Graphitic Carbon Nitride (C3N4) Reduces Cadmium and Arsenic Phytotoxicity and Accumulation in Rice (Oryza sativa L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. C3N4 Synthesis and Characterization
2.2. Hydroponic Experimental Design
2.3. Elemental Analysis of Rice Tissues
2.4. Real-Time Quantitative PCR Analysis of As and Cd Transporters in Rice
2.5. Random Amplified Polymorphic DNA (RAPD) Analysis
2.6. Statistical Analysis
3. Results and Discussion
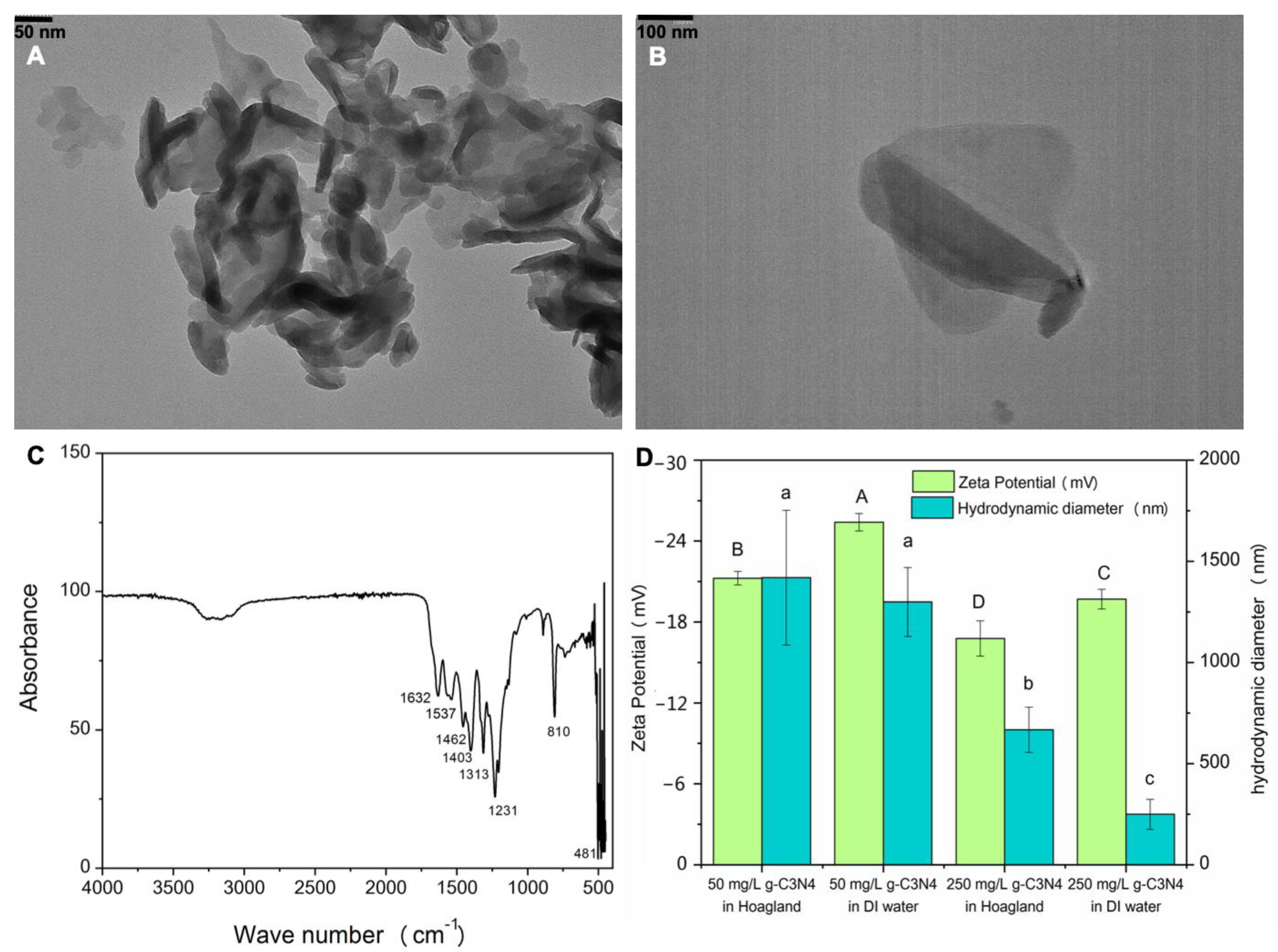
3.1. C3N4 Characterization
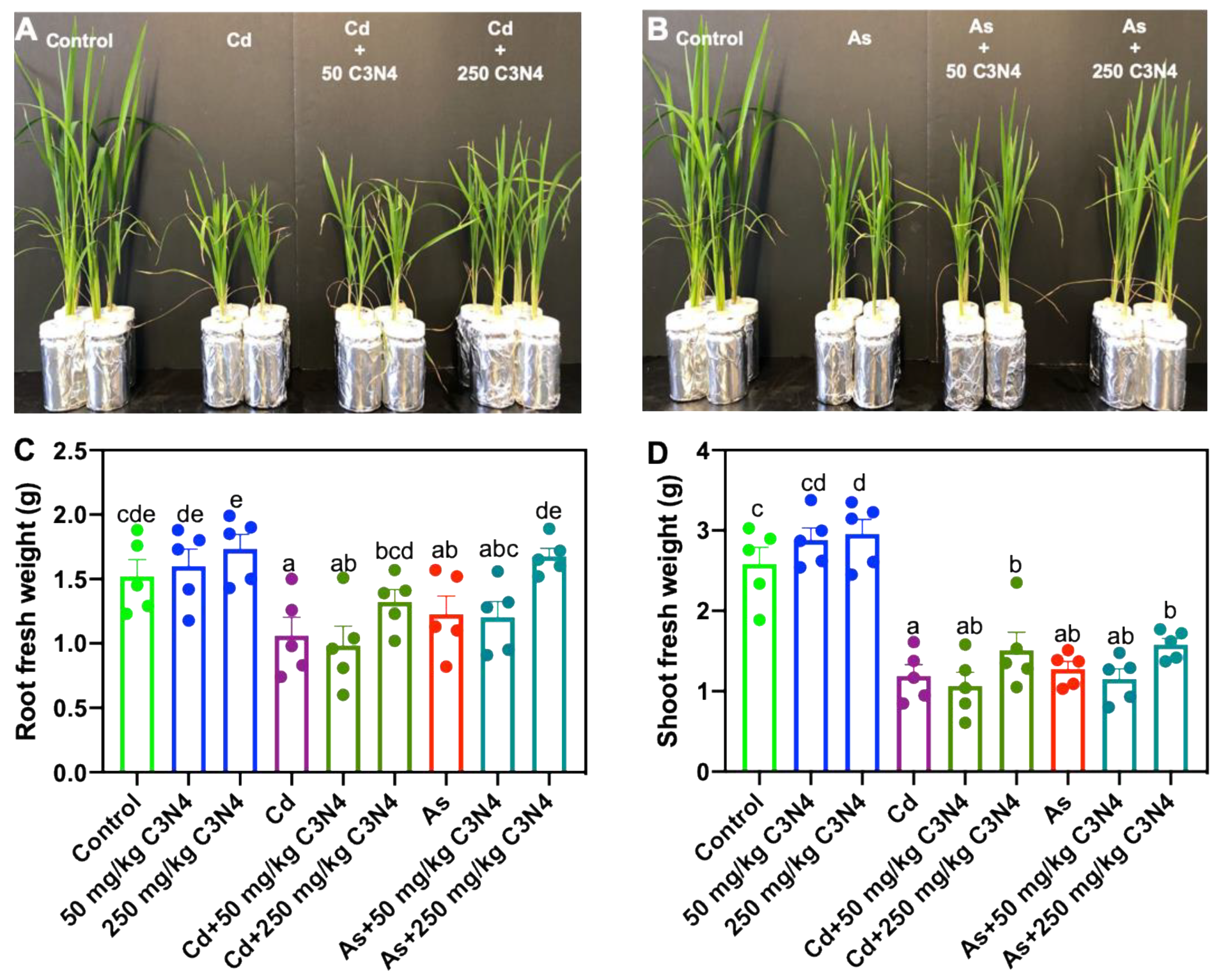
3.2. Fresh Biomass
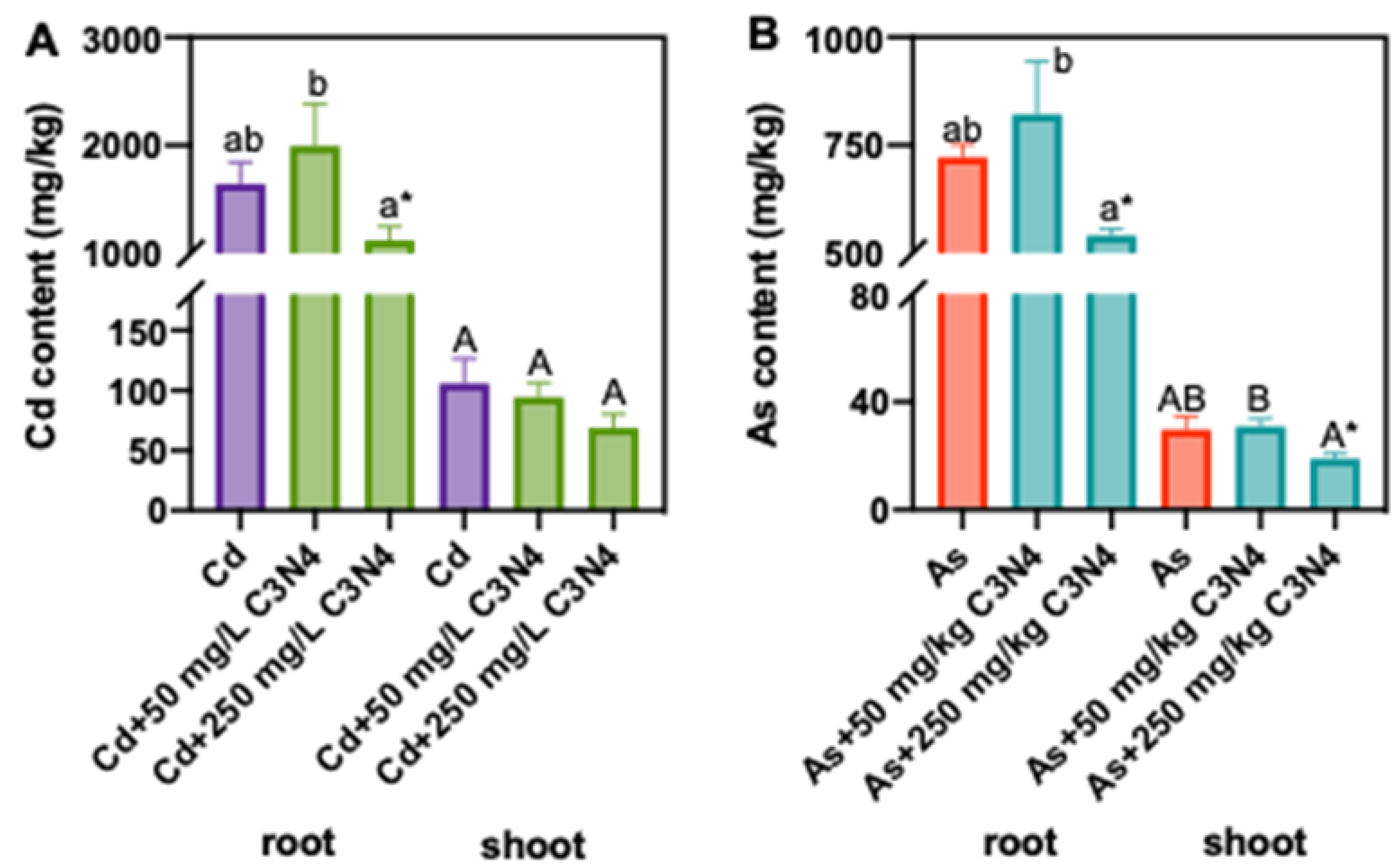
3.3. Cd and As Content in Rice Tissues
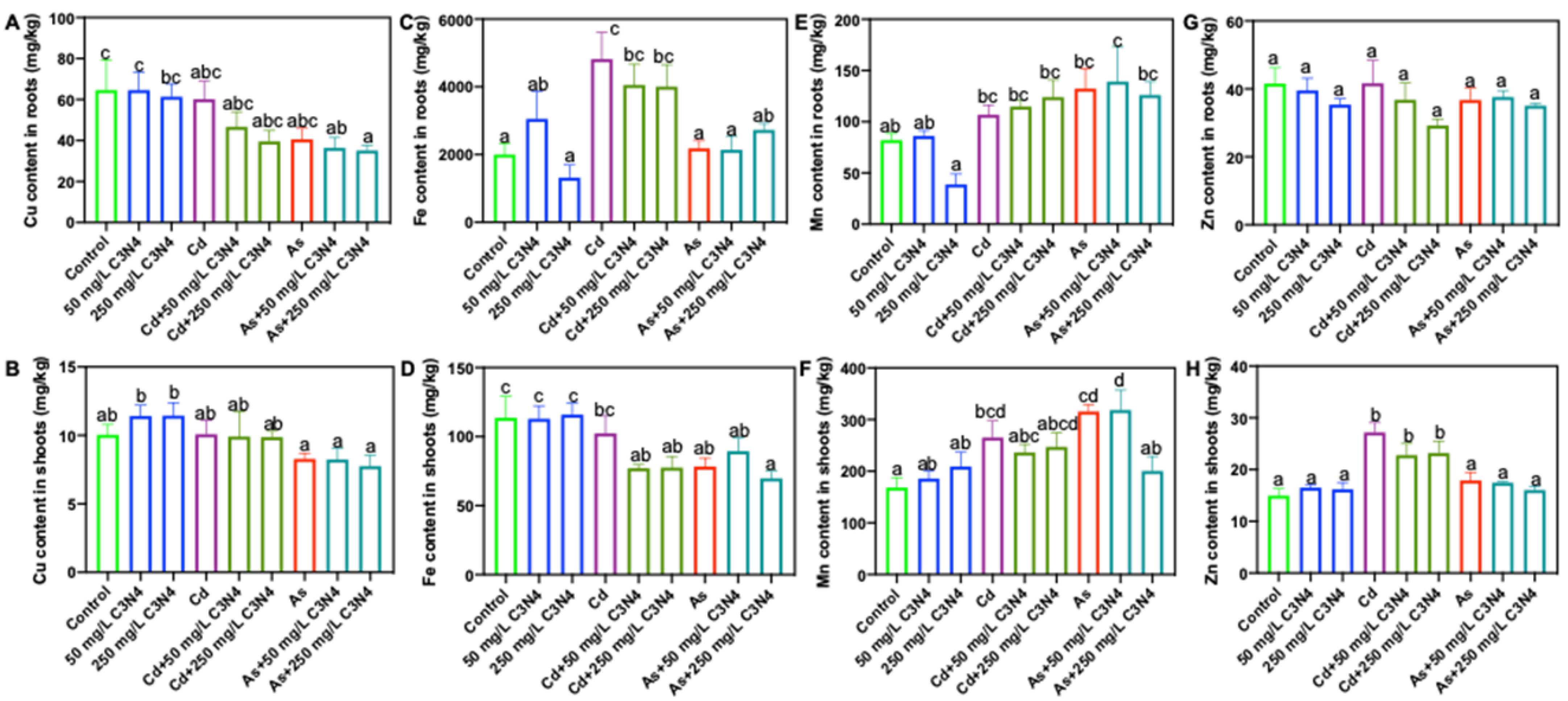
3.4. Macro- and Micronutrient Content Analysis in Rice Tissues
3.5. Plant Molecular Response
3.5.1. RAPD Analysis upon Exposure to C3N4 and Heavy Metals
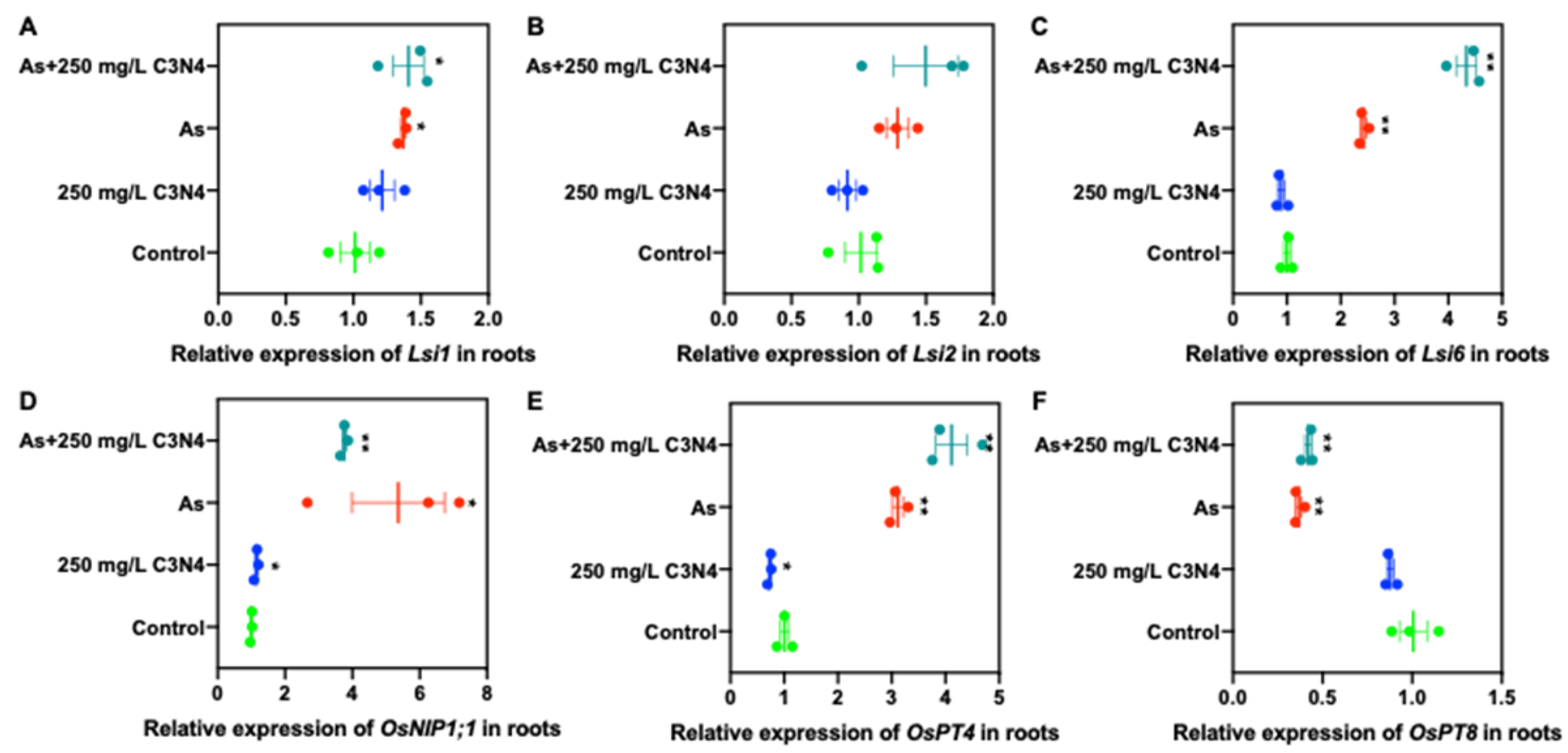
3.5.2. Relative Expression of Cd and As Transporters
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hou, D.; O’Connor, D.; Igalavithana, A.D.; Alessi, D.S.; Luo, J.; Tsang, D.C.W.; Sparks, D.L.; Yamauchi, Y.; Rinklebe, J.; Ok, Y.S. Metal contamination and bioremediation of agricultural soils for food safety and sustainability. Nat. Rev. Earth Environ. 2020, 1, 366–381. [Google Scholar] [CrossRef]
- Podgorski, J.; Berg, M.J.S. Global threat of arsenic in groundwater. Science 2020, 368, 845–850. [Google Scholar] [CrossRef]
- Li, S.W.; Sun, H.J.; Li, H.B.; Luo, J.; Ma, L.Q. Assessment of cadmium bioaccessibility to predict its bioavailability in contaminated soils. Environ. Int. 2016, 94, 600–606. [Google Scholar] [CrossRef]
- Marrugo-Negrete, J.; Pinedo-Hernández, J.; Díez, S. Assessment of heavy metal pollution, spatial distribution and origin in agricultural soils along the Sinú River Basin, Colombia. Environ. Res. 2017, 154, 380–388. [Google Scholar] [CrossRef]
- Shi, G.; Chen, Z.; Bi, C.; Li, Y.; Teng, J.; Wang, L.; Xu, S. Comprehensive assessment of toxic metals in urban and suburban street deposited sediments (SDSs) in the biggest metropolitan area of China. Environ. Pollut. 2010, 158, 694–703. [Google Scholar] [CrossRef]
