrochemical immunoassay. Anal. Chem. 1985, 75, 1321A–1331A. [Google Scholar]
- Lopez, G.A.; Estevez, M.C.; Soler, M.; Lechuga, L.M. Recent Advances in Nanoplasmonic Biosensors: Applications and Lab-on-a-Chip Integration. Nanophotonics 2017, 6, 123–136. [Google Scholar] [CrossRef]
- Song, Y.; Lin, B.; Tian, T.; Xu, X.; Wang, W.; Ruan, Q.; Guo, J.; Zhu, Z.; Yang, C. Recent Progress in Microfluidics-Based Biosensing. Anal. Chem. 2019, 91, 388–404. [Google Scholar] [CrossRef]
- Reverté, L.; Prieto-Simón, B.; Campàs, M. New Advances in Electrochemical Biosensors for the Detection of Toxins: Nanomaterials, Magnetic Beads and Microfluidics Systems. A Review. Anal. Chim. Acta 2016, 908, 8–21. [Google Scholar] [CrossRef]
- Pandey, C.M.; Augustine, S.; Kumar, S.; Kumar, S.; Nara, S.; Srivastava, S.; Malhotra, B.D. Microfluidics Based Point-of-Care Diagnostics. Biotechnol. J. 2018, 13, 1700047. [Google Scholar] [CrossRef]
- Lvov, Y.M.; Lu, Z.; Schenkman, J.B.; Rusling, J.F. Direct Electrochemistry of Myoglobin and Cytochrome P450cam in Alternate Polyion Layer-by-Layer Films with DNA and other polyions. J. Am. Chem. Soc. 1998, 120, 4073–4080. [Google Scholar] [CrossRef]
- Rusling, J.F. Electroactive and Enzyme-Active Protein-Polyion Films Assembled Layer-by-Layer. In Protein Architecture: Interfacing Molecular Assemblies and Immobilization Biotechnology; Marcel Dekker: New York, NY, USA, 2000; pp. 337–354. [Google Scholar]
- Tang, Z.; Wang, Y.; Podsiadlo, P.; Kotov, N.A. Biomedical Applications of Layer-by-Layer Assembly: From Biomimetics to Tissue Engineering. Adv. Mater. 2006, 18, 3203–3224. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Xu, J.J.; Chen, H.Y. Electrochemical Biosensors Based on Layer-by-Layer Assemblies. Electroanalysis 2006, 18, 1737–1748. [Google Scholar] [CrossRef]
- Koniev, O.; Wagner, A. Developments and Recent Advancements in the Field of Endogenous Amino Acid Selective Bond Forming Reactions for Bioconjugation. Chem. Soc. Rev. 2015, 44, 5495–5551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharafeldin, M.; McCaffrey, K.; Rusling, J.F. Influence of Antibody Immobilization Strategy on Carbon Electrode Immunoarrays. Analyst 2019, 144, 5108–5116. [Google Scholar] [CrossRef]
- Yang, H. Enzyme-Based Ultrasensitive Electrochemical Biosensors. Curr. Opin. Chem. Biol. 2012, 16, 422–428. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Sly, K.L.; Conboy, J.C. Comparison of the Energetics of Avidin, Streptavidin, NeutrAvidin, and Anti-Biotin Antibody Binding to Biotinylated Lipid Bilayer Examined by Second-Harmonic Generation. Anal. Chem. 2012, 84, 201–208. [Google Scholar] [CrossRef]
- Pierce, T.M. Antibody Biotinylation Kit for IP. Available online: https://www.thermofisher.com/order/catalog/product/90407#/90407 (accessed on 30 June 2020).
- Udeshi, N.D.; Pedram, K.; Svinkina, T.; Fereshetian, S.; Myers, S.A.; Aygun, O.; Krug, K.; Clauser, K.; Ryan, D.; Ast, T.; et al. Antibodies to Biotin Enable Large-Scale Detection of Biotinylation Sites on Proteins. Nat. Methods 2017, 14, 1167–1170. [Google Scholar] [CrossRef]
- Jain, A.; Cheng, K. The principles and applications of avidin-based nanoparticles in drug delivery and diagnosis. J. Control. Release 2017, 245, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Jain, A.; Barve, A.; Zhao, Z.; Jin, W.; Cheng, K. Comparison of avidin, neutravidin, and streptavidin as nanocarriers for efficient siRNA Delivery. Mol. Pharm. 2017, 14, 1517–1527. [Google Scholar] [CrossRef]
- Avidin and Streptavidin Conjugates—Section 7.6 | Thermo Fisher Scientific—US. Available online: https://www.thermofisher.com/us/en/home/references/molecular-probes-the-handbook/antibodies-avidins-lectins-and-related-products/avidin-streptavidin-neutravidin-and-captavidin-biotin-binding-proteins-and-affinity-matrices.html (accessed on 30 June 2020).
