The Nucleotide Capture Region of Alpha Hemolysin: Insights into Nanopore Design for DNA Sequencing from Molecular Dynamics Simulations
Abstract
:1. Introduction
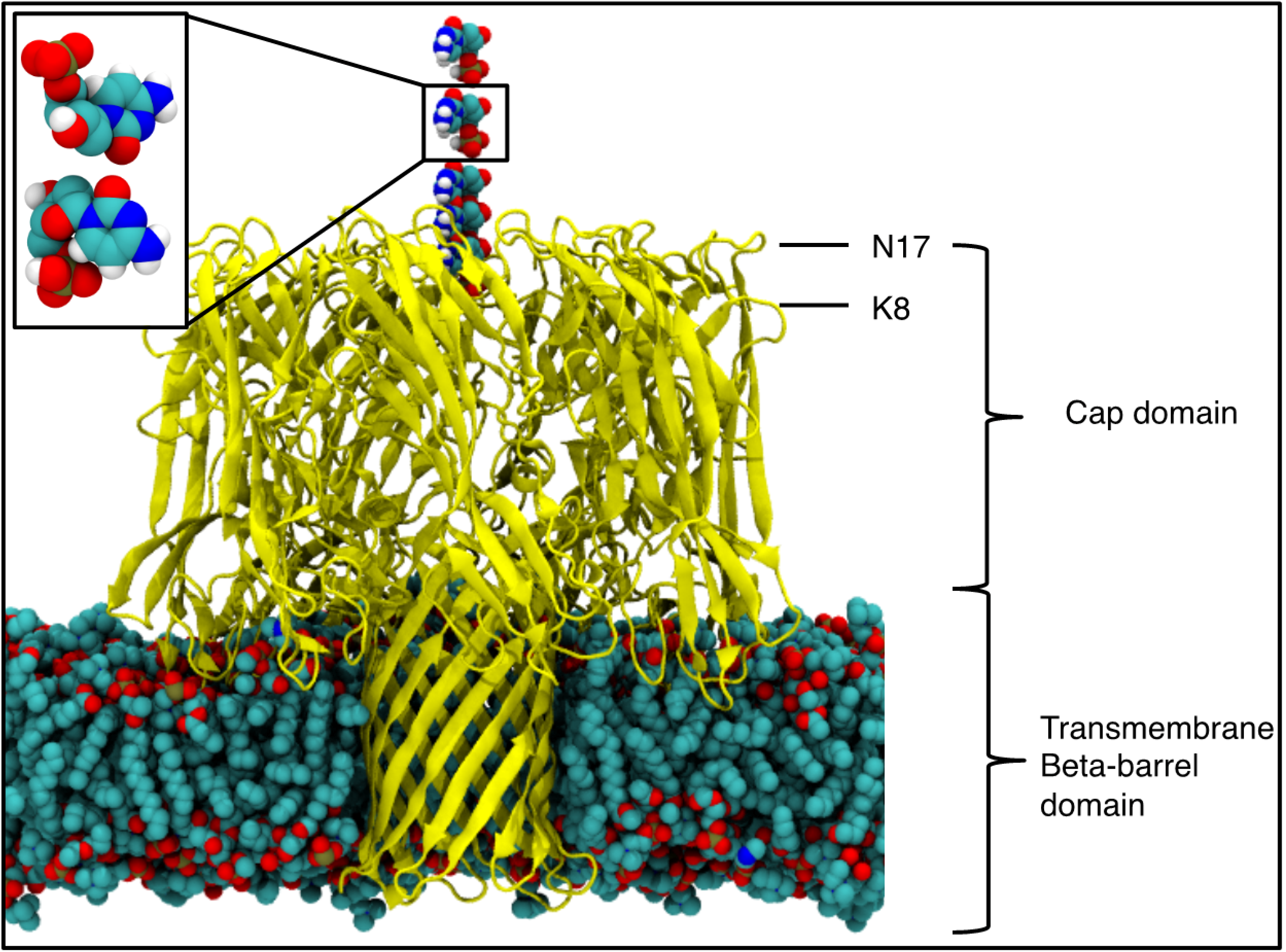
2. Results and Discussion
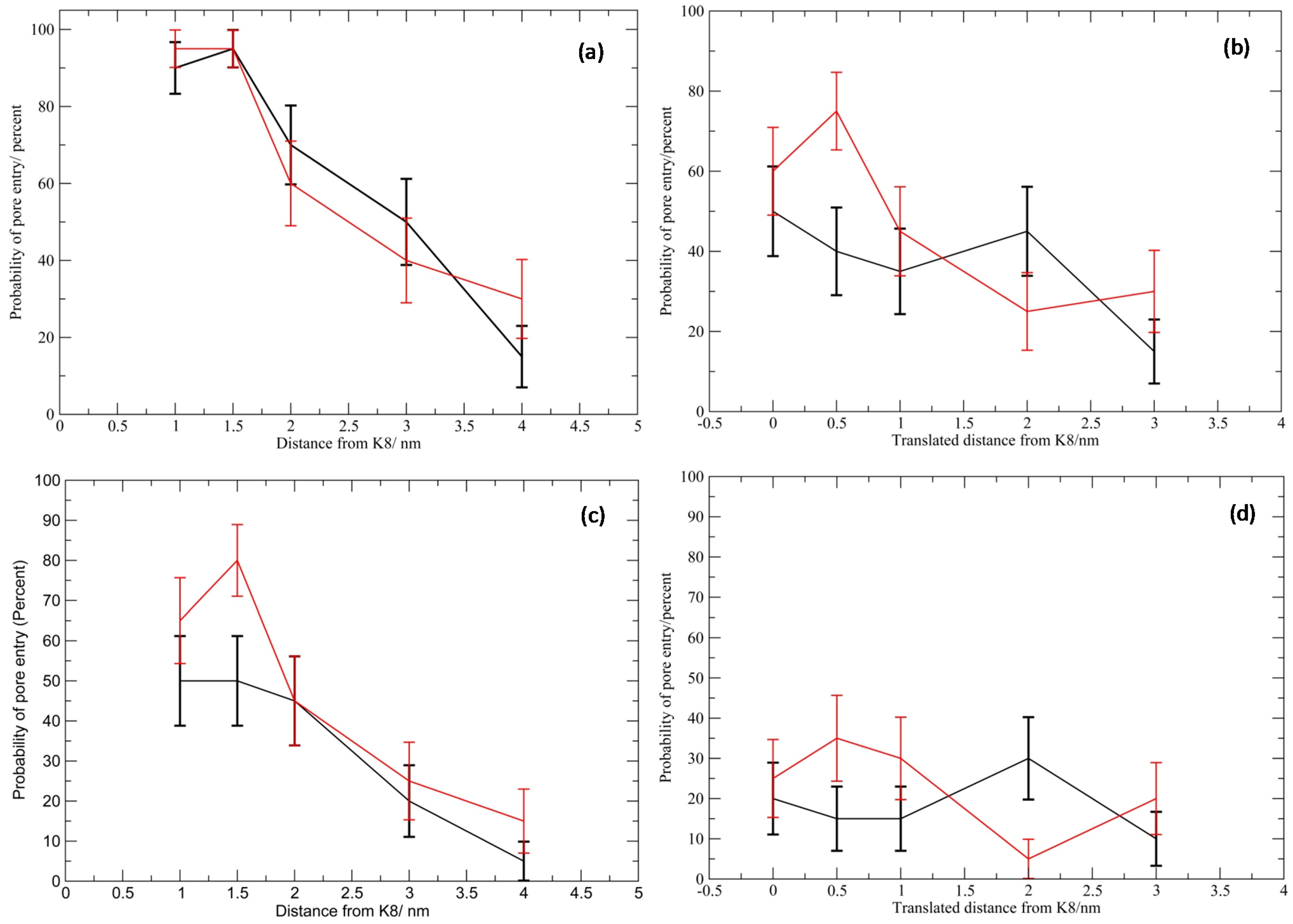
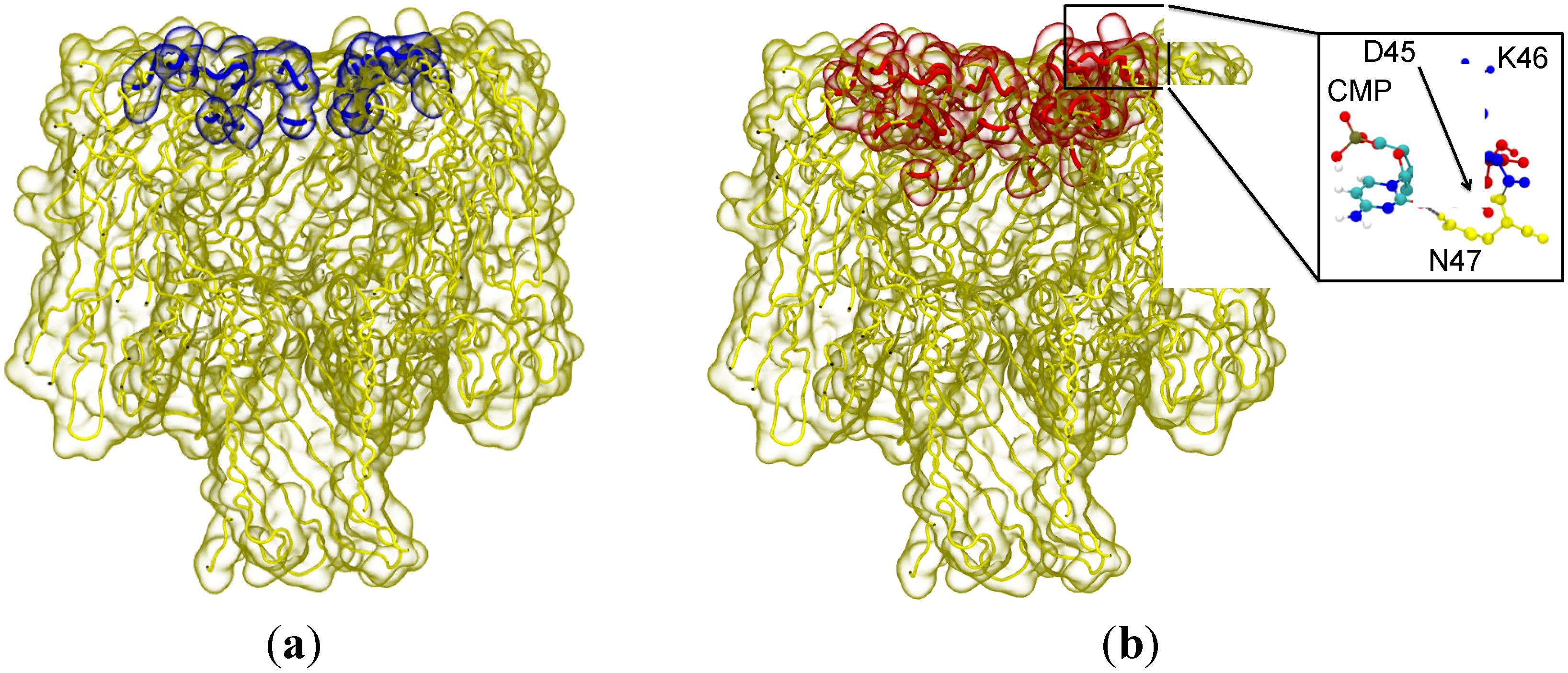

Sum Effects to CMP Entry into the Vestibule
3. Experimental Section
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schneider, G.F.; Dekker, C. DNA sequencing with nanopores. Nat. Biotechnol. 2012, 30, 326–328. [Google Scholar] [CrossRef] [PubMed]
- Rincon-Restrepo, M.; Milthallova, E.; Bayley, H.; Maglia, G. Controlled translocation of individual DNA molecules through protein nanopores with engineered molecular brakes. Nano Lett. 2011, 11, 746–750. [Google Scholar] [CrossRef] [PubMed]
- Derrington, I.M.; Butler, T.Z.; Collins, M.D.; Manrao, E.; Pavlenok, M.; Niederweis, M.; Gundlach, J.H. Nanopore DNA sequencing with mspa. Proc. Natl. Acad. Sci. USA 2010, 107, 16060–16065. [Google Scholar] [CrossRef] [PubMed]
- Wanunu, M. Nanopores: A journey towards DNA sequencing. Phys. Life Rev. 2012, 9, 125–158. [Google Scholar] [CrossRef] [PubMed]
- Dekker, C. Solid-state nanopores. Nat. Nanotechnol. 2007, 2, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Venkatesan, B.M.; Bashir, R. Nanopore sensors for nucleic acid analysis. Nat. Nanotechnol. 2011, 6, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Fyta, M.; Melchionna, S.; Succi, S. Translocation of biomolecules through solid-state nanopores: Theory meets experiments. J. Polym. Sci. Polym. Phys. 2011, 49, 985–1011. [Google Scholar] [CrossRef]
- Scheicher, R.H.; Grigoriev, A.; Ahuja, R. DNA sequencing with nanopores from an ab initio perspective. J. Mater. Sci. 2012, 47, 7439–7446. [Google Scholar] [CrossRef]