- Guadie, A.; Yesigat, A.; Gatew, S.; Worku, A.; Liu, W.; Ajibade, F.O.; Wang, A. Evaluating the health risks of heavy metals from vegetables grown on soil irrigated with untreated and treated wastewater in Arba Minch, Ethiopia. Sci. Total Environ. 2021, 761, 143302. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Sharifan, H.; Dou, F.; Sun, W. Simultaneous reduction of arsenic (As) and cadmium (Cd) accumulation in rice by zinc oxide nanoparticles. Chem. Eng. J. 2020, 384, 123802. [Google Scholar] [CrossRef]
- Kubier, A.; Wilkin, R.T.; Pichler, T. Cadmium in soils and groundwater: A review. Appl. Geochem. 2019, 108, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Arsenic Compounds. Available online: https://www.epa.gov/sites/production/files/2016-09/documents/arsenic-compounds.pdf (accessed on 20 March 2021).
- Kumarathilaka, P.; Seneweera, S.; Meharg, A.; Bundschuh, J. Arsenic speciation dynamics in paddy rice soil-water environment: Sources, physico-chemical, and biological factors—A review. Water Res. 2018, 140, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cui, X.; Zhang, X.; Liu, W.; Cui, Z. Combined toxicity and detoxification of lead, cadmium and arsenic in Solanum nigrum L. J. Hazard. Mater. 2020, 389, 121874. [Google Scholar] [CrossRef] [PubMed]
- Baderna, D.; Lomazzi, E.; Pogliaghi, A.; Ciaccia, G.; Lodi, M.; Benfenati, E. Acute phytotoxicity of seven metals alone and in mixture: Are Italian soil threshold concentrations suitable for plant protection? Environ. Res. 2015, 140, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Lan, P.; Ding, C.; Wang, J.; Zhang, T.; Wang, X. A new perspective on the toxicity of arsenic-contaminated soil: Tandem mass tag proteomics and metabolomics in earthworms. J. Hazard. Mater. 2020, 398, 122825. [Google Scholar] [CrossRef] [PubMed]
- Raiesi, F.; Sadeghi, E. Interactive effect of salinity and cadmium toxicity on soil microbial properties and enzyme activities. Ecotoxicol. Environ. Saf. 2019, 168, 221–229. [Google Scholar] [CrossRef]
- Toth, G.; Hermann, T.; Szatmari, G.; Pasztor, L. Maps of heavy metals in the soils of the European Union and proposed priority areas for detailed assessment. Sci. Total Environ. 2016, 565, 1054–1062. [Google Scholar] [CrossRef]