- Orelma, H.; Johansson, L.S.; Filpponen, I.; Rojas, O.J.; Laine, J. Generic Method for Attaching Biomolecules via Avidin—Biotin Complexes Immobilized on Films of Regenerated and Nanofibrillar Cellulose. Biomacromolecules 2012, 13, 2802–2810. [Google Scholar] [CrossRef]
- Contreras-Naranjo, J.E.; Aguilar, O. Suppressing Non-Specific Binding of Proteins onto Electrode Surfaces in the Development of Electrochemical Immunosensors. Biosensors 2019, 9, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iglesias-Mayor, A.; Amor-Gutiérrez, O.; Costa-García, A.; de la Escosura-Muñiz, A. Nanoparticles as Emerging Labels in Electrochemical Immunosensors. Sensors 2019, 19, 5137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Liu, B.; Yang, R.; Liu, J. Filling in the Gaps between Nanozymes and Enzymes: Challenges and Opportunities. Bioconjug. Chem. 2017, 28, 2903–2909. [Google Scholar] [CrossRef]
- Wu, J.; Wang, X.; Wang, Q.; Lou, Z.; Li, S.; Zhu, Y.; Qin, L.; Wei, H. Nanomaterials with Enzyme-like Characteristics (Nanozymes): Next-Generation Artificial Enzymes (II). Chem. Soc. Rev. 2019, 48, 1004–1076. [Google Scholar] [CrossRef] [PubMed]
- Pirsaheb, M.; Mohammadi, S.; Salimi, A. Current advances of carbon dots based biosensors for tumor marker detection, cancer cells analysis and bioimaging. TrAC—Trends Anal. Chem. 2019, 115, 83–99. [Google Scholar] [CrossRef]
- Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Carbon dots and graphene quantum dots in electrochemical biosensing. Nanomaterials 2019, 9, 634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karimi-Maleh, H. Electrochemical Sensors Based on Metal Nanoparticles, Carbon Based Nanomaterials or Ionic Liquids. Curr. Anal. Chem. 2017, 13, 4. [Google Scholar] [CrossRef]
- Dequaire, M.; Degrand, C.; Limoges, B. An Electrochemical Metalloimmunoassay Based on a Colloidal Gold Label. Anal. Chem. 2000, 72, 5521–5528. [Google Scholar] [CrossRef]
- Wang, J. Nanoparticle-Based Electrochemical Bioassays of Proteins. Electroanalysis 2007, 19, 769–776. [Google Scholar] [CrossRef]
- Liu, G.; Wang, J.; Kim, J.; Jan, M.R.; Collins, G.E. Electrochemical Coding for Multiplexed Immunoassays of Proteins. Anal. Chem. 2005, 76, 7126–7130. [Google Scholar] [CrossRef]
- Wang, J.; Liu, G.; Munge, B.; Lin, L.; Zhu, Q. DNA-Based Amplified Bioelectronic Detection and Coding of Proteins. Angew. Chem. 2004, 116, 2210–2213. [Google Scholar] [CrossRef]
- Chumbimuni-Torres, K.Y.; Dai, Z.; Rubinova, N.; Xiang, Y.; Pretsch, E.; Wang, J.; Bakker, E. Potentiometric Biosensing of Proteins with Ultrasensitive Ion-Selective Microelectrodes and Nanoparticle Labels. J. Am. Chem. Soc. 2006, 128, 13676–13677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Zhang, S.; Jia, Y.; Li, Y.; Wang, P.; Liu, Q.; Xu, Z.; Li, X. Sandwich-type electrochemical immunosensor for sensitive detection of CEA based on the enhanced effects of Ag NPs@CS spaced Hemin/rGO. Biosens. Bioelectron. 2019, 126, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ma, L.; Li, P.; Zheng, J. A novel electrochemical immunosensor based on nonenzymatic Ag@Au-Fe3O4 nanoelectrocatalyst for protein biomarker detection. Biosens. Bioelectron. 2016, 85, 343–350. [Google Scholar] [CrossRef] [Green Version]
- Lai, G.; Wang, L.; Wu, J.; Ju, H.; Yan, F. Electrochemical Stripping Analysis of Nanogold Label-Induced Silver Deposition for Ultrasensitive Multiplexed Detection of Tumor Markers. Anal. Chim. Acta 2012, 721, 1–6. [Google Scholar] [CrossRef]
- Li, F.; Li, Y.; Feng, J.; Gao, Z.; Lv, H.; Ren, X.; Wei, Q. Facile synthesis of MoS2@Cu2O-Pt nanohybrid as enzyme-mimetic label for the detection of the Hepatitis B surface antigen. Biosens. Bioelectron. 2018, 100, 512–518. [Google Scholar] [CrossRef]
- Feng, J.; Li, Y.; Li, M.; Li, F.; Han, J.; Dong, Y.; Chen, Z. A novel sandwich-type electrochemical immunosensor for PSA detection based on PtCu bimetallic hybrid (2D/2D) rGO/g-C3N4. Biosens. Bioelectron. 2017, 91, 441–448. [Google Scholar] [CrossRef]
- Liu, L.; Tian, L.; Zhao, G.; Huang, Y.; Wei, Q.; Cao, W. Ultrasensitive electrochemical immunosensor for alpha fetoprotein detection based on platinum nanoparticles anchored on cobalt oxide/graphene nanosheets for signal amplification. Anal. Chim. Acta 2017, 986, 138–144. [Google Scholar] [CrossRef]