- Laszlo, A.H.; Derrington, I.M.; Brinkerhoff, H.; Langford, K.W.; Nova, I.C.; Samson, J.M.; Bartlett, J.J.; Pavlenok, M.; Gundlach, J.H. Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore mspa. Proc. Natl. Acad. Sci. USA 2013, 110, 18904–18909. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.; Wu, H.C.; Jayasinghe, L.; Patel, A.; Reid, S.; Bayley, H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 2009, 4, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Song, L.Z.; Hobaugh, M.R.; Shustak, C.; Cheley, S.; Bayley, H.; Gouaux, J.E. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science 1996, 274, 1859–1866. [Google Scholar] [CrossRef] [PubMed]
- Stoddart, D.; Maglia, G.; Mikhailova, E.; Heron, A.J.; Bayley, H. Multiple base-recognition sites in a biological nanopore: Two heads are better than one. Angew. Chem. Int. Ed. 2010, 49, 556–559. [Google Scholar] [CrossRef] [Green Version]
- Branton, D.; Deamer, D.W.; Marziali, A.; Bayley, H.; Benner, S.A.; Butler, T.; di Ventra, M.; Garaj, S.; Hibbs, A.; Huang, X.H.; et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. [Google Scholar] [CrossRef] [PubMed]
- Reiner, J.E.; Balijepalli, A.; Robertson, J.W.F.; Drown, B.S.; Burden, D.L.; Kasianowicz, J.J. The effects of diffusion on an exonuclease/nanopore-based DNA sequencing engine. J. Chem. Phys. 2012, 137. [Google Scholar] [CrossRef]
- Bond, P.J.; Guy, A.T.; Heron, A.J.; Bayley, H.; Khalid, S. Molecular dynamics simulations of DNA within a nanopore: Arginine-phosphate tethering and a binding/sliding mechanism for translocation. Biochemistry 2011, 50, 3777–3783. [Google Scholar] [CrossRef] [PubMed]