- Rossi, L.; Sharifan, H.; Zhang, W.; Schwab, A.P.; Ma, X. Mutual effects and in planta accumulation of co-existing cerium oxide nanoparticles and cadmium in hydroponically grown soybean (Glycine max (L.) Merr.). Environ. Sci. Nano 2018, 5, 150–157. [Google Scholar] [CrossRef]
- Carey, A.M.; Scheckel, K.G.; Lombi, E.; Newville, M.; Choi, Y.; Norton, G.J.; Charnock, J.M.; Feldmann, J.; Price, A.H.; Meharg, A.A. Grain unloading of arsenic species in rice. Plant Physiol. 2010, 152, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Zhao, M.; Yu, Z.; Rong, H.; Zhang, C. Utilization of nanomaterials for in-situ remediation of heavy metal(loid) contaminated sediments: A review. Sci. Total Environ. 2019, 662, 205–217. [Google Scholar] [CrossRef]
- Riaz, M.; Kamran, M.; Fang, Y.; Wang, Q.; Cao, H.; Yang, G.; Deng, L.; Wang, Y.; Zhou, Y.; Anastopoulos, I.; et al. Arbuscular mycorrhizal fungi-induced mitigation of heavy metal phytotoxicity in metal contaminated soils: A critical review. J. Hazard. Mater. 2021, 402, 123919. [Google Scholar] [CrossRef]
- Suriyagoda, L.D.B.; Dittert, K.; Lambers, H. Arsenic in Rice Soils and Potential Agronomic Mitigation Strategies to Reduce Arsenic Bioavailability: A Review. Pedosphere 2018, 28, 363–382. [Google Scholar] [CrossRef]
- Vaculik, M.; Lukacova, Z.; Bokor, B.; Martinka, M.; Tripathi, D.K.; Lux, A. Alleviation mechanisms of metal(loid) stress in plants by silicon: A review. J. Exp. Bot. 2020, 71, 6744–6757. [Google Scholar] [CrossRef]
- Zhao, F.-J.; Wang, P. Arsenic and cadmium accumulation in rice and mitigation strategies. Plant Soil 2019, 446, 1–21. [Google Scholar] [CrossRef]
- Wu, J.; Li, Z.; Huang, D.; Liu, X.; Tang, C.; Parikh, S.J.; Xu, J. A novel calcium-based magnetic biochar is effective in stabilization of arsenic and cadmium co-contamination in aerobic soils. J. Hazard. Mater. 2020, 387, 122010. [Google Scholar] [CrossRef]
- Pramanik, K.; Mitra, S.; Sarkar, A.; Soren, T.; Maiti, T.K. Characterization of cadmium-resistant Klebsiella pneumoniae MCC 3091 promoted rice seedling growth by alleviating phytotoxicity of cadmium. Environ. Sci. Pollut. Res. Int. 2017, 24, 24419–24437. [Google Scholar] [CrossRef]