- Charoenkitamorn, K.; Tue, P.; Kawai, K.; Chailapakul, O.; Takamura, Y. Electrochemical Immunoassay Using Open Circuit Potential Detection Labeled by Platinum Nanoparticles. Sensors 2018, 18, 444. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Cong, Y.; Huang, Y.; Du, X. Nanomaterials-Based Electrochemical Immunosensors. Micromachines 2019, 10, 397. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Wang, H.; Wang, L.; Zhu, J.; Jiang, W. Cascade Signal Amplification Based on Copper Nanoparticle-Reported Rolling Circle Amplification for Ultrasensitive Electrochemical Detection of the Prostate Cancer Biomarker. ACS Appl. Mater. Interfaces 2016, 8, 2573–2581. [Google Scholar] [CrossRef] [PubMed]
- Rivas, L.; de la Escosura-Muñiz, A.; Pons, J.; Merkoçi, A. Alzheimer Disease Biomarker Detection through Electrocatalytic Water Oxidation Induced by Iridium Oxide Nanoparticles. Electroanalysis 2014, 26, 1287–1294. [Google Scholar] [CrossRef] [Green Version]
- Kalyoncu, D.; Buyuksunetci, Y.T.; Anık, Ü. Development of a Sandwich Immunosensor for concurrent detection of carcinoembryonic antigen (CEA), vascular endothelial growth factor (VEGF) and α-fetoprotein (AFP) biomarkers. Mater. Sci. Eng. C 2019, 101, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Putnin, T.; Ngamaroonchote, A.; Wiriyakun, N.; Ounnunkad, K.; Laocharoensuk, R. Dually functional polyethylenimine-coated gold nanoparticles: A versatile material for electrode modification and highly sensitive simultaneous determination of four tumor markers. Microchim. Acta 2019, 186, 305–317. [Google Scholar] [CrossRef]
- Tang, D.; Hou, L.; Niessner, R.; Xu, M.; Gao, Z.; Knopp, D. Multiplexed Electrochemical Immunoassay of Biomarkers Using Metal Sulfide Quantum Dot Nanolabels and Trifunctionalized Magnetic Beads. Biosens. Bioelectron. 2013, 46, 37–43. [Google Scholar] [CrossRef]
- Zhang, B.; Tang, D.; Goryacheva, I.Y.; Niessner, R.; Knopp, D. Anodic-Stripping Voltammetric Immunoassay for Ultrasensitive Detection of Low-Abundance Proteins Using Quantum Dot Aggregated Hollow Microspheres. Chemistry 2013, 19, 2496–2503. [Google Scholar] [CrossRef]
- Martín-Yerga, D.; González-García, M.B.; Costa-García, A. Electrochemical immunosensor for anti-tissue transglutaminase antibodies based on the in situ detection of quantum dots. Talanta 2014, 130, 598–602. [Google Scholar] [CrossRef]
- Martín-Yerga, D.; Costa-García, A. Towards a blocking-free electrochemical immunosensing strategy for anti-transglutaminase antibodies using screen-printed electrodes. Bioelectrochemistry 2015, 105, 88–94. [Google Scholar] [CrossRef]
- Merkoçi, A.; Marcolino-Junior, L.H.; Marín, S.; Fatibello-Filho, O.; Alegret, S. Detection of cadmium sulphide nanoparticles by using screen-printed electrodes and a handheld device. Nanotechnology 2007, 18, 035502–035507. [Google Scholar] [CrossRef]
- Wang, J.; Liu, G.; Jan, M.R. Ultrasensitive Electrical Biosensing of Proteins and DNA: Carbon-Nanotube Derived Amplification of the Recognition and Transduction Events. J. Am. Chem. Soc. 2004, 126, 3010–3011. [Google Scholar] [CrossRef]
- Malhotra, R.; Patel, V.; Vaqué, J.P.; Gutkind, J.S.; Rusling, J.F. Ultrasensitive Electrochemical Immunosensor for Oral Cancer Biomarker IL-6 Using Carbon Nanotube Forest Electrodes and Multilabel Amplification. Anal. Chem. 2010, 82, 3118–3123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munge, B.S.; Fisher, J.; Millord, L.N.; Krause, C.E.; Dowd, R.S.; Rusling, J.F. Sensitive Electrochemical Immunosensor for Matrix Metalloproteinase-3 Based on Single-Wall Carbon Nanotubes. Analyst 2010, 135, 1345–1350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Munge, B.; Patel, V.; Jensen, G.; Bhirde, A.; Gong, J.D.; Kim, S.N.; Gillespie, J.; Gutkind, J.S.; Papadimitrakopoulos, F.; et al. Carbon Nanotube Amplification Strategies for Highly Sensitive Immunodetection of Cancer Biomarkers. J. Am. Chem. Soc. 2006, 128, 11199–11205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.N.; Rusling, J.F.; Papadimitrakopoulos, F. Carbon Nanotubes for Electronic and Electrochemical Detection of Biomolecules. Adv. Mater. 2007, 19, 3214–3228. [Google Scholar] [CrossRef] [Green Version]
- Sharafeldin, M.; Bishop, G.W.; Bhakta, S.; El-Sawy, A.; Suib, S.L.; Rusling, J.F. Fe3O4 Nanoparticles on Graphene Oxide Sheets for Isolation and Ultrasensitive Amperometric Detection of Cancer Biomarker Proteins. Biosens. Bioelectron. 2017, 91, 359–366. [Google Scholar] [CrossRef] [Green Version]