- Guy, A.T.; Piggot, T.J.; Khalid, S. Single-stranded DNA within nanopores: Conformational dynamics and implications for sequencing; a molecular dynamics simulation study. Biophys. J. 2012, 103, 1028–1036. [Google Scholar] [CrossRef] [PubMed]
- Parkin, J.; Khalid, S. Atomistic molecular-dynamics simulations enable prediction of the arginine permeation pathway through occd1/oprd from pseudomonas aeruginosa. Biophys. J. 2014, 107, 1853–1861. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Derrington, I.M.; Pavlenok, M.; Niederweis, M.; Gundlach, J.H.; Aksimentiev, A. Molecular dynamics study of mspa arginine mutants predicts slow DNA translocations and ion current blockades indicative of DNA sequence. ACS Nano 2012, 6, 6960–6968. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.; Neidle, S. Molecular modeling and simulation of g-quadruplexes and quadruplex-ligand complexes. Methods Mol. Biol. 2010, 608, 17–37. [Google Scholar] [PubMed]
- Pronk, S.; Pall, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. Gromacs 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. Gromacs 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef]
- Oostenbrink, C.; Villa, A.; Mark, A.E.; van Gunsteren, W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: The gromos force-field parameter sets 53a5 and 53a6. J. Comput. Chem. 2004, 25, 1656–1676. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; Postma, J.P.M.; van Gunsteren, W.F.; Hermans, J. Interaction Models for Water in Relation to Protein Hydration. In Intermol. Forces; Pullman, B., Ed.; Springer: Berlin, Germany, 1981; pp. 331–342. [Google Scholar]