- Kamran, M.; Malik, Z.; Parveen, A.; Zong, Y.; Abbasi, G.H.; Rafiq, M.T.; Shaaban, M.; Mustafa, A.; Bashir, S.; Rafay, M.; et al. Biochar alleviates Cd phytotoxicity by minimizing bioavailability and oxidative stress in pak choi (Brassica chinensis L.) cultivated in Cd-polluted soil. J. Environ. Manag. 2019, 250, 109500. [Google Scholar] [CrossRef] [PubMed]
- White, J.C.; Gardea-Torresdey, J. Achieving food security through the very small. Nat. Nanotechnol. 2018, 13, 627–629. [Google Scholar] [CrossRef]
- Kah, M.; Tufenkji, N.; White, J.C. Nano-enabled strategies to enhance crop nutrition and protection. Nat. Nanotechnol. 2019, 14, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Servin, A.D.; White, J.C. Nanotechnology in agriculture: Next steps for understanding engineered nanoparticle exposure and risk. NanoImpact 2016, 1, 9–12. [Google Scholar] [CrossRef]
- Giraldo, J.P.; Wu, H.; Newkirk, G.M.; Kruss, S. Nanobiotechnology approaches for engineering smart plant sensors. Nat. Nanotechnol. 2019, 14, 541–553. [Google Scholar] [CrossRef]
- Venkatachalam, P.; Jayaraj, M.; Manikandan, R.; Geetha, N.; Rene, E.R.; Sharma, N.C.; Sahi, S.V. Zinc oxide nanoparticles (ZnONPs) alleviate heavy metal-induced toxicity in Leucaena leucocephala seedlings: A physiochemical analysis. Plant Physiol. Biochem. 2017, 110, 59–69. [Google Scholar] [CrossRef]
- Rizwan, M.; Ali, S.; ur Rehman, M.Z.; Malik, S.; Adrees, M.; Qayyum, M.F.; Alamri, S.A.; Alyemeni, M.N.; Ahmad, P. Effect of foliar applications of silicon and titanium dioxide nanoparticles on growth, oxidative stress, and cadmium accumulation by rice (Oryza sativa). Acta Physiol. Plant. 2019, 41, 35. [Google Scholar] [CrossRef]
- Lian, J.; Zhao, L.; Wu, J.; Xiong, H.; Bao, Y.; Zeb, A.; Tang, J.; Liu, W. Foliar spray of TiO2 nanoparticles prevails over root application in reducing Cd accumulation and mitigating Cd-induced phytotoxicity in maize (Zea mays L.). Chemosphere 2020, 239, 124794. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Dhungana, B.; Cobb, G.P. Copper oxide nanoparticles and arsenic interact to alter seedling growth of rice (Oryza sativa japonica). Chemosphere 2018, 206, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Fiyadh, S.S.; AlSaadi, M.A.; Jaafar, W.Z.; AlOmar, M.K.; Fayaed, S.S.; Mohd, N.S.; Hin, L.S.; El-Shafie, A. Review on heavy metal adsorption processes by carbon nanotubes. J. Clean. Prod. 2019, 230, 783–793. [Google Scholar] [CrossRef]
- Ihsanullah; Abbas, A.; Al-Amer, A.M.; Laoui, T.; Al-Marri, M.J.; Nasser, M.S.; Khraisheh, M.; Atieh, M.A. Heavy metal removal from aqueous solution by advanced carbon nanotubes: Critical review of adsorption applications. Sep. Purif. Technol. 2016, 157, 141–161. [Google Scholar] [CrossRef]
- Xu, J.; Cao, Z.; Zhang, Y.; Yuan, Z.; Lou, Z.; Xu, X.; Wang, X. A review of functionalized carbon nanotubes and graphene for heavy metal adsorption from water: Preparation, application, and mechanism. Chemosphere 2018, 195, 351–364. [Google Scholar] [CrossRef]
- Matos, M.P.S.R.; Correia, A.A.S.; Rasteiro, M.G. Application of carbon nanotubes to immobilize heavy metals in contaminated soils. J. Nanopart. Res. 2017, 19. [Google Scholar] [CrossRef]
- Yue, L.; Lian, F.; Han, Y.; Bao, Q.; Wang, Z.; Xing, B. The effect of biochar nanoparticles on rice plant growth and the uptake of heavy metals: Implications for agronomic benefits and potential risk. Sci. Total Environ. 2019, 656, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Ma, C.; Yin, M.; Sun, H.; Zhao, Q.; White, J.C.; Wang, C.; Xing, B. Accumulation of phenanthrene and its metabolites in lettuce (Lactuca sativa L.) as affected by magnetic carbon nanotubes and dissolved humic acids. Environ. Sci. Nano 2020, 7, 3759–3772. [Google Scholar] [CrossRef]
- Zheng, Y.; Lin, L.; Wang, B.; Wang, X. Graphitic Carbon Nitride Polymers toward Sustainable Photoredox Catalysis. Angew. Chem. Int. Ed. Engl. 2015, 54, 12868–12884. [Google Scholar] [CrossRef]
- Inagaki, M.; Tsumura, T.; Kinumoto, T.; Toyoda, M. Graphitic carbon nitrides (g-C3N4) with comparative discussion to carbon materials. Carbon 2019, 141, 580–607. [Google Scholar] [CrossRef]
- Praus, P.; Svoboda, L.; Ritz, M.; Troppová, I.; Šihor, M.; Kočí, K. Graphitic carbon nitride: Synthesis, characterization and photocatalytic decomposition of nitrous oxide. Mater. Chem. Phys. 2017, 193, 438–446. [Google Scholar] [CrossRef]