- Serafín, V.; Valverde, A.; Garranzo-Asensio, M.; Barderas, R.; Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Simultaneous amperometric immunosensing of the metastasis-related biomarkers IL-13Rα2 and CDH-17 by using grafted screen-printed electrodes and a composite prepared from quantum dots and carbon nanotubes for signal amplification. Microchim. Acta 2019, 186, 411. [Google Scholar] [CrossRef]
- Otieno, B.A.; Krause, C.E.; Rusling, J.F. Bioconjugation of Antibodies and Enzyme Labels onto Magnetic Beads. Methods Enzymol. 2016, 571, 135–150. [Google Scholar]
- Mani, V.; Chikkaveeraiah, B.V.; Patel, V.; Gutkind, J.S.; Rusling, J.F. Ultrasensitive Immunosensor for Cancer Biomarker Proteins Using Gold Nanoparticle Film Electrodes and Multienzyme-Particle Amplification. ACS Nano 2009, 3, 585–594. [Google Scholar] [CrossRef] [Green Version]
- Munge, B.S.; Coffey, A.L.; Doucette, J.M.; Somba, B.K.; Malhotra, R.; Patel, V.; Gutkind, J.S.; Rusling, J.F. Nanostructured Immunosensor for Attomolar Detection of Cancer Biomarker Interleukin-8 Using Massively Labeled Superparamagnetic Particles. Angew. Chem. 2011, 123, 8061–8064. [Google Scholar] [CrossRef]
- Chikkaveeraiah, B.V.; Mani, V.; Patel, V.; Gutkind, J.S.; Rusling, J.F. Microfluidic Electrochemical Immunoarray for Ultrasensitive Detection of Two Cancer Biomarker Proteins in Serum. Biosens. Bioelectron. 2011, 26, 4477–4483. [Google Scholar] [CrossRef] [Green Version]
- Malhotra, R.; Patel, V.; Chikkaveeraiah, B.V.; Munge, B.S.; Cheong, S.C.; Zain, R.B.; Abraham, M.T.; Dey, D.K.; Gutkind, J.S.; Rusling, J.F. Ultrasensitive Detection of Cancer Biomarkers in the Clinic by Use of a Nanostructured Microfluidic Array. Anal. Chem. 2012, 84, 6249–6255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, V.; Martin, D.; Malhotra, R.; Marsh, C.A.; Doçi, C.L.; Veenstra, T.D.; Nathan, C.A.O.; Sinha, U.K.; Singh, B.; Molinolo, A.A.; et al. DSG3 as a Biomarker for the Ultrasensitive Detection of Occult Lymph Node Metastasis in Oral Cancer Using Nanostructured Immunoarrays. Oral Oncol. 2013, 49, 93–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krause, C.E.; Otieno, B.A.; Latus, A.; Faria, R.C.; Patel, V.; Gutkind, J.S.; Rusling, J.F. Rapid Microfluidic Immunoassays of Cancer Biomarker Proteins Using Disposable Inkjet-Printed Gold Nanoparticle Arrays. ChemistryOpen 2013, 2, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Otieno, B.A.; Krause, C.E.; Latus, A.; Chikkaveeraiah, B.V.; Faria, R.C.; Rusling, J.F. On-Line Protein Capture on Magnetic Beads for Ultrasensitive Microfluidic Immunoassays of Cancer Biomarkers. Biosens. Bioelectron. 2014, 53, 268–274. [Google Scholar] [CrossRef] [Green Version]
- Otieno, B.A.; Krause, C.E.; Jones, A.L.; Kremer, R.B.; Rusling, J.F. Cancer Diagnostics via Ultrasensitive Multiplexed Detection of Parathyroid Hormone-Related Peptides with a Microfluidic Immunoarray. Anal. Chem. 2016, 88, 9269–9275. [Google Scholar] [CrossRef]
- Krause, C.E.; Otieno, B.A.; Bishop, G.W.; Phadke, G.; Choquette, L.; Lalla, R.V.; Peterson, D.E.; Rusling, J.F. Ultrasensitive Microfluidic Array for Serum Pro-Inflammatory Cytokines and C-Reactive Protein to Assess Oral Mucositis Risk in Cancer Patients. Anal. Bioanal. Chem. 2017, 7239–7243. [Google Scholar] [CrossRef] [Green Version]
- Phadke, G.S.; Satterwhite-Warden, J.E.; Choudhary, D.; Taylor, J.A.; Rusling, J.F. A Novel and Accurate Microfluidic Assay of CD62L in Bladder Cancer Serum Samples. Analyst 2018, 143, 5505–5511. [Google Scholar] [CrossRef]
- Mercer, C.; Jones, A.; Rusling, J.F.; Leech, D. Multiplexed electrochemical cancer diagnostics with automated microfluidics. Electroanalysis 2019, 31, 208–211. [Google Scholar] [CrossRef] [Green Version]
- Uliana, C.V.; Peverari, C.R.; Afonso, A.S.; Cominetti, M.R.; Faria, R.C. Fully disposable microfluidic electrochemical device for detection of estrogen receptor alpha breast cancer biomarker. Biosens. Bioelectron. 2018, 99, 156–162. [Google Scholar] [CrossRef]
- Fukuda, T.; Tani, Y.; Kobayashi, T.; Hirayama, Y.; Hino, O. A New Western Blotting Method Using Polymer Immunocomplexes: Detection of Tsc1 and Tsc2 Expression in Various Cultured Cell Lines. Anal. Biochem. 2000, 285, 274–276. [Google Scholar] [CrossRef]