- Pongprayoon, P.; Beckstein, O.; Wee, C.L.; Sansom, M.S.P. Simulations of anion transport through oprp reveal the molecular basis for high affinity and selectivity for phosphate. Proc. Natl. Acad. Sci. USA 2009, 106, 21614–21618. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. Vmd: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.C.; Postma, J.P.M.; Vangunsteren, W.F.; Dinola, A.; Haak, J.R. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126. [Google Scholar] [CrossRef]
- Nosé, S.; Klein, M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983, 50, 1055–1076. [Google Scholar] [CrossRef]
- He, Y.H.; Tsutsui, M.; Scheicher, R.H.; Fan, C.; Taniguchi, M.; Kawai, T. Mechanism of how salt-gradient-induced charges affect the translocation of DNA molecules through a nanopore. Biophys. J. 2013, 105, 776–782. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manara, R.M.A.; Tomasio, S.; Khalid, S. The Nucleotide Capture Region of Alpha Hemolysin: Insights into Nanopore Design for DNA Sequencing from Molecular Dynamics Simulations. Nanomaterials 2015, 5, 144-153. https://doi.org/10.3390/nano5010144
Manara RMA, Tomasio S, Khalid S. The Nucleotide Capture Region of Alpha Hemolysin: Insights into Nanopore Design for DNA Sequencing from Molecular Dynamics Simulations. Nanomaterials. 2015; 5(1):144-153. https://doi.org/10.3390/nano5010144
Chicago/Turabian StyleManara, Richard M. A., Susana Tomasio, and Syma Khalid. 2015. "The Nucleotide Capture Region of Alpha Hemolysin: Insights into Nanopore Design for DNA Sequencing from Molecular Dynamics Simulations" Nanomaterials 5, no. 1: 144-153. https://doi.org/10.3390/nano5010144
APA StyleManara, R. M. A., Tomasio, S., & Khalid, S. (2015). The Nucleotide Capture Region of Alpha Hemolysin: Insights into Nanopore Design for DNA Sequencing from Molecular Dynamics Simulations. Nanomaterials, 5(1), 144-153. https://doi.org/10.3390/nano5010144