- Xiao, G.; Wang, Y.; Xu, S.; Li, P.; Yang, C.; Jin, Y.; Sun, Q.; Su, H. Superior adsorption performance of graphitic carbon nitride nanosheets for both cationic and anionic heavy metals from wastewater. Chin. J. Chem. Eng. 2019, 27, 305–313. [Google Scholar] [CrossRef]
- Shorie, M.; Kaur, H.; Chadha, G.; Singh, K.; Sabherwal, P. Graphitic carbon nitride QDs impregnated biocompatible agarose cartridge for removal of heavy metals from contaminated water samples. J. Hazard. Mater. 2019, 367, 629–638. [Google Scholar] [CrossRef]
- Hao, Y.; Lv, R.; Ma, C.; Adeel, M.; Zhao, Z.; Rao, Y.; Rui, Y. Graphitic carbon nitride (g-C3N4) alleviates cadmium-induced phytotoxicity to rice (Oryza sativa L.). Environ. Sci. Pollut. Res. Int. 2021. [Google Scholar] [CrossRef]
- Introduction to Rice. Available online: https://archive.gramene.org/species/oryza/rice_intro.html (accessed on 20 March 2021).
- Qi, Y.; Liang, Q.; Lv, R.; Shen, W.; Kang, F.; Huang, Z.H. Synthesis and photocatalytic activity of mesoporous g-C3N4/MoS2 hybrid catalysts. R. Soc. Open Sci. 2018, 5, 180187. [Google Scholar] [CrossRef] [PubMed]
- Shang, H.; Guo, H.; Ma, C.; Li, C.; Chefetz, B.; Polubesova, T.; Xing, B. Maize (Zea mays L.) root exudates modify the surface chemistry of CuO nanoparticles: Altered aggregation, dissolution and toxicity. Sci. Total Environ. 2019, 690, 502–510. [Google Scholar] [CrossRef]
- Ma, C.; Liu, H.; Guo, H.; Musante, C.; Coskun, S.H.; Nelson, B.C.; White, J.C.; Xing, B.; Dhankher, O.P. Defense mechanisms and nutrient displacement in Arabidopsis thaliana upon exposure to CeO2 and In2O3 nanoparticles. Environ. Sci. Nano 2016, 3, 1369–1379. [Google Scholar] [CrossRef]
- Ma, J.F.; Yamaji, N.; Mitani, N.; Xu, X.-Y.; Su, Y.-H.; McGrath, S.P.; Zhao, F.-J. Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc. Natl. Acad. Sci. USA 2008, 105, 9931–9935. [Google Scholar] [CrossRef]
- Ye, Y.; Li, P.; Xu, T.; Zeng, L.; Cheng, D.; Yang, M.; Luo, J.; Lian, X. OsPT4 Contributes to Arsenate Uptake and Transport in Rice. Front. Plant Sci. 2017, 8, 2197. [Google Scholar] [CrossRef]