- Dhawan, S. Design and Construction of Novel Molecular Conjugates for Signal Amplification (I): Conjugation of Multiple Horseradish Peroxidase Molecules to Immunoglobulin via Primary Amines on Lysine Peptide Chains. Peptides 2002, 23, 2091–2098. [Google Scholar] [CrossRef]
- Mishra, M.; Tiwari, S.; Gunaseelan, A.; Li, D.; Hammock, B.D.; Gomes, A.V. Improving the Sensitivity of Traditional Western Blotting via Streptavidin Containing Poly-horseradish Peroxidase (PolyHRP). Electrophoresis 2019, 40, 12–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, C.K.; Vaze, A.; Rusling, J.F. Fabrication of Immunosensor Microwell Arrays from Gold Compact Discs for Detection of Cancer Biomarker Proteins. Lab. Chip 2012, 12, 281–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojeda, I.; Moreno-Guzmán, M.; González-Cortés, A.; Yáñez-Sedeño, P.; Pingarrón, J.M. Electrochemical Magnetoimmunosensor for the Ultrasensitive Determination of Interleukin-6 in Saliva and Urine Using Poly-HRP Streptavidin Conjugates as Labels for Signal Amplification. Anal. Bioanal. Chem. 2014, 406, 6363–6371. [Google Scholar] [CrossRef]
- Sánchez-Tirado, E.; Martínez-García, G.; González-Cortés, A.; Yáñez-Sedeño, P.; Pingarrón, J.M. Electrochemical Immunosensor for Sensitive Determination of Transforming Growth Factor (TGF)—Β1 in Urine. Biosens. Bioelectron. 2017, 88, 9–14. [Google Scholar] [CrossRef]
- Sánchez-Tirado, E.; Salvo, C.; González-Cortés, A.; Yáñez-Sedeño, P.; Langa, F.; Pingarrón, J.M. Electrochemical immunosensor for simultaneous determination of interleukin-1 beta and tumor necrosis factor alpha in serum and saliva using dual screen printed electrodes modified with functionalized double–walled carbon nanotubes. Anal. Chim. Acta 2017, 959, 66–73. [Google Scholar] [CrossRef]
- Pastucha, M.; Farka, Z.; Lacina, K.; Mikušová, Z.; Skládal, P. Magnetic Nanoparticles for Smart Electrochemical Immunoassays: A Review on Recent Developments. Microchim. Acta 2019, 186, 1–26. [Google Scholar] [CrossRef]
- Kanyong, P.; Patil, A.V.; Davis, J.J. Functional Molecular Interfaces for Impedance-Based Diagnostics. Annu. Rev. Anal. Chem. 2020, 13, 183–200. [Google Scholar] [CrossRef]
- Luo, X.; Davis, J.J. Electrical Biosensors and the Label Free Detection of Protein Disease Biomarkers. Chem. Soc. Rev. 2013, 42, 5944–5962. [Google Scholar] [CrossRef]
- Johnson, A.; Song, Q.; Ko Ferrigno, P.; Bueno, P.R.; Davis, J.J. Sensitive Affimer and Antibody Based Impedimetric Label-Free Assays for C-Reactive Protein. Anal. Chem. 2012, 84, 6553–6560. [Google Scholar] [CrossRef]
- Fernandes, F.C.B.; Góes, M.S.; Davis, J.J.; Bueno, P.R. Label Free Redox Capacitive Biosensing. Biosens. Bioelectron. 2013, 50, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Xu, M.; Freeman, C.; James, T.; Davis, J.J. Ultrasensitive Label Free Electrical Detection of Insulin in Neat Blood Serum. Anal. Chem. 2013, 85, 4129–4134. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Fan, X.; Xu, S.; Davis, J.J.; Luo, X. Low Fouling Label-Free DNA Sensor Based on Polyethylene Glycols Decorated with Gold Nanoparticles for the Detection of Breast Cancer Biomarkers. Biosens. Bioelectron. 2015, 71, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Bueno, P.R.; Davis, J.J. A Dual Marker Label Free Electrochemical Assay for Flavivirus Dengue Diagnosis. Biosens. Bioelectron. 2018, 100, 519–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanyong, P.; Davis, J.J. Homogeneous Functional Self-Assembled Monolayers: Faradaic Impedance Baseline Signal Drift Suppression for High-Sensitivity Immunosensing of C-Reactive Protein. J. Electroanal. Chem. 2020, 856, 113675. [Google Scholar] [CrossRef]
- Kanyong, P.; Catli, C.; Davis, J.J. Ultrasensitive Impedimetric Immunosensor for the Detection of C-Reactive Protein in Blood at Surface-Initiated-Reversible Addition–Fragmentation Chain Transfer Generated Poly (2-Hydroxyethyl Methacrylate) Brushes. Anal. Chem. 2020, 92, 4707–4710. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Han, H.; Ma, Z. Improved Screen-Printed Carbon Electrode for Multiplexed Label-Free Amperometric Immuniosensor: Addressing Its Conductivity and Reproducibility Challenges. Biosens. Bioelectron. 2018, 101, 304–310. [Google Scholar] [CrossRef]
- Khoshroo, A.; Mazloum-Ardakani, M.; Forat-Yazdi, M. Enhanced Performance of Label-Free Electrochemical Immunosensor for Carbohydrate Antigen 15-3 Based on Catalytic Activity of Cobalt Sulfide/Graphene Nanocomposite. Sens. Actuators B Chem. 2018, 255, 580–587. [Google Scholar] [CrossRef]
- Shafaat, A.; Faridbod, F.; Ganjali, M.R. Label-Free Detection of Cytochrome: C by a Conducting Polymer-Based Impedimetric Screen-Printed Aptasensor. New J. Chem. 2018, 42, 6034–6039. [Google Scholar] [CrossRef]
- Srivastava, M.; Nirala, N.R.; Srivastava, S.K.; Prakash, R. A Comparative Study of Aptasensor vs. Immunosensor for Label-Free PSA Cancer Detection on GQDs-AuNRs Modified Screen-Printed Electrodes. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Mollarasouli, F.; Serafín, V.; Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M.; Asadpour-Zeynali, K. Ultrasensitive Determination of Receptor Tyrosine Kinase with a Label-Free Electrochemical Immunosensor Using Graphene Quantum Dots-Modified Screen-Printed Electrodes. Anal. Chim. Acta 2018, 1011, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.U.; Hossain, M.M.; Safavieh, M.; Wong, Y.L.; Rahman, I.A.; Zourob, M.; Tamiya, E. Toward the development of smart and low cost point-of-care biosensors based on screen printed electrodes. Crit. Rev. Biotechnol. 2016, 36, 495–505. [Google Scholar] [CrossRef] [PubMed]
- DuPont Conductive Inks for Digital Printing. Available online: https://www.dupont.com/products/inkjet-silver-conductor-inks.html (accessed on 15 May 2020).
- A Range of Conductive Inks That Are Optimized for Various Applications on Multiple Substrates. Available online: https://www.sunchemical.com/product/conductive-inks/ (accessed on 15 May 2020).
- Acheson | Electrodag | Lumidag | Minico | Flexible Circuit | PCB |. Available online: https://www.materialtech.biz/index.php/brands/acheson (accessed on 15 May 2020).
- Conductive Silver Inks | Conductive Carbon Inks | Tekra, LLC. Available online: https://www.tekra.com/products/conductive-inks (accessed on 15 May 2020).
- Metrohm DropSens Screen-Printed Electrodes. Available online: http://www.dropsens.com/en/screen_printed_electrodes_pag.html (accessed on 15 May 2020).
- Carbon Screen-Printed Electrodes. Available online: https://pineresearch.com/shop/products/electrodes/screen-printed-electrodes/carbon-spes (accessed on 15 May 2020).
- Screen Printed Electrodes. Available online: https://www.palmsens.com/products/sensors/screen-printed-electrodes (accessed on 15 May 2020).
- Screen Printed Electrodes. Available online: https://www.basinc.com/products/ec/screen-printed-electrodes (accessed on 15 May 2020).
- Kanichi Research Services. Available online: http://www.gmclifesciencesfund.com/Medical-Devices/kanichi-research-services/30980 (accessed on 15 May 2020).
- Tallapragada, S.D.; Layek, K.; Mukherjee, R.; Mistry, K.K.; Ghosh, M. Development of screen-printed electrode based immunosensor for the detection of HER2 antigen in human serum samples. Bioelectrochemistry 2017, 118, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Mistry, K.K.; Layek, K.; Chell, T.N.; Chaudhuri, C.R.; Saha, H. Design and development of an amperometric immunosensor based on screen-printed electrodes. Anal. Methods 2016, 8, 3096–3101. [Google Scholar] [CrossRef]
- Das, J.; Kelley, S.O. Protein detection using arrayed microsensor chips: Tuning sensor footprint to achieve ultrasensitive readout of CA-125 in serum and whole blood. Anal. Chem. 2011, 83, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.F.; Lim, H.N.; Shams, N.; Jayabal, S.; Pandikumar, A.; Huang, N.M. Fabrication of graphene/gold-modified screen-printed electrode for detection of carcinoembryonic antigen. Mater. Sci. Eng. C 2016, 58, 666–674. [Google Scholar] [CrossRef]
- Suresh, L.; Brahman, P.K.; Reddy, K.R.; Bondili, J.S. Development of an electrochemical immunosensor based on gold nanoparticles incorporated chitosan biopolymer nanocomposite film for the detection of prostate cancer using PSA as biomarker. Enzyme Microb. Technol. 2018, 112, 43–51. [Google Scholar] [CrossRef]
- Giannetto, M.; Bianchi, M.V.; Mattarozzi, M.; Careri, M. Competitive amperometric immunosensor for determination of p53 protein in urine with carbon nanotubes/gold nanoparticles screen-printed electrodes: A potential rapid and noninvasive screening tool for early diagnosis of urinary tract carcinoma. Anal. Chim. Acta 2017, 991, 133–141. [Google Scholar] [CrossRef]