- Jiang, M.; Jiang, J.; Li, S.; Li, M.; Tan, Y.; Song, S.; Shu, Q.; Huang, J. Glutamate alleviates cadmium toxicity in rice via suppressing cadmium uptake and translocation. J. Hazard. Mater. 2020, 384, 121319. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Chhikara, S.; Xing, B.; Musante, C.; White, J.C.; Dhankher, O.P. Physiological and Molecular Response of Arabidopsis thaliana (L.) to Nanoparticle Cerium and Indium Oxide Exposure. ACS Sustain. Chem. Eng. 2013, 1, 768–778. [Google Scholar] [CrossRef]
- Cao, D.; Oard, J.H. Pedigree and RAPD-based DNA analysis of commercial US rice cultivars. Crop Sci. 1997, 37, 1630–1635. [Google Scholar] [CrossRef]
- Yu, H.; Chen, F.; Chen, F.; Wang, X. In situ self-transformation synthesis of g-C3N4-modified CdS heterostructure with enhanced photocatalytic activity. Appl. Surf. Sci. 2015, 358, 385–392. [Google Scholar] [CrossRef]
- Deng, X.; Chen, Y.; Yang, Y.; Lu, L.; Yuan, X.; Zeng, H.; Zeng, Q. Cadmium accumulation in rice (Oryza sativa L.) alleviated by basal alkaline fertilizers followed by topdressing of manganese fertilizer. Environ. Pollut. 2020, 262, 114289. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Ma, C.; Chen, G.; White, J.C.; Wang, Z.; Xing, B.; Dhankher, O.P. Titanium Dioxide Nanoparticles Alleviate Tetracycline Toxicity to Arabidopsis thaliana (L.). ACS Sustain. Chem. Eng. 2017, 5, 3204–3213. [Google Scholar] [CrossRef]
- Ma, C.; Liu, H.; Chen, G.; Zhao, Q.; Eitzer, B.; Wang, Z.; Cai, W.; Newman, L.A.; White, J.C.; Dhankher, O.P.; et al. Effects of titanium oxide nanoparticles on tetracycline accumulation and toxicity in Oryza sativa (L.). Environ. Sci. Nano 2017, 4, 1827–1839. [Google Scholar] [CrossRef]
- Sharifan, H.; Ma, X.; Moore, J.M.; Habib, M.R.; Evans, C. Zinc Oxide Nanoparticles Alleviated the Bioavailability of Cadmium and Lead and Changed the Uptake of Iron in Hydroponically Grown Lettuce (Lactuca sativa L. var. Longifolia). ACS Sustain. Chem. Eng. 2019, 7, 16401–16409. [Google Scholar] [CrossRef]
- Cai, F.; Wu, X.; Zhang, H.; Shen, X.; Zhang, M.; Chen, W.; Gao, Q.; White, J.C.; Tao, S.; Wang, X. Impact of TiO2 nanoparticles on lead uptake and bioaccumulation in rice (Oryza sativa L.). NanoImpact 2017, 5, 101–108. [Google Scholar] [CrossRef]