- Marques, R.C.B.; Costa-Rama, E.; Viswanathan, S.; Nouws, H.P.A.; Costa-García, A.; Delerue-Matos, C.; González-García, M.B. Voltammetric immunosensor for the simultaneous analysis of the breast cancer biomarkers CA 15-3 and HER2-ECD. Sens. Actuators B Chem. 2018, 255, 918–925. [Google Scholar] [CrossRef] [Green Version]
- Fanjul-Bolado, P.; Hernández-Santos, D.; González-García, M.B.; Costa-García, A. Alkaline phosphatase-catalyzed silver deposition for electrochemical detection. Anal. Chem. 2007, 79, 5272–5277. [Google Scholar] [CrossRef]
- Selvolini, G.; Marrazza, G. MIP-based sensors: Promising new tools for cancer biomarker determination. Sensors 2017, 17, 718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozal-Palabiyik, B.; Lettieri, M.; Uslu, B.; Marrazza, G. Electrochemical Detection of Vascular Endothelial Growth Factor by Molecularly Imprinted Polymer. Electroanalysis 2019, 31, 1458–1464. [Google Scholar] [CrossRef]
- Gomes, R.S.; Moreira, F.T.C.; Fernandes, R.; Sales, M.G.F. Sensing CA 15-3 in point-of-care by electropolymerizing O-phenylenediamine (oPDA) on Au-screen printed electrodes. PLoS ONE 2018, 13, e0196656. [Google Scholar] [CrossRef] [PubMed]
- Ronkainen-Matsuno, N.J.; Thomas, J.H.; Halsall, H.B.; Heineman, W.R. Electrochemical immunoassay moving into the fast lane. TrAC—Trends Anal. Chem. 2002, 21, 213–225. [Google Scholar] [CrossRef]
- Bange, A.; Halsall, H.B.; Heineman, W.R. Microfluidic immunosensor systems. Biosens. Bioelectron. 2005, 20, 2488–2503. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.W.; Phillips, S.T.; Butte, M.J.; Whitesides, G.M. Patterned Paper as a Platform for Inexpensive, Low-Volume, Portable Bioassays. Angew. Chem. Int. Ed. 2007, 46, 1318–1320. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-Capitán, M.; Baldi, A.; Fernández-Sánchez, C. Electrochemical Paper-Based Biosensor Devices for Rapid Detection of Biomarkers. Sensors 2020, 20, 967. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Shi, S.; Ma, J.; Guo, Y. A paper-based electrochemical immunosensor with reduced graphene oxide/thionine/gold nanoparticles nanocomposites modification for the detection of cancer antigen 125. Biosens. Bioelectron. 2019, 135, 1–7. [Google Scholar] [CrossRef]
- Tortorich, R.; Shamkhalichenar, H.; Choi, J.-W. Inkjet-Printed and Paper-Based Electrochemical Sensors. Appl. Sci. 2018, 8, 288. [Google Scholar] [CrossRef] [Green Version]
- Huther Da Costa, T.; Song, E. A Paper-Based Electrochemical Sensor Using Inkjet-Printed Carbon Nanotube Electrodes. ECS J. Solid State Sci. Technol. 2015, 4, S3044–S3047. [Google Scholar] [CrossRef]
- Tortorich, R.P.; Song, E.; Choi, J.-W. Inkjet-Printed Carbon Nanotube Electrodes with Low Sheet Resistance for Electrochemical Sensor Applications. J. Electrochem. Soc. 2014, 161, B3044–B3048. [Google Scholar] [CrossRef]
- Määttänen, A.; Vanamo, U.; Ihalainen, P.; Pulkkinen, P.; Tenhu, H.; Bobacka, J.; Peltonen, J. A low-cost paper-based inkjet-printed platform for electrochemical analyses. Sens. Actuators B Chem. 2013, 177, 153–162. [Google Scholar] [CrossRef]
- Shamkhalichenar, H.; Choi, J.-W. An Inkjet-Printed Non-Enzymatic Hydrogen Peroxide Sensor on Paper. J. Electrochem. Soc. 2017, 164, B3101–B3106. [Google Scholar] [CrossRef]
- Carvajal, S.; Fera, S.N.; Jones, A.L.; Baldo, T.A.; Mosa, I.M.; Rusling, J.F.; Krause, C.E. Disposable inkjet-printed electrochemical platform for detection of clinically relevant HER-2 breast cancer biomarker. Biosens. Bioelectron. 2018, 104, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Fernández-la-Villa, A.; Pozo-Ayuso, D.F.; Castaño-Álvarez, M. Microfluidics and Electrochemistry: An Emerging Tandem for next-Generation Analytical Microsystems. Curr. Opin. Electrochem. 2019, 15, 175–185. [Google Scholar] [CrossRef]
- Sanjay, S.T.; Zhou, W.; Dou, M.; Tavakoli, H.; Ma, L.; Xu, F.; Li, X.J. Recent Advances of Controlled Drug Delivery Using Microfluidic Platforms. Adv. Drug Deliv. Rev. 2018, 128, 3–28. [Google Scholar] [CrossRef]
- Shi, H.; Nie, K.; Dong, B.; Long, M.; Xu, H.; Liu, Z. Recent Progress of Microfluidic Reactors for Biomedical Applications. Chem. Eng. J. 2019, 361, 635–650. [Google Scholar] [CrossRef]
- van Heeren, H.; Tantra, R.; Salomon, P. Microfluidic Devices: A Road Forward by Standardization of Interconnects and Classification. Microfluidics Nanofluidics 2015, 19, 1203–1207. [Google Scholar] [CrossRef]
- Sharafeldin, M.; Kadimisetty, K.; Bhalero, K.R.; Chen, T.; Rusling, J.F. 3D-printed Immunosensor arrays for cancer diagnostics. Sensors 2020, 20, 4514. [Google Scholar] [CrossRef]
- Iliescu, C.; Taylor, H.; Avram, M.; Miao, J.; Franssila, S. A Practical Guide for the Fabrication of Microfluidic Devices Using Glass and Silicon. Biomicrofluidics 2012, 6, 16505. [Google Scholar] [CrossRef] [Green Version]
- Gale, B.K.; Jafek, A.R.; Lambert, C.J.; Goenner, B.L.; Moghimifam, H.; Nze, U.C.; Kamarapu, S.K.A. Review of Current Methods in Microfluidic Device Fabrication and Future Commercialization Prospects. Inventions 2018, 3, 60. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.H.; Lee, J.K.; Kim, J.H.; Choi, H.S.; Kim, J.; Lee, S.H.; Lee, H.Y. Single Microfluidic Electrochemical Sensor System for Simultaneous Multi-Pulmonary Hypertension Biomarker Analyses. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Oliveira, R.A.G.; Materon, E.M.; Melendez, M.E.; Carvalho, A.L.; Faria, R.C. Disposable Microfluidic Immunoarray Device for Sensitive Breast Cancer Biomarker Detection. ACS Appl. Mater. Interfaces 2017, 9, 27433–27440. [Google Scholar] [CrossRef] [PubMed]














| Biomarker | Sample | Cancer Type | Clinical Use | Assay | |
|---|---|---|---|---|---|
| Abbreviation | Name | ||||
| Free PSA/fPSa | Free Prostate Specific Antigen | Serum | Prostate | Benign Hyperplasia vs. Cancer diagnosis | Immunoassay |
| tPSA | Total PSA | Serum | Prostate | S, M | Immunoassay |
| cPSA | Complex PSA | Serum | Prostate | S, M | Immunoassay |
| p63 | Transformation-related protein 63 | FFPE tissue† | Prostate | S, M | Immunohistochemistry |
| TG | Thyroglobulin | Serum | Thyroid | S, M | Immunoassay |
| EGFR | Epidermal growth factor receptor | Colon tissue | Colon | Pre | Immunoassay |
| CEA | Carcinoembryonic antigen | Serum | Colon | M | Immunoassay |
| MW CEA | High molecular weight CEA | Urine | Bladder | M | Immunofluorescence |
| FDP (AMDL-ELISA DR-70) | Fibrin/fibrinogen degradation products | Urine/Serum | Bladder | M | Immunoassay |
| NMP/22 | Nuclear matrix protein 22 | Urine | Bladder | S, M | Immunoassay |
| BTA | Bladder tumor antigen | Urine | Bladder | M | Immunoassay |
| HER2 | Human epidermal growth factor receptor | Serum | Breast | M | Immunohistochemistry |
| CA15-3 * | Carbohydrate antigen 15-3 | Serum, plasma | Breast | M | Immunoassay |
| CA27-29 * | Carbohydrate antigen 27-29 | Serum | Breast | M | Immunoassay |
| HER/NEU | Human epidermal growth factor receptor 2 | FFPE tissue† | Breast | P, Pre | Immunohistochemistry |
| ER | Estrogen factor | FFPE tissue† | Breast | P, Pre | Immunohistochemistry |
| PR | Progesterone factor | FFPE tissue† | Breast | P, Pre | Immunohistochemistry |
| AFP* | α-fetoprotein | Serum, plasma, amniotic fluid | Testicular | St | Immunoassay |
| β-hGC * | Human chorionic gonadotropin-β | Serum | Testicular | St | Immunoassay |
| AFP-L3% | α-fetoprotein L3% isoform | Serum | Hepatocellular | P | HPLC, microfluidic capillary electrophoresis |
| KIT | Receptor Tyrosine Kinase | FFPE tissue† | Gastrointestinal stromal tumors | Pre | Immunohistochemistry |
| CA 19-9 * | Carbohydrate antigen 19-9 | Serum | Pancreatic | M | Immunoassay |
| CA 125 * | Carbohydrate antigen 125 | Serum | Ovarian | M | Immunoassay |
| HE4 | Human epididymis protein 4 | Serum | Ovarian | M | Immunoassay |
| OVA1 (Multiprotein test | CA125, Apolipoprotein A1, Beta-2 microglobulin, Transferrin, Pre-albumin | Ovarian | Serum | P | Immunoassay |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhanapala, L.; Krause, C.E.; Jones, A.L.; Rusling, J.F. Printed Electrodes in Microfluidic Arrays for Cancer Biomarker Protein Detection. Biosensors 2020, 10, 115. https://doi.org/10.3390/bios10090115
Dhanapala L, Krause CE, Jones AL, Rusling JF. Printed Electrodes in Microfluidic Arrays for Cancer Biomarker Protein Detection. Biosensors. 2020; 10(9):115. https://doi.org/10.3390/bios10090115
Chicago/Turabian StyleDhanapala, Lasangi, Colleen E. Krause, Abby L. Jones, and James F. Rusling. 2020. "Printed Electrodes in Microfluidic Arrays for Cancer Biomarker Protein Detection" Biosensors 10, no. 9: 115. https://doi.org/10.3390/bios10090115