- Marschner, H.; Römheld, V.; Horst, W.J.; Martin, P. Root-induced changes in the rhizosphere: Importance for the mineral nutrition of plants. Z. Pflanzenernähr. Bodenkd. 1986, 149, 441–456. [Google Scholar] [CrossRef]
- Ma, C.; Chhikara, S.; Minocha, R.; Long, S.; Musante, C.; White, J.C.; Xing, B.; Dhankher, O.P. Reduced Silver Nanoparticle Phytotoxicity in Crambe abyssinica with Enhanced Glutathione Production by Overexpressing Bacterial gamma-Glutamylcysteine Synthase. Environ. Sci. Technol. 2015, 49, 10117–10126. [Google Scholar] [CrossRef] [PubMed]
- Mosa, K.A.; El-Naggar, M.; Ramamoorthy, K.; Alawadhi, H.; Elnaggar, A.; Wartanian, S.; Ibrahim, E.; Hani, H. Copper Nanoparticles Induced Genotoxicty, Oxidative Stress, and Changes in Superoxide Dismutase (SOD) Gene Expression in Cucumber (Cucumis sativus) Plants. Front. Plant Sci. 2018, 9, 872. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Ma, C.; Chen, F.; Luo, X.; Musante, C.; White, J.C.; Zhao, X.; Wang, Z.; Xing, B. New insight into the mechanism of graphene oxide-enhanced phytotoxicity of arsenic species. J. Hazard. Mater. 2020, 410, 124959. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, C.; Hao, Y.; Zhao, J.; Zuverza-Mena, N.; Meselhy, A.G.; Dhankher, O.P.; Rui, Y.; White, J.C.; Xing, B. Graphitic Carbon Nitride (C3N4) Reduces Cadmium and Arsenic Phytotoxicity and Accumulation in Rice (Oryza sativa L.). Nanomaterials 2021, 11, 839. https://doi.org/10.3390/nano11040839
Ma C, Hao Y, Zhao J, Zuverza-Mena N, Meselhy AG, Dhankher OP, Rui Y, White JC, Xing B. Graphitic Carbon Nitride (C3N4) Reduces Cadmium and Arsenic Phytotoxicity and Accumulation in Rice (Oryza sativa L.). Nanomaterials. 2021; 11(4):839. https://doi.org/10.3390/nano11040839
Chicago/Turabian StyleMa, Chuanxin, Yi Hao, Jian Zhao, Nubia Zuverza-Mena, Ahmed G. Meselhy, Om Parkash Dhankher, Yukui Rui, Jason C. White, and Baoshan Xing. 2021. "Graphitic Carbon Nitride (C3N4) Reduces Cadmium and Arsenic Phytotoxicity and Accumulation in Rice (Oryza sativa L.)" Nanomaterials 11, no. 4: 839. https://doi.org/10.3390/nano11040839






